Abstract
We have incorporated an RNA binding site for the biological amino acid tryptophan within an RNA complex with affinity for phospholipid bilayer membranes. The resulting RNA (9:10Trp) creates a selective route through the bilayer for the amino acid. Binding and enhanced tryptophan permeability are nonlinear in RNA concentration, suggesting that RNA aggregation is required for both. Tryptophan permeability saturates with increased concentration, though at ~1000-fold greater level than when binding a free aptamer. The RNA (9:10Trp) complex, bound at a mean of two per liposome, halves the activation energy for tryptophan transport (to 46 kJ/mole), specifically increasing tryptophan entry to a maximal velocity of 0.5 sec-1 per liposome with little or no accompanying increase in general permeability. Individual RNAs turn over tens of thousands of times at high tryptophan concentration. Thus, a specific passive membrane transporter whose properties overlap those of single-molecule transporter proteins, can be made of RNA alone. Permeability changes probably rely on disturbances in lipid conformation as well as on an advantageous low free energy position for tryptophan at the membrane. Other RNA activities may yield other RNA-membrane nanosystems via this route.
Keywords: carrier, selection, amino acid, nucleic acid, phospholipids, ribocyte
INTRODUCTION
RNA molecules readily bind phospholipid bilayers (Khvorova et al. 1999; Vlassov et al. 2001; Janas and Yarus 2003), probably residing within one of the two polar surface zones that comprise about half the membrane thickness (Wiener and White 1992). By selecting RNAs that chromatograph with liposomes, we have found many RNA complexes composed of ~120 nucleotide monomers which bind to dioleoylphosphatidylcholine bilayers (Vlassov et al. 2001). Such membrane RNAs oligomerize when free and, even more strikingly, within the membrane (Janas and Yarus 2003). RNAs we use here bind optimally as a 2:1 complex of RNA 9:RNA 10 with RNA contact via kissing loops (Vlassov et al. 2001). The crucial design principle is based on the observation that the two RNAs differ in function within the membrane complex. RNA 9 alone shows weak but detectable membrane affinity, suggesting that it incorporates the phospholipid site. In contrast, RNA 10 has no such activity, and may only be a linker (Janas and Yarus 2003) allowing the specific oligomerization of RNA 9.
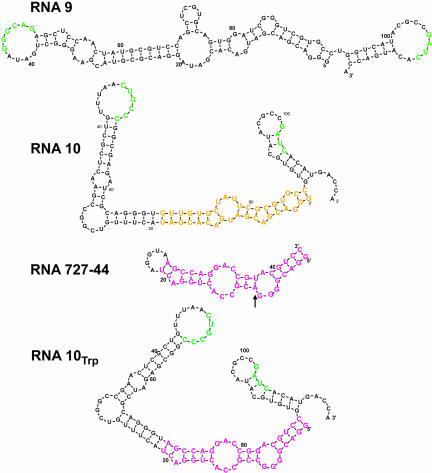
These functional ideas are the basis for the transformation of the RNA (9:10) complex into a transporter. We combined two structures to form a bifunctional RNA 10Trp (Fig. 1 ▶) in which the hypothetically dispensable center of RNA 10 is substituted by a stereo-specific binding site for the amino acid tryptophan. The central (Fig. 1 ▶, yellow) nucleotides of RNA 10 were replaced by RNA 727–44 sequences (magenta). These latter sequences contain an internal bulge motif selected for tryptophan binding (I. Majerfeld and M. Yarus, in prep.). The Trp binding site is thereby flanked by loops containing sequences needed for complex formation and membrane affinity (Fig. 1 ▶, green). A base paired U-A inessential for amino acid affinity (Fig. 1 ▶, arrow) was changed to G-C to better favor the local structure of the tryptophan binding fold within the larger RNA.
FIGURE 1.
Proposed secondary structures for RNA 9, RNA 10, RNA 10Trp. Drawings are the output of BayesFold, a Bayesian RNA folding program (Knight et al. 2004; http://jaynes.colorado.edu/Bayes/). In RNA 10Trp a central region (yellow) has been replaced with a tryptophan binding site (RNA 727–44, magenta; I. Majerfeld and M. Yarus, in prep.). An arrow marks a sequence change (AU →CG; nucleotides 9–82) introduced to increase the free-energy advantage of the correct fold for the tryptophan binding site. Flanking regions contain essential complementary loop sequences (Vlassov et al. 2001; green) for RNA (9:10) complex formation and membrane binding.
The construction exploits the fact that these RNAs are expected to perturb regular lipid structures (Janas and Yarus 2003). Thus, they should bring bound tryptophan into apposition with a region of perturbed membrane lipid. Below we test the idea that this combination of actions will provide a specific route across the phospholipid bilayer for the RNA ligand tryptophan.
RESULTS
Testing amino acid binding and complex formation
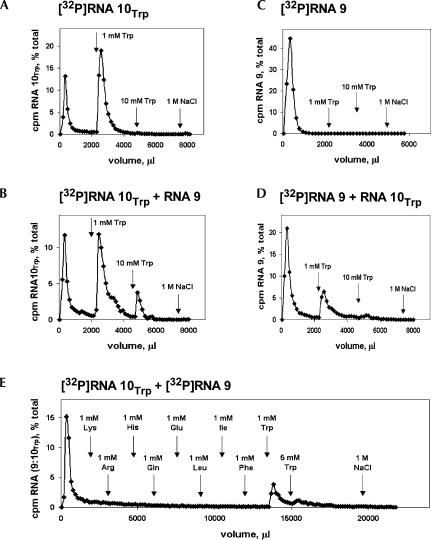
To test the continued function of the Trp binding site within RNA 10Trp (Fig. 1 ▶), we performed affinity chromatography on Trp-Sepharose using both RNA 10Trp (Fig. 2A ▶) and the radiolabeled hybrid complex, RNA (9:10Trp; Fig 2B ▶). Both free and complexed RNA 10Trp is mostly folded into an active conformation, judged from the observation that two-thirds of both free and complexed RNA 10Trp bind to Trp-Sepharose and are eluted by a 1-mM Trp solution.
FIGURE 2.
RNA affinity for L-tryptophan: Affinity elution from tryptophan-Sepharose using (A) [32P]RNA 10Trp, (B) RNA (9:[32P]RNA 10Trp), (C) [32P]RNA 9, (D) ([32P]RNA 9:RNA 10Trp), (E) amino acid selectivity assay by successive elution of affinity-immobilized RNA.
Radiolabeled RNA 9 alone, as expected, shows no affinity for the amino acid (Fig. 2C ▶). On the other hand, radiolabeled RNA 9 in the presence of unlabeled RNA10Trp binds Trp-Sepharose and is affinity eluted by tryptophan (Fig 2D ▶). Therefore, complex formation with RNA 9 has also survived construction of RNA 10Trp. This can be confirmed by native gel electrophoresis (Vlassov et al. 2001) showing stable complex formation between RNA 9 and RNA 10Trp with and without 1 mM tryptophan (not shown). By integrating the total counts in RNA 9 and RNA 10 eluted by tryptophan in Figure 2, B and D ▶, we calculate that the molar composition of RNA (9:10Trp) is 1.2:1, somewhat less (Vlassov et al. 2001) than the 2:1 ratio in the parental RNA (9:10).
We wished to exploit the specificity of the tryptophan binding site in permeability experiments, so we tested free RNA (9:10Trp) by elution from Trp-Sepharose with 1 mM of nine different amino acids (Fig. 2E ▶). Five prior, consistent isocratic affinity elution experiments (Ciesiolka et al. 1996) using 20 to 50μM L-Trp suggest that the KD of RNA (9:10Trp) for L-Trp in solution is 7 ± 1 μM (±SEM; data not shown), close to the dissociation constant of the original Trp site in RNA 727–44 (I. Majerfeld and M. Yarus, in prep.). Therefore, in Figure 2E ▶, the RNA slowly elutes, even in the absence of ligand for the Trp site, as expected from its finite affinity for Trp-Sepharose. Nevertheless, several amino acids can be serially tested to see if they increase elution over the spontaneously dissociating tail. Millimolar L-Lys, Arg, His, Gln, Glu, Leu, Ile, and Phe are negligible eluants (Fig. 2E ▶). Conversely, L-Trp works as expected, eluting remaining RNA counts per minute quantitatively. Recovery of RNA approximates 100% and a subsequent gentle column sweep without divalents (using 1 M NaCl) elutes no RNA (Fig. 2E ▶).
Thus, RNA (9:10Trp) in solution is very specific for tryptophan and does not respond to other basic, acidic, polar, aliphatic, and aromatic amino acids. Taking these data conservatively as an indication that KD > 1 mM for the other amino acids (Ciesiolka et al. 1996), discrimination in favor of Trp is 150-fold or greater.
Binding to phospholipid bilayers tested
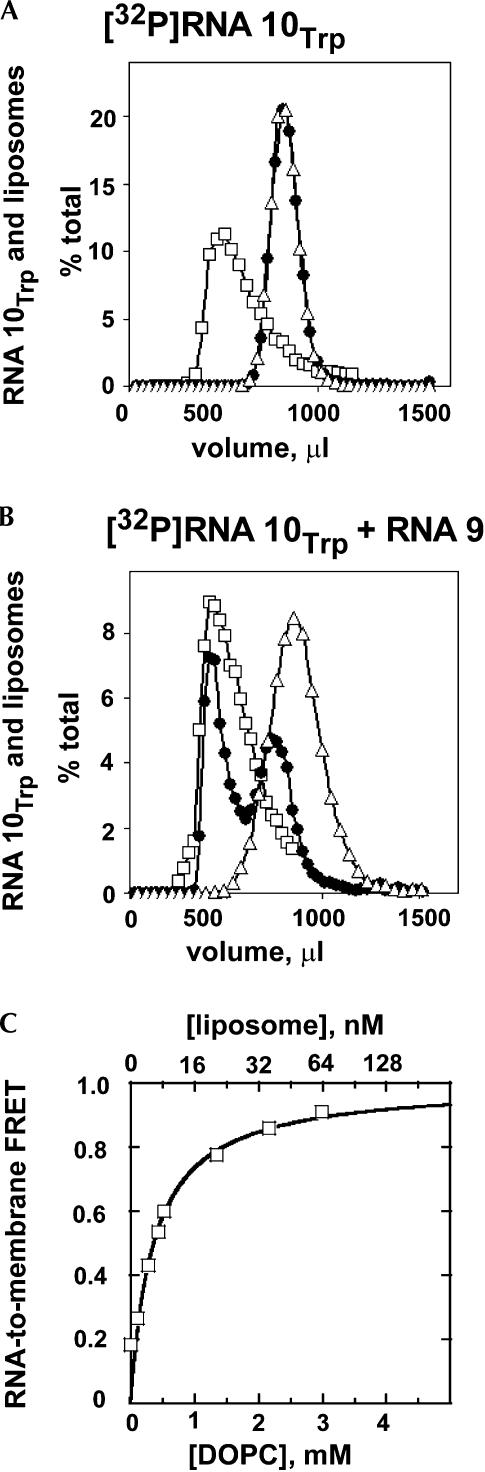
Above we show that tryptophan affinity and RNA complex formation are intact. To now show that RNA (9:10Trp) binds phosphatidylcholine membranes, we first used gel filtration on a column that voids liposomes but includes RNAs of this size (Khvorova et al. 1999). Figure 3A ▶ shows that free RNA 10Trp is retarded by Sephacryl S-1000 as expected. Furthermore, RNA 10Trp is not displaced toward the void when exposed to phospholipid liposomes (Fig. 3A ▶). Therefore, RNA 10Trp shows no detectable membrane interaction. However, the inclusion of RNA 9 displaces labeled RNA 10Trp into the void, tracking the liposome peak (Fig. 3B ▶). Thus, though neither RNA alone binds well, RNA (9:10Trp) efficiently binds phosphatidylcholine liposomes.
FIGURE 3.
RNA affinity for phosphatidylcholine liposomes measured by gel filtration on Sephacryl S-1000. (A) [32P]RNA 10Trp un-reacted (triangles) and reacted (circles) with liposomes (as OD320, squares). (B) (RNA 9:[32P]RNA 10Trp) complex unreacted (triangles) and reacted (circles) with liposomes. Membrane affinity by RNA-to-membrane FRET: (C) Membrane containing Texas Red titrated with YOYO-labeled RNA (9:10Trp) complex.
To confirm this essential quality of RNA (9:10Trp), we did FRET measurements between RNA labeled by the intercalated green dye, YOYO-1, and membranes labeled with the intercalated red dye, Texas Red (Janas and Yarus 2003). FRET is highly distance-dependent and becomes efficient only when the RNA enters the membrane, bringing the two bound fluors together. The data (Fig. 3C ▶, fitted line) suggest a normal binding isotherm as the number of liposomes is increased, corresponding to KD = 6 ± 1 nM, considering one of our 100 nm diameter liposomes as a “site.” Thus RNA (9:10Trp) binds phospholipid membranes by both qualitative and quantitative indices. To summarize the activities of RNA (9:10Trp), the two RNAs are complexed, the tryptophan site is active, and affinity for phospholipid membranes is demonstrable.
Permeability: Response to tryptophan and RNA concentration
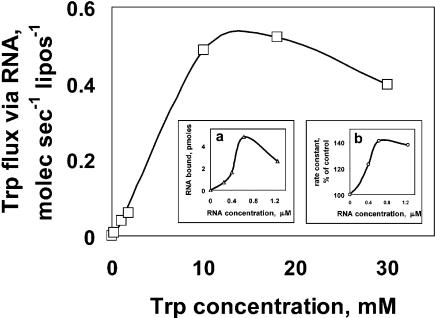
Figure 4 ▶ shows the result of varying external tryptophan concentration from 0.18 μM up to the closest approach we can make to the solubility limit, 30 mM L-tryptophan. At low concentrations the increase in uptake velocity due to RNA (liposome data subtracted) is accurately linear (low concentration points are not resolved in a linear plot like Figure 4 ▶). Saturation is observable, as expected for a limited number of amino acid binding sites, at an RNA-dependent velocity ≅0.5 molecule sec-1 liposome-1 at 4°C. However, half-saturation occurs at unexpectedly high L-tryptophan, about 5 mM. This approaches 1000-fold more concentrated tryptophan than required to saturate the free RNA 10Trp (above) so we suspect the action of the RNA binding site is altered on the phospholipid membrane.
FIGURE 4.
Membrane activities of RNA (9:10Trp) complex versus tryptophan concentration and versus RNA concentration. (Main panel) Concentration dependence of zero-trans Trp uptake due to liposome-bound RNA. (Inset a) Liposome-bound RNA versus RNA concentration. (Inset b) Relative tryptophan uptake rate as a function of RNA concentration.
The insets in Figure 4 ▶ show data for variation of the RNA concentration versus liposome binding (inset a), as well as RNA concentration versus RNA-dependent tryptophan permeation (inset b). We consider these independent experiments identical within experimental error. Roughly speaking, at low RNA concentration there is an upward-curving trend in both binding and permeability, then a point of maximal effectiveness at ~0.6 μM RNA. The data at lower RNA concentrations suggest that RNA molecules collaborate by complex formation to bind membranes and alter permeability. Such complex formation was documented previously using gel electrophoresis atomic force microscopy (Vlassov et al. 2001; Janas and Yarus 2003). Above 0.6 μM, it is possible that RNA aggregates become too large for optimal function. Below we have usually used the RNA concentration giving peak activity, 0.6 μM.
Kinetics of the permeability effect
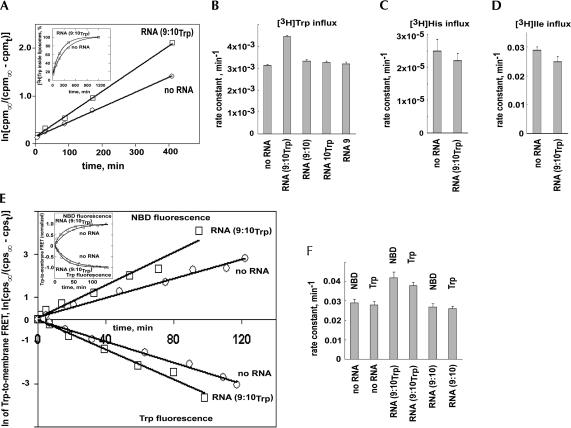
We measured entry of external 0.18 μM [3H]Trp into liposomes, using gel filtration in the presence of external non-isotopic tryptophan to remove external [3H]tryptophan (see Materials and Methods). Trp has a very hydrophobic amino acid side chain, and so it has an easily detectable spontaneous entry rate. Our measured rates for this spontaneous permeation agree closely with previous literature determinations, thus supporting our permeability measurements (Chakrabarti 1994). As expected, final levels were always the same inside ±RNA (Fig. 5A ▶ inset), so only rate constants vary between experiments. Because RNA increased the inward rate without change in final steady-state Trp level, the implication is that RNA increased outward rate by the same factor, as required by the principle of Microscopic Reversibility.
FIGURE 5.
Kinetics of amino acid influx into phosphatidylcholine liposomes. Using scintillation counting of liposomes: (A) representative [3H]Trp experiment at 4°C showing convergence (inset) and slope proportional to rate, k; cpmt = cpm∞(1-e-kt); (B) average of seven rate constants with SEM bars for tryptophan; (C) average of five rate constants with SEM bars for histidine; (D) average of three rate constants with SEM bars for isoleucine. Using optical techniques: (E) liposomal entry of tryptophan measured by Trp →NBD-PE FRET at 4°C showing convergence (inset) and slope proportional to rate. (F) Average of three FRET Trp uptake experiments with SEM bars.
Total tritium, as well as the much smaller amount of tritium within liposomes, co-chromatographed entirely with a sharp peak of non-isotopic L-tryptophan absorbance on HPLC (not shown). Therefore these 3H measurements characterize membrane transit by bona fide tryptophan, not a radioactive contaminant or decay product.
Liposomes in the absence of RNA fill with Trp at a rate of 3.14 ± 0.03 × 10-3 min-1 at 4°C (±SEM; details available from yarus@colorado.edu). When RNA (9:10Trp) is present, this rate increases to 4.44 ± 0.04 × 10-3 min-1; the rate is 41% ± 2% faster with RNA. To see what this overall change implies for the small number of bound RNAs, we must take a molecular viewpoint (below). However, a t-test shows that (Fig. 5B ▶; details available from yarus@colorado.edu) the probability of this difference or greater, if normal populations have equal means, is only 1.8 × 10-12. Thus Trp entry is certainly stimulated by RNA (9:10Trp). Similarly with no Trp binding site, the rate of Trp entry in the presence of RNA (9:10) is measured to be 3.32 × 10-3 min-1. This rate difference is also overwhelmingly significant; the probability that RNA (9:10) liposomes and RNA (9:10Trp) liposomes are the same is 3.4 × 10-8. Therefore, RNA not only stimulates Trp entry, but stimulation is unmistakably dependent on the Trp binding site.
Furthermore, no significant Trp permeability effect occurs for RNA 10Trp alone and RNA 9 alone (Fig. 5B ▶). Thus enhanced permeability is not evoked by an arbitrary RNA, but instead requires both RNAs of the complex and also the Trp site of the RNA (9:10Trp) complex. The RNA complex appears to have little or no effect on general Trp permeability (probability of the no RNA versus RNA (9:10) difference = 0.016). The absence of a significant general permeability effect is discussed further below.
Specificity of the permeability effect
The effect of the RNA (9:10Trp) complex was specific for tryptophan; no significant increase of the influx rate was observed for [3H]histidine or [3H]isoleucine (Fig. 5C,D ▶). Observations (available by e-mail from yarus@colorado.edu) actually show a non-significant decrease of permeability: 87 ± 15% (±SEM) for both histidine in the presence of RNA (9:10Trp) and 87 ± 7% for isoleucine. This correlates with the observation (Fig. 2E ▶, above) that these amino acids are not bound by the Trp site. Thus, RNA (9:10Trp) does not generally increase membrane permeability to amino acids (as would an open aqueous conduit) but instead must use a more selective mechanism.
The presence of 0.2 mM nonisotopic Trp inside liposomes has no effect on the rate of external [3H]Trp entry in the presence of 0.6 μM RNA; the ratio of Trp entry rates ± internal Trp was 1.000 ± 0.067 (±SEM; a counterflow experiment). Thus, the membrane-active RNA does not perceive internal ligand, even at 1000-fold the concentration of external Trp. Finally, external tritiated His does not enter more readily when external 0.2 mM Trp is also present. Thus, Trp does not open a route for other amino acids; instead, it is uniquely transported. These observations together are as expected for simple, one-site facilitated diffusion via RNA (9:10Trp), in which transported amino acids must pass, one by one, through the unique external Trp binding site.
Because RNA transporter activity is unprecedented, we confirmed it using an independent technique, tryptophan-to-membrane FRET (Fluorescence Resonance Energy Transfer) spectroscopy (Cajal et al. 1997). The intrinsic fluorescence of tryptophan was used as the origin of FRET. To make FRET dependent on importation of the amino acid into a vesicle, we measured tryptophan FRET to a fluor (NBD-PE; Cajal et al. 1997) incorporated specifically into the inner leaflet of a phospholipid liposome (McIntire and Sleight 1991). With internal NBD-PE only (external dye has been destroyed; see Materials and Methods), FRET from tryptophan to NBD requires crossing the membrane (Fig. 5E,F ▶; data available by e-mail from yarus@colorado.edu).
RNA (9:10) without a Trp binding site did not increase membrane permeability to tryptophan (93 ± 8% of a control without RNA using Trp emission and 92 ± 8% using NBD-PE emission). Therefore, the insignificant effect of RNA on general permeability is once again supported. However, tryptophan permeability with RNA (9:10Trp) was increased 144 ± 14%, using the rise in NBD-PE fluorescence, and increased 134 ± 10%, (±SEM) using the decrease in Trp emission. Thus FRET confirms the dependence on the amino acid binding site and quantitatively confirms permeability enhancements measured using [3H]Trp. Because FRET is an optical method entirely independent of the above isotopic permeability measurements (for example, no purification of liposomes occurs), these congruent results strongly support RNA-mediated increases in membrane permeability to tryptophan of the magnitude cited.
Activation energy
In order to estimate the activation energy for tryptophan influx, experiments were performed at 4°C, 14°C and 24°C (description available by e-mail from yarus@colorado.edu). This [3H]tryptophan assay showed that the activation energy for tryptophan influx into unmodified phosphatidylcholine liposomes was 91 ± 5 kJ/mole, whereas the value for influx via RNA (9:10Trp) was 46 ± 3 kJ/mole. RNA (9:10Trp) therefore halves the activation energy for membrane passage by Trp. This reduction is greater than the binding free energy of the parental tryptophan site (29 kJ/mole; calculated from KD), so it may require contributions from both ligand binding and RNA-induced lipid rearrangement, as posited above.
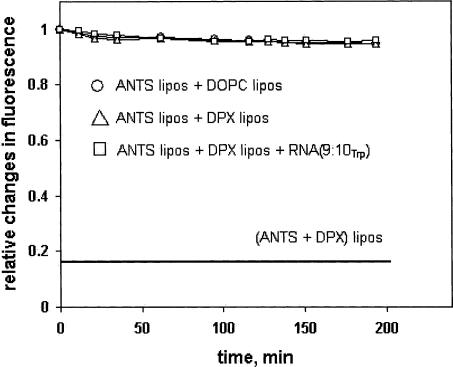
No role for liposome fusion
We wished to be sure that the permeability effect of RNA (9:10Trp) did not depend on liposome fusion. This is improbable even at the start, because it would require a special explanation for the observed amino acid selectivity, as well as for the measured differences between RNA (9:10) and RNA (9:10Trp). However, we determined membrane fusion directly by encapsulating a fluor, aminonaphthalene trisulfonic acid (ANTS), in one set of dioleoylphosphatidylcholine liposomes and an effective fluorescence quencher, p-xylene-bis-pyridinium (DPX), in another set (Ellens et al. 1985). Mixed liposomes detect vesicle fusion by quenching of ANTS fluorescence at 514 nm. The lower solid line in Figure 6 ▶ shows the level of fluorescence expected for complete mixing of the fluor and quencher (ca. complete fusion); thus, mixing of liposome contents would be measurable. In fact, since a fusion hypothesis would require the involvement of most liposomes to explain permeation of the total liposome volume at an enhanced rate (Fig. 5A ▶), the absence of any observable RNA (9:10Trp) effect in this experiment (Fig. 6 ▶) effectively rules out liposomal fusion as an essential and pervasive step in Trp permeation.
FIGURE 6.
Fluorometric liposomal fusion assay measuring the interaction of liposomes containing a fluor (ANTS) with others containing a quenching agent (DPX).
The magnitude of the increase in permeability per RNA
We now narrow our focus to discuss transport at individual membrane RNA sites. At the optimal RNA concentration usually used, 12 RNA molecules were added per liposome. Measured binding (Fig. 3B ▶) is 40%, so 4.8 RNA per liposome are stably bound, on average. If the stoichiometry of these complexes is 1 molecule of RNA 10Trp per 1.2 molecules of RNA 9, then there are about two Trp binding sites per liposome on average. Given the virtually complete dependence of increased permeability on the Trp binding site, most RNA-provoked phenomena likely occur at or very near the two Trp binding centers.
To gain an approximate microscopic view of Trp transport, we first assume that all RNAs are equally active. If they differ or only some are active, then we will calculate a minimum RNA effect. Conservatively, the cross sectional area of the RNA 10Trp is 16 nm2. This estimate maximizes the membrane surface required by considering RNA10Trp an A-form helix. A more realistic globular shape would be smaller in cross section, and give a higher apparent activity. A 200-nm diameter liposome has a surface of 1.26 × 105 nm2. Two RNA 10Trp therefore occupy 0.025% or 1/4000 of the surface, where the increased permeability takes place. Using our most precise data, a 41% increase in tryptophan influx implies a permeability increase due to RNA of greater than 0.41 × 4000 ≈1640-fold, or about 820-fold for each RNA 10Trp /tryptophan binding site.
Turnover
Tryptophan uptake increases with tryptophan concentration outside. Unmodified liposomes show an approximately linear increase in total tryptophan uptake as the concentration of Trp increases. After subtraction of this background uptake, the maximal RNA-induced uptake per liposome was ~0.5 sec-1 (Fig. 4 ▶), so the true first order rate is ≅0.25 sec-1 at 4°C for each RNA10Trp. At high, saturating Trp concentrations, in the 1700 min of the usual kinetic experiment, each transporter site turns over in the inward direction more than 25,000 times. Thus the RNA transporter performs many identical permeation cycles, as also do proteins. Multiple turnover occurs at a constant rate (Fig. 5 ▶); this suggests that the RNA porter has a stable, consistent membrane conformation.
Finally, in order to pass tryptophan only, a Trp carrier is apparently logically required to have two functional orientations, one in which it acquires Trp from bulk solution and the other for release of bound Trp toward the membrane. Transport may therefore be mediated by repetitive molecular motions at 0.25 sec-1 involving the Trp binding site of RNA (9:10Trp).
Known RNA properties can explain increases in specific permeability
A mechanism for facilitated tryptophan permeation is evident from prior data. We found that membrane RNAs prefer the edges of the lipid bilayer and other sites where irregular lipid conformations occur (Janas and Yarus 2003). This suggests that membrane RNAs prefer and stabilize irregular lipid conformations, perhaps including those required to form transmembrane defects. There is experimental evidence of the existence of transient, aqueous defects in membranes (Smith et al. 1984; Paula et al. 1996; Sandre et al. 1999), as well as evidence that polar molecules must traverse such routes crossing the bilayer (Paula et al. 1996). The observed 800-fold increase in permeability would require ≈2,000-fold increase in defect lifetime (Raphael 2001). This seems plausible because it requires only about 20 kJ/mole stabilization of the defect, corresponding to a few secondary interactions. The defect density of a phosphatidylcholine bilayer is otherwise very small—defects occupy only 2 × 10-7 of an unperturbed membrane (Smith et al. 1984). Thus, we suggest that RNA (9:10Trp) is the stabilizing nucleus of an aqueous, membrane-crossing defect.
However, the stabilized defect must be mostly blocked by RNA (9:10Trp) to explain its relative inaccessibility to free tryptophan, isoleucine, and histidine. Only RNA-bound tryptophan has access, and thereby, increased permeability. This difference between previous observations (Khvorova et al. 1999; Vlassov et al. 2001) of increased general permeability and these present results may be explained by the use of 10- to 15-fold fewer RNA molecules per liposome here, or by unanticipated molecular changes accompanying the introduction of the Trp site.
Comparison to proteins
Turnover numbers of protein transporters (Stein 1990) are usually in the range of 0.1 to 1000 sec-1. Protein transporters that use specific binding sites to move one molecule per cycle have rates at the slower end of this range. A mitochondrial ADP/ATP carrier works at 0.05–0.1 sec-1 at 25°C (Schonfeld et al. 1996), while the glucose transporter in human erythrocytes turns over at 600 sec-1 at 20°C (Stein 1990). The activation energy for protein transmembrane porters is usually in the range 40 to 130 kJ/mole, for example, sulfate ion transport by band 3 protein in human erythrocytes is at EA = 131 kJ/mole (Janas et al. 1988) and, for the Glossina morsitans proline transporter, EA = 46 kJ/mole (Njagi et al. 1992). In this context, RNA (9:10Trp) at ≥0.25 sec-1 and 46 kJ/mole is both quantitatively and, by mechanism, comparable to slower protein transporters, and therefore appropriately called a tryptophan transporter made of RNA.
Potential role in cellular life
These findings suggest that RNA may have been underestimated as an active agent in biological processes. In particular, RNA-mediated transport might have been essential to early cellular life. Amphipathic lipids should have been freely available on the early Earth (Deamer et al. 2002) and growth of membrane-bound compartments via lipid assimilation is facilitated by common clay minerals (Hancyzc et al. 2003). However, a lipid bilayer enclosing an RNA cell (ribocyte) must exhibit specific molecular permeabilities in order to retain intermediates and import substrates. For a ribocyte living before the advent of templated peptide synthesis (Yarus 2002), specific permeability modification probably requires membrane RNAs. By whatever means RNA accomplishes the reaction, it is clear from this work that specific membrane transport is within the RNA repertoire. Following Bayes’ Theorem, such confirmed predictions necessarily make an RNA world and its ribocytes more probable (Yarus 2002).
Extension of the method to other RNA activities
Finally, these data taken together corroborate the initial assumptions used to design RNA (9:10Trp). As we originally supposed, RNA 10 can be substituted with an independently derived RNA motif, x, without disrupting the ability of the RNA (9:10x) complex to bind phospholipid bilayers or disrupting the activity of the substituted motif. This suggests that other nanoconstructions would be successful. For example, using protein aptamers, any protein could be made a peripheral membrane protein, or using ribozymes, any catalytic acceleration in the RNA repertoire could be conferred on membranes.
MATERIALS AND METHODS
Affinity chromatography
A total of 10 pmole RNA (6.7 pmole RNA 9, 3.3 pmole RNA10Trp) in water was heated for 3 min at 65°C, brought to 1× buffer (50 mM Hepes at pH 7.0, 250 mM NaCl, 5 mM MgCl2, 2 mM CaCl2), allowed 10 min at room temperature, loaded onto the 0.2-mL column at 23°C, and eluted with 1× buffer. Trp-Sepharose contained about 1.2 mM L-tryptophan. Fractions contained about 150 μL.
Gel filtration
A total of 25 pmole RNA (in Fig. 3 ▶; molar ratio 2 RNA 9:1 RNA 10) in water was held for 3 min at 65°C, brought to 1× buffer H (50 mM Hepes at pH 7.0, 50 mM NaCl, 5 mM MgCl2, 2 mM CaCl2) and cooled to room temperature over 10 min. DOPC liposomes (10 mg/ml) were incubated with RNA (0.6 μM) for 5 min at room temperature and applied to a 1 mL Sephacryl S-1000 column at 23°C and eluted with buffer H. Fractions contained about 30 μL.
Binding by FRET
RNA-to-membrane FRET (Fluorescence Resonance Energy Transfer) between RNA-bound YOYO and membrane-bound Texas Red PE (molecular probes) was measured by exciting YOYO while monitoring its emission decrease due to FRET (Kohout et al. 2003). RNA (0.6 μM in buffer H) was mixed with the cationic cyanide fluorescence dye YOYO-1 iodide (final concentration 6.6 μM). The liposomes were added and RNA-to-membrane FRET was monitored by decrease in YOYO emission; λex = 466 nm and λem = 511 nm (Janas and Yarus 2003). FRET is highly distance dependent, and becomes efficient only when the RNA enters the membrane, bringing the fluors together.
Permeability using tritiated Trp
Tritiated amino acids were added (to 0.18 μM) at 4°C to a suspension of DOPC vesicles (6.3 mM lipid) and RNA (0.6 μM) in buffer H where required. Where amino acid concentration was varied, cold tryptophan was also added. Above 1 mM tryptophan, the increased osmotic strength was compensated by increases in internal NaCl in the liposome preparation. Unilamellar liposomes were prepared from phosphatidylcholine (DOPC; 1,2-dioleoyl-snglycero-3-phosphocholine; Avanti Polar Lipids) at a final concentration of 20 mg/ml, by using the Avanti MiniExtruder with a pore filter diameter of 200 nm. Aliquots were taken at intervals and free amino acid was removed at 4°C by two serial passages through ProbeQuant G-50 Sephadex Micro Columns (Amersham) pre-equilibrated with cold amino acid (0.2 mM) in buffer H. Typically, eight points over about 28 h were fit with a least-squares exponential to get the rate constant.
Permeability using FRET
Tryptophan-to-membrane FRET between tryptophan and membrane bound NBD-PE (Cajal et al. 1997) was measured in buffer H by exciting tryptophan while monitoring both its emission decrease due to FRET (λex = 300 nm and λem = 351 nm) and the NBD-PE emission increase due to FRET (λex = 300 nm and λem = 531 nm). In these measurements, a fluor is selectively placed on the inner surface of 100 nm phospholipid liposomes by forming vesicles with the dye N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoylsn-glycero-3-phosphoethanolamine (NBD-PE; molecular probes) in both inner and outer membrane leaflets, then destroying the external fluorophore with an aqueous reductant (McIntyre and Sleight 1991). The resulting asymmetrically labeled liposome must be penetrated by Trp in order for the intrinsically fluorescent Trp molecule to be within range for efficient FRET to dye molecules in the inner membrane leaflet. Accordingly, FRET can be used to measure Trp entry.
Liposome fusion assay
The following liposomal pools were prepared: DOPC liposomes containing buffer H (50 mM Hepes at pH 7.0, 50 mM NaCl, 5 mM MgCl2, 2 mM CaCl2) inside; DPX liposomes containing buffer H and 90 mM quencher DPX (p-xylene-bis-pyridinium bromide, Sigma) inside; ANTS liposomes containing buffer H and 25 mM fluor ANTS (8-aminonaphthalene-1,3,5-trisulfonic acid, disodium salt, Sigma) inside; control (ANTS + DPX) liposomes containing both ANTS and DPX. The liposomes were extruded in the presence of appropriate solution, and then the liposomes were separated from untrapped material using two subsequent passages through ProbeQuant G-50 Sephadex Micro Columns. The external buffer was adjusted to match the osmotic pressures. Quenching of ANTS fluorescence was monitored at 25°C using excitation and emission at λex = 354 nm and λem = 514 nm, respectively.
Acknowledgments
We thank the members of our lab for comments on a draft and also Dr. Joseph J. Falke (University of Colorado, Biochemistry) for use of his spectrofluorometer. This work was supported by NIH Research Grant GM 30881 and NASA Center NCC2–1052.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7112704.
REFERENCES
- Cajal, Y., Boggs, J.M., and Jain, M.K. 1997. Salt-triggered intermembrane exchange of phospholipids and hemifusion by myelin basic protein. Biochemistry 36: 2566–2576. [DOI] [PubMed] [Google Scholar]
- Chakrabarti, A.C. 1994. Permeability of membranes to amino acids and modified amino acids: Mechanisms involved in translocation. Amino Acids 6: 213–229. [PubMed] [Google Scholar]
- Ciesiolka, J., Illangasekare, M., Majerfeld, I., Nickles, T., Welch, M., Yarus, M., and Zinnen, S. 1996. Affinity selection-amplification from randomized ribooligonucleotide pools. Methods Enzymol. 267: 315–335. [DOI] [PubMed] [Google Scholar]
- Deamer, D., Dworkin, J.P., Sandford, S.A., Bernstein, M.P., and Allamandola, L.J. 2002. The first cell membrane. Astrobiology 2: 371–381. [DOI] [PubMed] [Google Scholar]
- Ellens, H., Bentz, J., and Szoka, F.C. 1985. H+ and Ca+-induced fusion and destabilization of liposomes. Biochemistry 24: 3099. [DOI] [PubMed] [Google Scholar]
- Hanczyc, M.M., Fujikawa, S.M., and Szostak, J.W. 2003. Experimental models of primitive cellular compartments: Encapsulation, growth, and division. Science 302: 618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas, T. and Yarus, M. 2003. Visualization of membrane RNAs. RNA 9: 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas, T., Janas, T., Kilian, M., and Przestalski, S. 1988. Activation energy of sulfate ion transport across methylated human erythrocyte membranes. FEBS Lett. 236: 93–94. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Kwak, Y.-G., Tamkun, M., Majerfeld, I., and Yarus, M. 1999. RNAs that bind and change the permeability of phospholipid membranes. Proc. Natl. Acad. Sci. 96: 10649–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, R., Birmingham, A., and Yarus, M. 2004. BayesFold: Rational 2° folds that combine thermodynamic, covariation, and chemical data for aligned RNA sequences. RNA 10: 1323–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout, S.C., Corbalan-Garcia, S., Gomez-Fernandez, J.C., and Falke, J.J. 2003. C2 domain of protein kinase C α: Elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry 42: 1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre, J.C. and Sleight, R.G. 1991. Fluorescence assay for phospholipids membrane asymmetry. Biochemistry 30: 11819– 11827. [DOI] [PubMed] [Google Scholar]
- Njagi, E.N.M., Olembo, N.K., and Pearson, D.J. 1992. Proline transport by tsetse fly Glossina morsitans flight muscle mitochondria. Comp. Biochem. Physiol. B 102: 579–584. [DOI] [PubMed] [Google Scholar]
- Paula, S., Volkov, A.G., Van Hoek, A.N., Haines, T.H., and Deamer, D.W. 1996. Permeation of protons, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys. J. 70: 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raphael, R.M. 2001. Fractional occurrence of defects in membranes and mechanically driven interleaflet phospholipid transport. Phys Rev. E Stat. Nonlin. Soft Matter Phys. 64: 051913/1–051913/10 [DOI] [PubMed] [Google Scholar]
- Sandre, O., Moreaux, L., and Brochard-Wyart, F. 1999. Dynamics of transient pores in stretched vesicles. Proc. Natl. Acad. Sci. 96: 10591–10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonfeld, P., Schild, L., and Bohnensack, R. 1996. Expression of the ADP/ATP carrier and expansion of the mitochondrial (ATP + ADP) pool contribute to postnatal maturation of the rat heart. Eur. J. Biochem. 241: 895–900. [DOI] [PubMed] [Google Scholar]
- Smith, J.R., Laver, D.R., and Coster, H.G.L. 1984. The conductance of lecithin bilayers: The dependence upon temperature. Chem. Phys. Lipids 34: 227–236. [Google Scholar]
- Stein, W.D. 1990. Channels, Carriers, and Pumps: An introduction to membrane transport. Academic Press, San Diego, CA.
- Vlassov, A., Khvorova, A., and Yarus, M. 2001. Binding and disruption of phospholipid bilayers by supramolecular RNA complexes. Proc. Natl. Acad. Sci. 98: 7706–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener, W.C. and White, S.H. 1992. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. Biophys. J. 61: 433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus, M. 2002. Primordial genetics: Phenotype of the ribocyte. Annu. Rev. Genet. 36: 125–151. [DOI] [PubMed] [Google Scholar]