Abstract
Adenosine deaminases that act on RNA (ADARs) catalyze adenosine to inosine conversion in RNA that is largely double stranded. Human ADAR2 (hADAR2) contains two double-stranded RNA binding motifs (dsRBMs), separated by a 90-amino acid linker, and these are followed by the C-terminal catalytic domain. We assayed enzymatic activity of N-terminal deletion constructs of hADAR2 to determine the role of the dsRBMs and the intervening linker peptide. We found that a truncated protein consisting of one dsRBM and the deaminase domain was capable of deaminating a short 15-bp substrate. In contrast, full-length hADAR2 was inactive on this short substrate. In addition, we observed that the N terminus, which was deleted from the truncated protein, inhibits editing activity when added in trans. We propose that the N-terminal domain of hADAR2 contains sequences that cause auto-inhibition of the enzyme. Our results suggest activation requires binding to an RNA substrate long enough to accommodate interactions with both dsRBMs.
Keywords: ADAR, deamination, RNA editing, dsRBM, inosine
INTRODUCTION
RNA editing by adenosine deamination is catalyzed by members of an enzyme family known as adenosine deaminases that act on RNA (ADARs; Bass 2002). The RNA substrates for ADARs are largely double stranded, and the product of the reaction is RNA that contains inosine. Cellular enzymes recognize inosine as guanosine, and thus, ADARs change the primary sequence information of RNA. In addition, ADARs are capable of altering RNA structure by changing AU base pairs to IU mismatches, or AC mismatches to IC base pairs. In vivo, human ADARs (hADARs) deaminate adenosines in coding regions of glutamate and serotonin receptor pre-mRNAs, in the hepatitis delta virus antigenome, and in noncoding regions of mRNAs such as introns and UTRs (Sommer et al. 1991; Lomeli et al. 1994; Polson et al. 1996; Burns et al. 1997; Morse et al. 2002). Purified enzyme preparations deaminate the same adenosines as observed in vivo, indicating that specificity is intrinsic to the enzyme and does not require other factors (Maas et al. 1996; Polson et al. 1996).
All ADARs have a conserved C-terminal deaminase domain and contain from one to three double-stranded RNA binding motifs (dsRBMs) that comprise the N-terminal RNA binding domain. Different ADARs exhibit varying distances between the dsRBMs. For example, the distance between the two dsRBMs in hADAR2 is ~90 residues, more than twice as long as the analogous sequence in hADAR1. The N termini of several ADARs have been deleted, and the resulting truncated proteins show varying levels of activity. For example, when all three of the dsRBMs of rat ADAR1 are deleted from the N terminus, the resulting protein retains deaminase activity in vivo (Herbert and Rich 2001). In contrast, when the first two dsRBMs of hADAR1 are deleted, leaving dsRBM3 and the catalytic domain, the resulting protein is not active in vitro (Liu et al. 2000). In addition, when the extreme N terminus of Drosophila melanogaster ADAR (dADAR) is deleted, leaving both of its two dsRBMs intact, the protein is inactive in vitro, but retains the ability to bind dsRNA (Gallo et al. 2003).
We conducted a study to characterize the contribution of the dsRBMs, and the linker sequence between them, to deamination by hADAR2 in vitro. We constructed several N-terminal deletion mutants of hADAR2 and determined the ability of each to deaminate RNA. From our results, we suggest hADAR2 is subject to auto-inhibition by sequences in the N-terminal domain. Activation requires binding to dsRNA and does not occur with dsRNA of very short length, raising the possibility that auto-inhibition functions to minimize deamination of short helices within endogenous RNA. In our model, activation involves binding of dsRBM1 to an extended RNA duplex, which then allows for binding of dsRBM2 and deamination of the substrate by the catalytic domain.
RESULTS
Limited proteolysis of hADAR2
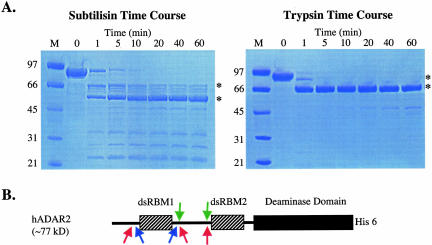
We subjected purified hADAR2 to limited proteolysis using subtilisin and trypsin to find stable domains that retained deaminase activity. Subtilisin nonspecifically cleaves peptide bonds in solvent-accessible regions, and trypsin is specific for accessible peptide bonds that are C terminal to basic amino acids. A 1-h time course showed that both subtilisin and trypsin rapidly degrade the full-length ADAR2 into smaller fragments within 1 min (Fig. 1A ▶). However, some of the smaller polypeptides were exceptionally stable and remained intact for the full hour (Fig. 1A ▶). Subtilisin digestion produced numerous fragments, two of which were prominent (~61 kDa and ~55 kDa), whereas trypsin gave one prominent fragment (~61 kDa). Chymotrypsin showed a proteolysis profile similar to that of subtilisin (data not shown). The N termini of several of the larger fragments were sequenced to determine the sites of proteolysis, and the results are presented in schematic form in Figure 1B ▶. All protease sites were clustered in two regions outside of the characterized domains of hADAR2, and the majority of the sites were in the linker between the two dsRBMs.
FIGURE 1.
Limited proteolysis of hADAR2. (A) hADAR2 was treated with either subtilisin or trypsin for the indicated times. The products were run on denaturing polyacrylamide gels and visualized by staining with Coomassie brilliant blue. Peptides for which the N terminus was sequenced are labeled with an asterisk. (B) A schematic diagram of hADAR2 showing the approximate sites of proteolysis as determined by N-terminal sequencing for trypsin (blue arrows), subtilisin (green arrows), and chymotrypsin (pink arrows; primary data not shown for chymotrypsin). His 6 denotes a C-terminal tag of six histidine residues.
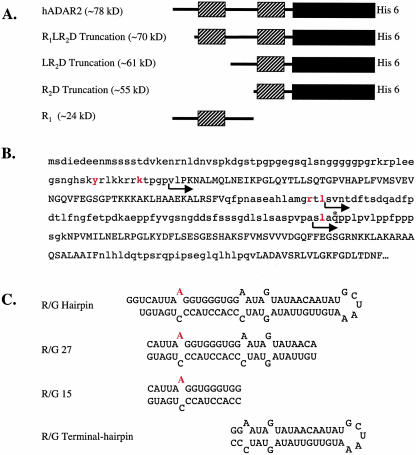
On the basis of the proteolysis results, we cloned and overexpressed three truncations of hADAR2 (Fig. 2A ▶). The constructs are named from the motifs that remain after deletion of the N terminus. Truncation R1LR2D, in which the N terminus up to the first dsRBM is deleted, retains amino acids 77–701 including dsRBM1, the linker between the dsRBMs, dsRBM2, and the deaminase domain; this truncation is similar to a recently described truncation of ADAR from D. melanogaster (Gallo et al. 2003). In the LR2D truncation, the N terminus, including the first dsRBM and the first 16 amino acids of the 92 residue linker between the two dsRBMs, is deleted, leaving residues 160–701, which include most of the linker, dsRBM2, and the deaminase domain. The R2D truncation encompasses residues 216–701 and leaves only dsRBM2 and the deaminase domain intact.
FIGURE 2.
Protein constructs and RNA substrates. (A) Schematic representation of hADAR2 constructs with rectangles showing dsRBMs (striped) and the catalytic domain (solid). (B) Sequence of the N-terminal RNA binding domain of hADAR2. Sequences corresponding to the two dsRBMs and catalytic domain are capitalized, amino acids N terminal to the protease cleavage sites are in red, and the N termini of the truncations that were cloned are denoted by arrows. The R1 construct begins at the initiator methionine and ends at the glutamine labeled by the asterisk four residues from the start of R2D. (C) R/G editing site constructs showing the edited adenosine in red. (R/G hairpin) Intramolecular stem–loop of 71 nucleotides; (R/G 27 and R/G 15) intermolecular duplexes formed from complementary strands of 27 and 15 nucleotides, respectively; (R/G terminal hairpin) a 41-nucleotide-long intramolecular stem–loop representing the terminal stem and loop of the R/G hairpin that does not include the sequence of R/G 15.
Expression and deaminase activity of N-terminal deletion constructs
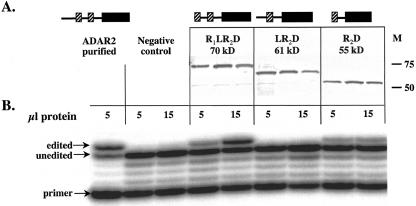
The three truncations of hADAR2, each containing a 6×-histidine tag at the C terminus, were expressed in the yeast Saccharomyces cerevisiae. To test for expression efficiency, we analyzed clarified cell extracts of cultures expressing the different truncations by Western blot using an anti-His antibody (Fig. 3A ▶). Because we often observe variation in protein expression from different clones, we analyzed extracts from three different clones for each truncation. Each of the three clones for each truncation were expressed at strong levels, with the expected sizes of ~70, 61, and 55 kDa (Fig. 3A ▶).
FIGURE 3.
Expression and deamination activity of the ADAR truncations. (A) A Western blot using an anti-His antibody to detect the three tagged truncations of hADAR2 (in triplicate) expressed in S. cerevisiae. (M) Molecular weight markers. (B) The deaminase activity of various amounts of extract protein on 2.5-nM R/G hairpin substrate (see Fig. 2C ▶) was determined with a limited primer extension assay (Öhman et al. 2000).
A natural substrate of hADAR2 is the R/G editing site in the mRNA encoding the B subunit of the glutamate receptor (Lomeli et al. 1994). This adenosine is efficiently deaminated by hADAR2 in vitro when in the context of a small RNA hairpin that mimics the structure of the pre-mRNA (Fig. 2C ▶; Öhman et al. 2000). Using the same extracts (Fig. 3A ▶), we assayed whether the truncated versions of hADAR2 were capable of deaminating this RNA at the R/G site. The R/G hairpin was incubated with either purified hADAR2 or extracts containing one of the three truncations, and editing was monitored with a limited primer extension assay (Öhman et al. 2000). As expected, the purified hADAR2 efficiently deaminated the R/G site adenosine, whereas extract from yeast harboring an empty expression vector did not (Fig. 3B ▶, negative control). The R1LR2D and R2D truncations also deaminated the R/G site adenosine (Fig. 3B ▶). Curiously, the LR2D truncation, which retains most of the linker sequence between the two dsRBMs as well as the second dsRBM and the catalytic domain, showed no deaminase activity when activity was assayed in a cell extract (Fig. 3B ▶) but was active when purified.
Purification of the smallest ADAR truncation
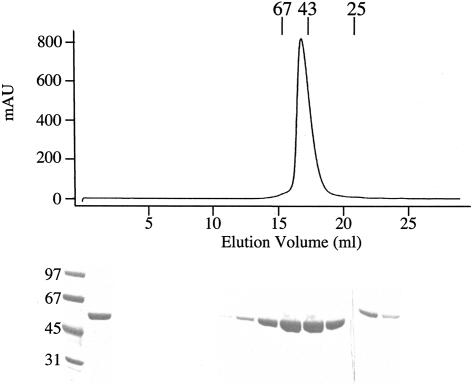
Because the R2D truncation, which contains only dsRBM2 and the catalytic domain, retained deaminase activity, we decided to characterize its activity in detail. To this end, we engineered a 10×-histidine tag at the N terminus that was immediately followed by the tobacco etch virus (TEV) protease recognition domain, to facilitate removal of nonnative sequences. The R2D truncation eluted as a monomer from a gel filtration column at the expected size of 54 kDa (Fig. 4B ▶) and had a high degree of purity (>95%) as assessed by Coomassie staining of an SDS-polyacrylamide gel (Fig. 4B ▶).
FIGURE 4.
Purification of the R2D truncation. In the final step of purification, the protein was chromatographed using Sephadex 200 gel filtration resin. The UV trace from this column is shown with the elution peaks of molecular weight standards (25, 43, and 67 kDa) labeled. The Coomassie-stained SDS-PAGE analysis of fractions is shown below the trace. The lanes under 22–25 mL show the trailing off of the main hADAR2 peak; these bands are offset because they were electrophoresed on a different gel.
Comparison of editing efficiency of hADAR2 and the R2D truncation
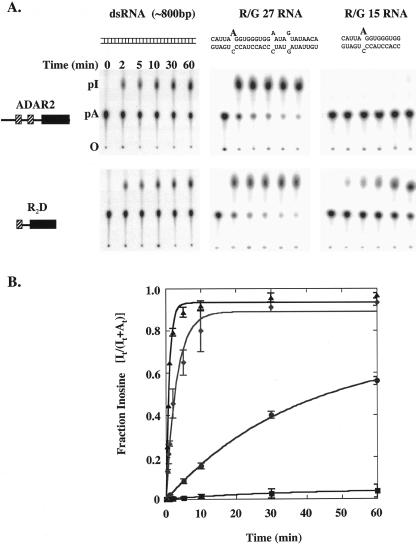
We directly compared the activities of truncation R2D and hADAR2 in vitro, using RNAs of several different lengths. We found both proteins deaminated a long, uniformly labeled dsRNA with approximately equal efficiency over the course of 1 h (Fig. 5A ▶, left panels). To monitor deamination of a specific adenosine, we prepared intermolecular duplexes corresponding to sequences surrounding the R/G editing site by hybridizing complementary strands of 27 (R/G 27) or 15 (R/G 15) nucleotides long. These molecules contained one 32P labeled adenosine at the R/G site (Fig. 2C ▶, red). Both proteins deaminated R/G 27 at the R/G editing site efficiently, with the reaction using the full-length protein being slightly faster than that of the R2D truncation (Fig. 5A ▶, center panels). Rather unexpectedly, we observed that the R2D truncation deaminated R/G 15 far more efficiently than full-length hADAR2 (Fig. 5A ▶, right panels).
FIGURE 5.
Comparison of the deaminase activity of full-length hADAR2 and the R2D truncation on RNA substrates of different lengths. One hundred nanomolar hADAR2 or the R2D truncation were incubated with 1 nM of an internally labeled ~800-bp dsRNA substrate or 5 nM of the R/G 27 or R/G 15 substrates (see Fig. 2C ▶) specifically labeled at the 5′ phosphate of the edited adenosine. After incubation for the times indicated, the reactions were stopped, and the RNA was extracted with phenol and chloroform and digested with Nuclease P1. The resulting 5′ mononucleotides were separated by TLC. (A) From left to right: TLC plates of the reaction of full-length hADAR2 (top panels) or R2D truncation (lower panels) with each of the three substrates. Positions of the origin (O), 5′ AMP (pA), and 5′ IMP (pI) are labeled. (B) The plot shows the fraction of IMP produced as a function of time for the reaction of 100-nM enzyme and 5-nM substrate. (Triangles) hADAR2 with R/G 27; (diamonds) R2D truncation with R/G 27; (circles) R2D truncation with R/G 15; (squares) hADAR2 with R/G 15. The data are the average of three independent experiments and were fit to the equation Ft = Fend(1 − e−kt), where Ft is the fraction of inosine at time t, Fend is the fraction of inosine at the endpoint, and k is the fitted rate constant.
To quantify these observations, we performed a kinetic analysis under single-turnover conditions (100 nM enzyme, 5 nM RNA). R/G 27 or R/G 15 RNA, specifically labeled at the editing site, was incubated with hADAR2 or the R2D truncation, and product formation was followed using the TLC assay. The rate of deamination of R/G 27 by hADAR2 was 1.1 min−1 and only slightly faster than the rate of 0.3 min−1 for the R2D truncation (Fig. 5B ▶; Table 1 ▶). However, a striking difference in the reaction rates of the two proteins was observed when using the smaller R/G 15 substrate. The rate of deamination of this RNA by hADAR2 was very slow, with a value of 0.00073 min−1, and the rate with the R2D truncation was 0.016 min−1, ~20-fold faster than the full-length enzyme (Fig. 5B ▶; Table 1 ▶).
TABLE 1.
Single-turnover kinetic parameters
| Reaction contents | kobs, min −1 | Fend |
| ADAR + R/G 27 | 1.1 ± 0.07 | 0.93 ± .012 |
| R2D + R/G 27 | 0.3 ± 0.03 | 0.89 ± 0.03 |
| R2D + R/G 15 | 0.016 ± 0.007 | 0.71 ± 0.05 |
| ADAR + R/G 15 | 0.00073 ± 0.0006 | 0.04 ± 0.0019 |
Reconstitution of the hADAR2 reaction in trans
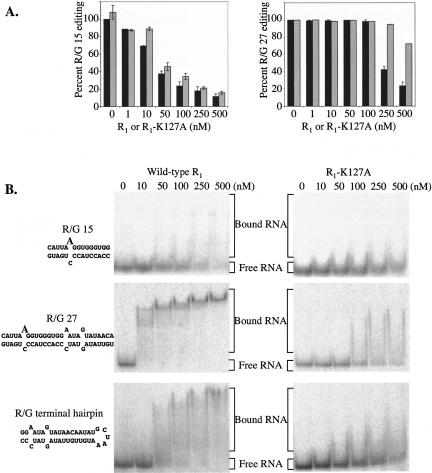
Because R2D, but not the full-length hADAR2, edited the R/G 15 substrate, we wondered whether the full-length protein contained an inhibitory domain not present in the R2D truncated protein. To explore this possibility, we investigated the effect of adding the N terminus of hADAR2 (construct R1; see Fig. 2 ▶) in trans to the R2D editing reaction. Indeed, we found that R1 inhibited editing of R/G 15 by the R2D truncation (Fig. 6A ▶, left panel, black bars). In contrast, but consistent with the observation that both proteins edited R/G 27, the reaction of R2D with R/G 27 was only inhibited when the R1 was added at very high concentrations (Fig. 6A ▶, right panel, black bars). Thus, we were able to mimic the activity of the full-length hADAR2 on both R/G 15 and R/G 27, in trans, by mixing the R1 and R2D proteins.
FIGURE 6.
The N terminus of hADAR2 inhibits R2D editing of R/G RNA when added in trans. (A) Increasing concentrations of the isolated N terminus of hADAR2 (R1, residues 1–219) or the mutant R1-K127A was added to reactions containing 100 nM R2D and 20 nM R/G 15 or R/G 27 labeled at the edited adenosine. The RNA was digested and editing was analyzed by TLC as described in Materials and Methods. (Left panel) Editing of R/G 15 substrate; (right panel) editing of R/G 27 substrate; (black bars) wild-type R1; (gray bars) R1-K127A. (B) Binding of the R1 constructs to R/G 15, R/G 27, or the R/G terminal hairpin RNA. Increasing concentrations of the wild-type R1 (left panels) or the R1-K127A mutant (right panels) were mixed with 32P end-labeled RNA, incubated at room temperature for 20 min, and analyzed by native gel electrophoresis. The top panels show binding to R/G 15, middle panels show binding to R/G 27, and lower panels show binding to the R/G terminal loop.
We considered the possibility that R1, which contains dsRBM1, inhibited the reaction of R2D with R/G 15 simply by competing for binding to the substrate. To test this idea, we prepared an N-terminal fragment (R1-K127A) that contained a point mutation in dsRBM1. The analogous mutation in a dsRBM of hADAR1 (Liu and Samuel 1996), as well as a dsRBM of protein kinase activated by RNA (PKR; Milligan et al. 1987; Patel et al. 1996), inhibits dsRNA binding. The K127A mutant R1 inhibited editing activity of R2D on the R/G 15 substrate (Fig. 6A ▶, left panel, gray bars) at levels similar to the wild-type R1, suggesting that this inhibition does not result from a simple competition for RNA binding. However, the inhibition of the R/G 27 reaction that occurs at very high concentrations of the wild-type R1 was not observed with the mutant R1 (Fig. 6A ▶, right panel, gray bars). Thus, this inhibition may occur by a simple competition of the dsRBM motifs (see Discussion).
Finally, we directly tested the ability of the isolated R1 and the K127A mutant to bind RNA in a gel-shift assay. The wild-type R1 bound the R/G 15 substrate only poorly, but binding was reproducibly decreased with the K127A mutant, as evident by an increase in the free, unbound RNA at high concentrations (Fig. 6B ▶). The wild-type R1 bound the R/G 27 substrate, as well as a stem–loop that mimicked the remaining portion of the R/G hairpin, not included in R/G 15, relatively tightly, with a Kd of <25 nM; the K127A mutant bound these RNAs only weakly, with a Kd >500 nM (Fig. 6B ▶).
DISCUSSION
The modular structure of ADARs
Limited proteolysis often provides information that is useful for structure–function studies. With this in mind, we performed limited proteolysis on hADAR2. We found a strong resistance of the known domains to proteolysis, whereas the region between the two dsRBMs was extremely susceptible to cleavage. In this study, we addressed two questions. First, do the proteolytic fragments of hADAR2 edit RNA, and second, what role do the dsRBMs and the linker between them play in catalysis? To answer these questions, we cloned, expressed, and purified several N-terminal deletion constructs and assayed the ability of each construct to deaminate RNA in vitro.
N-terminal deletions of hADAR2
From our proteolysis data, we generated three N-terminal deletions of hADAR2. The largest truncated protein (Truncation R1LR2D) contained a deletion that removed the first 75 residues; this protein began two residues before the start of dsRBM1. In vitro, R1LR2D retained deaminase activity on a naturally occurring substrate (Fig. 3B ▶). A similar deletion in the context of the D. melanogaster ADAR binds dsRNA but is catalytically inactive (Gallo et al. 2003). Within the deleted region of the fly protein is a sequence necessary for editing and proposed to be involved in RNA-dependent dimerization. The apparent difference in activities between our truncation and the D. melanogaster truncation could be due to the existence of an alternative dimerization sequence in the human enzyme, or a different mechanism of activation.
The N terminus of truncation LR2D began 16 residues after the first dsRBM. This protein contained most of the linker between the two dsRBMs as well as the entire dsRBM2 and catalytic domain. Interestingly, LR2D did not edit the R/G site RNA when assayed in a crude extract but did edit this RNA after further purification (data not shown). Because further deletion of the linker to produce the R2D truncation (see following) resulted in an active protein, even in a crude extract, one explanation for this result is that the linker of LR2D is associating with a protein in the extract that inhibits activity.
The third truncated protein, R2D, contained a deletion that removed the rest of the linker between the two dsRBMs, leaving only dsRBM2 and the catalytic domain. To our surprise, this protein retained the ability to deaminate both long dsRNA and R/G site RNA substrates with only a slightly reduced rate compared with the full-length protein (Fig. 5 ▶; Table 1 ▶).
An auto-inhibition model for hADAR2
We found that deletion of nearly half of hADAR2 (as in R2D) caused only a three-fold reduction in activity, as assayed with the R/G 27 RNA, a mimic of the naturally occurring R/G editing site (Fig. 5 ▶; Table 1 ▶). However, when the full-length hADAR2 and R2D were compared using the shorter R/G 15 RNA, or a similar molecule, R/G 19 (data not shown), the differences in activity were much more dramatic. R2D efficiently deaminated these shorter substrates, whereas the activity of the full-length enzyme was almost undetectable. We also tested the specificity of R2D by mapping inosines using a mutant form of ribonuclease T1 (E64Q, Polson and Bass 1994) and found that the editing pattern was identical to that produced by the full-length hADAR2 (data not shown). As previously reported for ADAR2 from rat (Kallman et al. 2003), we observed minor editing by both R2D and hADAR2 at the adenosine immediately 5′ to the R/G site.
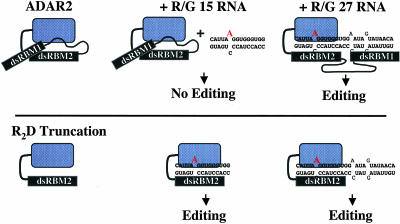
Figure 7 ▶ shows a model to explain the differences between the activities of full-length protein and R2D on short substrates. The model assumes that in the absence of RNA, the full-length hADAR2 is in an inactive conformation and subject to auto-inhibition. Activation can only occur in the presence of an RNA substrate that is long enough to accommodate binding of both dsRBMs. On very short substrates such as the R/G 15 RNA, there is no binding site for dsRBM1 and no deamination of the R/G site. In contrast, the R2D truncation is constitutively active and will deaminate RNA substrates regardless of their length.
FIGURE 7.
A model for RNA-mediated hADAR2 activation. The two horizontal panels illustrate the full-length hADAR2 and the R2D truncation, in the absence of RNA (left), in the presence of a short RNA (center), or in the presence of a long RNA (right). The full-length enzyme is inactive in the absence of RNA or in the presence of short RNA because the N terminus inhibits association of dsRBM2 and the catalytic domain with the substrate. When a longer RNA is present, dsRBM1 binds to a specific site, causing a conformational change that allows dsRBM2 and the catalytic domain to bind as well, resulting in an active enzyme. The R2D truncation does not contain the inhibitory region and is constitutively active on substrates of any length.
The model implies that there are inhibitory sequences in full-length hADAR2 that are absent in the R2D truncation. Indeed, we found that addition of the N terminus of hADAR2 (R1) to a reaction containing the R2D protein, in trans, mimicked the reaction of hADAR2; that is, activity was only observed with longer RNA substrates. Reconstitution of the hADAR2 reaction in trans was observed even with an R1 fragment mutated so as to eliminate dsRNA binding (Fig. 6A ▶). Thus, we do not think that inhibition occurs simply because the two dsRBMs compete for RNA binding. Rather, we speculate that inhibition in trans involves a protein–protein interaction between components of the N terminus that exist in R1 and the C terminus (dsRBM2/catalytic domain) that prevents editing of short duplexes. We tested whether the linker peptide (containing amino acids 144–234) by itself could inhibit the R2D−R/G 15 reaction by adding it in trans. This peptide did not inhibit the reaction (data not shown) suggesting the inhibitory region is N terminal to the linker, either within dsRBM1 or the extreme N terminus.
The model of Figure 7 ▶ implies that the transition from an inactive to active hADAR2 involves binding of dsRBM1. Our gel-shift analyses of Figure 6B ▶ show that the R1 fragment binds very poorly to R/G 15 but tightly to the R/G terminal hairpin, which mimics the stem–loop region of the natural R/G hairpin. The tighter binding to the R/G terminal hairpin could be because this molecule is slightly longer than R/G 15. However, we propose that the terminal portion of the R/G hairpin is a preferred site for dsRBM1. As shown in our model, we speculate that binding of dsRBM1 induces a conformational change that effectively “unmasks” dsRBM2 and the catalytic domain. We also note that the R1 fragment binds to R/G 27 nearly as well as the full-length ADAR2 (see Fig. 6 ▶; Stephens et al. 2000). This suggests that binding of the full-length protein is predominantly a function of dsRBM1. Consistent with this idea, the R2D truncation, which only has dsRBM2, exhibits only minimal RNA binding in a gel-shift analysis (data not shown).
The data of Figure 6A ▶ show that R1 inhibits the reaction of R2D with R/G 27 (black bars, right panel) only when it is present at very high concentrations. In particular, inhibition by R1 is observed when the R1 concentration exceeds the concentration of R2D (100 nM). One explanation for this observation is that the in trans association of R1 and R2D is very favorable; when R1 is present at concentrations less than or equal to that of R2D, it is quantitatively associated with R2D and mediates binding to R/G 27 and relief of auto-inhibition. According to this hypothesis, at high concentrations of R1 that exceed the amount of R2D in the reaction, free R1, which lacks a catalytic domain, would begin to compete with the catalytically active R1−R2D molecule for RNA binding. Consistent with the idea that this inhibition involves a competition for dsRNA binding, it is not observed with the R1-K127A mutant, which cannot bind dsRNA.
Several laboratories have reported that activation of certain ADARs involves RNA-dependent dimerization (Jaikaran et al. 2002; Cho et al. 2003; Gallo et al. 2003). Our studies have not addressed dimerization, but it is possible that the activation we observe involves dimerization. The dimerization domain in the D. melanogaster enzyme is in the extreme N terminus, before dsRBM1, and deletion of this sequence abolishes deaminase activity (Gallo et al. 2003). Our R1LR2D truncation of the human enzyme is similar to this dADAR truncation, yet it retains a significant amount of activity. Thus, if dimerization is involved in hADAR2 activation, the dimerization motif in hADAR2 must be in a different region than that of the dADAR. Activation by dimerization could also explain why hADAR2 does not deaminate R/G 15. If dimerization is required for activity as proposed, R/G 15 could be too short to accommodate an ADAR2 dimer. As discussed earlier, our experiments suggest the in trans association of R1 and R2D is very favorable, and possibly this is because the association involves dimerization.
Our model for activation of hADAR2 is reminiscent of those proposed for another dsRNA binding protein, PKR. It has been proposed that the cooperative binding of PKR’s dsRBMs to dsRNA is responsible for activation, and perhaps dimerization, and requires a substrate of at least 30 bp (Manche et al. 1992; Carpick et al. 1997). In our model, we propose that it is a sequence in the N-terminal fragment (R1), which contains dsRBM1 but not dsRBM2, that prevents hADAR2 from editing short RNAs. NMR data for PKR, in the absence of RNA, suggest an interaction between dsRBM2 and the kinase domain. From these data, it has been suggested that dsRBM2 of PKR promotes auto-inhibition by masking the kinase domain (Nanduri et al. 2000). In the presence of dsRNA, dsRBM1 of PKR is proposed to induce the cooperative binding of dsRBM2, which promotes the active conformation, causing dimerization and activation of the kinase. Although our data suggest dsRBM1 of hADAR2 is more important than dsRBM2 for activation, as yet we cannot rule out an interaction between dsRBM2 and the catalytic domain. Although a construct similar to R2D has not been analyzed for PKR, when both dsRBMs are deleted from PKR, the resulting protein is constitutively active, consistent with auto-inhibition (Wu and Kaufman 1997).
In summary, our data suggest hADAR2 is inactive in the absence of dsRNA long enough to bind both dsRBMs. Our results are consistent with, and help explain, the previous observation that hADAR2 requires an extended duplex in the HDV antigenomic RNA in order to edit the amber/W site (Sato et al. 2001). One reason that hADAR2 might have evolved such a regulatory mechanism is to ensure that shorter dsRNA helixes, such as those found in endogenous RNAs like rRNA, tRNA, and pre-miRNA, are not deaminated. Indeed, all of the edited sequences found in vivo are in longer base-paired regions (Bass 2002; Maas et al. 2003).
MATERIALS AND METHODS
Limited proteolysis of hADAR2
hADAR2 was overexpressed and purified as described (Ley 2001). Proteases (Sigma) were freshly prepared before use. Lyophilized trypsin was resuspended in 50 mM acetic acid; lyophilized subtilisin was resuspended in 50 mM Tris-HCl (pH 8.0) and 150 mM NaCl. For a 10 μL reaction, 10 μg purified hADAR2 was mixed with 0.01 μg of trypsin or 0.005 μg subtilisin in 50 mM NaHCO3. Reactions proceeded at room temperature for varying times and were quenched by adding 1 μL of 25 mM phenylmethylsulfonyl fluoride and 10 μL SDS-PAGE loading buffer. Samples were boiled (5 min), then electrophoresed in minigels containing a 4%–15% polyacrylamide gradient. Peptide fragments were visualized with Coomassie blue, or blotted onto a PVDF membrane, stained with Coomassie blue, and submitted for N-terminal sequencing by Edmann degradation according to published methods (Matsudaira 1987).
Construction, expression, and purification of hADAR2 deletion mutants
N-terminal deletions of hADAR2 were constructed by PCR amplification of the cDNA of hADAR2 in the expression vector YEpTOP2PGAL1 (Ley 2001) using primers designed to produce an ORF encoding a truncated hADAR2 of desired length. BsmBI restriction sites, which when cut leave overhangs for BamHI at the 5′ end of the gene and XhoI at the 3′ end of the gene, were included in the forward and reverse primers and allowed for ligation of the PCR product into the expression vector YEpTOP2PGAL1 (Giaever et al. 1988; Ley 2001). The following DNA oligonucleotides were used as upstream primers for amplifying the truncated hADAR2 genes: RLRD-UP: CCCCCGTCTCGGATCCGTAACCATGTCAGTCCTCCCCAAGAACGCCCTGATG; LRD-UP: CCCCCGTCTCGGATCCGTAACCATGTCAGTCAACACGGACTTCACATCTGAC; RD-UP: CCCCCGTCTCGGATCCGTAACCATGTCAGGGAAGAATCCCGTGATGATCTTG. The downstream primer for amplifying and adding six histidine residues to these three constructs was as follows: A2His6-Down: CCCCCGTCTCCTCGAGTCAATGGTGATGGTGATGGTGGACTAGGGGCGTGA. For amplifying the RD truncation with a TEV-cleavable 10×-histidine, the following upstream and downstream primers were used. H10RD-UP: GGGCCCCGTCTCGGATCCGTAACCATGTCACACCATCACCATCACCATCACCATCACCATGAGAACCTCTATTTCCAGGGAGCCAGCCTAGCCCAGCCTGCTCTCCCTGTC and A2-DOWN: CCCCCGTCTCCTCGAGTCAGGGCGTGAGTGAGAACTGGTCCTG.
Truncated proteins were expressed in S. cerevisiae as described for full-length ADAR2 (Ley 2001). To screen for expression, we lysed 0.1 g of cells by vortexing with glass beads (30 min) at 4°C in 20 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.5 mM DTT, and 0.01% Nonidet P-40. Lysate was clarified by centrifugation at 14,000 rpm in a microfuge, and expressed proteins were detected by Western blotting using the Penta-His antibody (Qiagen). The clarified lysate was analyzed for deaminase activity (see following), and the remainder stored at −80°C. Truncation R2D was purified as follows, with all manipulations carried out at 4°C: 25 g (wet wt) of yeast cells were resuspended in 100 mL of Buffer A (20 mM Tris-HCl at pH 8.0, 5% glycerol, 1 mM 2-mercaptoethanol) containing 750 mM NaCl, 35 mM imidazole, and 0.01% Nonidet P-40. Cells were lysed by three passes through a Gaulin homogenizer at 20,000 psi, and lysate was centrifuged at 100,000g for 1 h. The supernatant was removed and mixed with 2.5 mL of Ni-NTA agarose (Qiagen) equilibrated with lysis buffer, adsorbed for 10 min with gentle rocking, and poured into a column. The column was washed in three steps with Buffer A containing 35 mM imidazole, and NaCl concentrations of 1 M, 500 mM, or 100 mM, respectively. Protein was eluted with Buffer A containing 100 mM NaCl and 400 mM imidazole and loaded onto a 5 mL Hi-Trap Heparin column (Pharmacia) equilibrated with Buffer B (Buffer A containing 100 mM NaCl). The column was washed with 50 mL of Buffer B and developed with a 25 mL gradient of 100 mM–1 M NaCl. Fractions were analyzed by electrophoresis on a 4%–15% polyacrylamide gradient gel stained with Coomassie blue and those containing truncated protein pooled and subjected to proteolysis by the addition of 100 units of TEV protease (Invitrogen). The reaction was dialyzed against Buffer B overnight, then bound to 1 mL of Ni-NTA agarose for 1 h with gentle rocking, and poured into a column. The resin was washed with Buffer B containing 15 mM imidazole, and the flowthrough and washes containing the cleaved R2D truncation were pooled. The uncleaved protein, TEV protease, and 10×-His tag were eluted by washing with Buffer B containing 400 mM imidazole and discarded. The protein was loaded on a Superdex 200 gel filtration column (Pharmacia) developed in Buffer B; it eluted as one peak at a molecular weight of 55 kDa, relative to the standards bovine serum albumin (67 kDa), ovalbumin (43 kDa), and trypsin inhibitor (25kDa). Fractions were analyzed by electrophoresis as described earlier, and those containing R2D were pooled. The protein was dialyzed into storage buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 20% glycerol, and 1 mM 2-mercaptoethanol; concentrated using an Amicon Ultra-free 30 device; and quantified by the Bradford method. The protein was stored in aliquots at −80°C and typical yields were 2–3 mg of pure protein per liter of yeast culture.
The cDNA coding for the N terminus of hADAR2 spanning residues 1–218 was amplified by PCR using the hADAR2 cDNA template as described earlier. The product was ligated into the expression vector pKM263 via the NcoI and BamHI restriction sites (Melcher 2000). The translated protein is fused to the C terminus of a histidine-tagged GST and is separated from GST by a TEV protease recognition sequence. The K127A mutant R1 was constructed using the QuickChange PCR mutagenesis kit (Biocrest) according to the manufacturer’s instructions. Protein was expressed in Escherichia coli for 3 h at room temperature after induction with 0.2 mM IPTG. Cells were lysed by two passes through a French pressure cell at 20,000 psi and the protein was purified from the cleared lysate by nickel chromatography and cleaved from GST by TEV protease as described earlier.
RNA preparation
R/G site hairpin and the R/G terminal hairpin were transcribed from partially single-stranded DNA templates (Milligan et al. 1987; Lehmann and Bass 1999) and purified on a 12% denaturing gel. CAT duplex RNA was labeled internally by transcribing with α-32P ATP as previously described (Bass and Weintraub 1987). The following DNA oligonucleotides were used for transcribing the 71 nucleotide R/G hairpin: BB1BB: TAATACGACTCACTATAG and R/G hairpin: ACATCAGGGTAGGTGGGATACTATAACAACATTTAGCATATTGTTATACTATTCCACCCACCTTAATGACCTATAGTGAGTCGTATTA. To specifically label R/G 27 and R/G 15 RNA at the R/G editing site, we ligated two RNA “half molecules” using a DNA splint and T4 DNA ligase as previously described (Stephens et al. 2000). For the gel-shift assay, the R/G terminal hairpin was transcribed using the BB1BB primer and R/G-term: GGGATACTATAACAACATTTAGCATATTGTTATACTATTCCTATAGTGAGTCGTATT. The RNA was dephosphorylated with calf intestinal phosphatase (New England Biolabs) before gel purification. The 5′ ends of the R/G terminal hairpin and the top strand of R/G 15 were labeled with 32P using polynucleotide kinase (New England Biolabs) and gamma 32P-ATP (Amersham).
Deamination assays
Five or 15 μL of yeast extracts (prepared as described earlier) were incubated with R/G hairpin RNA (final concentration 2.5 nM) for 1 h at 30°C in assay buffer (20 mM Tris-HCl at pH 7.5, 40 mM KCl, 5% glycerol, 0.5 mM DTT, 0.01% Nonidet P-40, and 1 U/μL RNasin (Promega). The RNA was extracted, and editing was monitored by limited primer extension as described (Öhman et al. 2000). Purified hADAR2 (final concentration, 15 nM) and extract prepared from yeast harboring an expression vector lacking the ADAR gene were also reacted with R/G hairpin RNA as positive and negative controls.
Editing of internally labeled RNAs (CAT duplex, R/G 27 and R/G 15) by either purified hADAR2 or the R2D truncation was monitored by TLC. One nanomolar CAT duplex RNA or 5 nM R/G 27 or R/G 15 RNA was incubated with 100 nM enzyme in assay buffer at 30°C for various times. At each time point, an aliquot was removed and quenched by the addition of 0.5% SDS and heating at 95°C (1 min). RNA was extracted and digested with Nuclease P1 and the 5′ nucleoside monophosphates resolved by TLC as described (Lehmann and Bass 1999). For the single-turnover kinetic analysis, reactions were performed as described earlier, except the time points were 0, 0.33, 0.67, 1, 2, 5, 10, 30, and 60 min. For the R1 inhibition experiments, 100 nM R2D was mixed with R1 (at the final concentrations indicated in Fig. 6 ▶) and incubated with 20 nM R/G RNA for 45 min at 30°C. The reactions were processed as described earlier.
Gel mobility shift assay
Gel-shift assays were performed essentially as described (Öhman et al. 2000), except for the following modifications. Either R2D or R1 (at the concentrations indicated in Fig. 6 ▶) were incubated with RNA in a buffer containing 20 mM Tris-HCl (pH 8.0), 40 mM KCl, 15% glycerol, 0.5 mM DTT, 0.01% Nonidet P-40, and 1 U/μL RNasin (Promega).
Acknowledgments
We thank Robert Shackmann for prompt synthesis of RNA and DNA oligonucleotides and for N-terminal sequencing of proteins. We also thank members of the Bass lab and the lab of Peter Beal for helpful discussions and Sabine Hellwig for critically reading this manuscript. The S. cerevisiae expression vector and strain BCY123 were generous gifts from Janet Lindsley and Brad Cairns, respectively, of the University of Utah. This work was supported by funds to B.L.B. from the National Institute of General Medical Sciences (GM44073). B.L.B. is a Howard Hughes Medical Institute investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7920904.
REFERENCES
- Bass, B.L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71: 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L. and Weintraub, H. 1987. A developmentally regulated activity that unwinds RNA duplexes. Cell 48: 607–613. [DOI] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., and Emeson, R.B. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308. [DOI] [PubMed] [Google Scholar]
- Carpick, B.W., Graziano, V., Schneider, D., Maitra, R.K., Lee, X., and Williams, B.R. 1997. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J. Biol. Chem. 272: 9510–9516. [DOI] [PubMed] [Google Scholar]
- Cho, D.S., Yang, W., Lee, J.T., Shiekhattar, R., Murray, J.M., and Nishikura, K. 2003. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 278: 17093–17102. [DOI] [PubMed] [Google Scholar]
- Gallo, A., Keegan, L.P., Ring, G.M., and O’Connell, M.A. 2003. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 22: 3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G.N., Snyder, L., and Wang, J.C. 1988. DNA supercoiling in vivo. Biophys. Chem. 29: 7–15. [DOI] [PubMed] [Google Scholar]
- Herbert, A. and Rich, A. 2001. The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc. Natl. Acad. Sci. 98: 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaikaran, D.C., Collins, C.H., and MacMillan, A.M. 2002. Adenosine to inosine editing by ADAR2 requires formation of a ternary complex on the GluR-B R/G site. J. Biol. Chem. 277: 37624–37629. [DOI] [PubMed] [Google Scholar]
- Kallman, A.M., Sahlin, M., and Ohman, M. 2003. ADAR2 A→I editing: Site selectivity and editing efficiency are separate events. Nucleic Acids Res. 31: 4874–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, K.A. and Bass, B.L. 1999. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 291: 1–13. [DOI] [PubMed] [Google Scholar]
- Ley, H.L.I. 2001. “Editing of hepatitis delta virus antigenomic RNA by recombinant human adenosine deaminases that act on RNA.” Ph.D. thesis, University of Utah, Salt Lake City.
- Liu, Y. and Samuel, C.E. 1996. Mechanism of interferon action: Functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J. Virol. 70: 1961–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Lei, M., and Samuel, C.E. 2000. Chimeric double-stranded RNA-specific adenosine deaminase ADAR1 proteins reveal functional selectivity of double-stranded RNA-binding domains from ADAR1 and protein kinase PKR. Proc. Natl. Acad. Sci. 97: 12541–12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J.R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., and Seeburg, P.H. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266: 1709–1713. [DOI] [PubMed] [Google Scholar]
- Maas, S., Melcher, T., Herb, A., Seeburg, P.H., Keller, W., Krause, S., Higuchi, M., and O’Connell, M.A. 1996. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem. 271: 12221–12226. [DOI] [PubMed] [Google Scholar]
- Maas, S., Rich, A., and Nishikura, K. 2003. A-to-I RNA editing: Recent news and residual mysteries. J. Biol. Chem. 278: 1391–1394. [DOI] [PubMed] [Google Scholar]
- Manche, L., Green, S.R., Schmedt, C., and Mathews, M.B. 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12: 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262: 10035–10038. [PubMed] [Google Scholar]
- Melcher, K. 2000. A modular set of prokaryotic and eukaryotic expression vectors. Anal. Biochem. 277: 109–120. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, D.P., Aruscavage, P.J., and Bass, B.L. 2002. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. 99: 7906–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri, S., Rahman, F., Williams, B.R., and Qin, J. 2000. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 19: 5567–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhman, M., Källman, A.M., and Bass, B.L. 2000. In vitro analysis of the binding of ADAR2 to the pre-mRNA encoding the GluR-B R/G site. RNA 6: 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, R.C., Stanton, P., and Sen, G.C. 1996. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J. Biol. Chem. 271: 25657–25663. [DOI] [PubMed] [Google Scholar]
- Polson, A.G. and Bass, B.L. 1994. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 13: 5701–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson, A.G., Bass, B.L., and Casey, J.L. 1996. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380: 454–456. [DOI] [PubMed] [Google Scholar]
- Sato, S., Wong, S.K., and Lazinski, D.W. 2001. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J. Virol. 75: 8547–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, B., Kohler, M., Sprengel, R., and Seeburg, P.H. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19. [DOI] [PubMed] [Google Scholar]
- Stephens, O.M., Yi-Brunozzi, H.Y., and Beal, P.A. 2000. Analysis of the RNA-editing reaction of ADAR2 with structural and fluorescent analogues of the GluR-B R/G editing site. Biochemistry 39: 12243–12251. [DOI] [PubMed] [Google Scholar]
- Wu, S. and Kaufman, R.J. 1997. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem. 272: 1291–1296. [DOI] [PubMed] [Google Scholar]