Abstract
A spliced leader contributes the mature 5′ends of many mRNAs in trans-splicing organisms. Trans-spliced metazoan mRNAs acquire an m32,2,7GpppN cap from the added spliced leader exon. The presence of these caps, along with the typical m7GpppN cap on non-trans-spliced mRNAs, requires that cellular mRNA cap-binding proteins and mRNA metabolism deal with different cap structures. We have developed and used an in vitro system to examine mRNA degradation and decapping activities in nematode embryo extracts. The predominant pathway of mRNA decay is a 3′ to 5′ pathway with exoribonuclease degradation of the RNA followed by hydrolysis of resulting mRNA cap by a scavenger (DcpS-like) decapping activity. Direct decapping of mRNA by a Dcp1/Dcp2-like activity does occur, but is ~15-fold less active than the 3′ to 5′ pathway. The DcpS-like activity in nematode embryo extracts hydrolyzes both m7GpppG and m32,2,7GpppG dinucleoside triphosphates. The Dcp1/Dcp2-like activity in extracts also hydrolyzes these two cap structures at the 5′ ends of RNAs. Interestingly, recombinant nematode DcpS differs from its human ortholog in its substrate length requirement and in its capacity to hydrolyze m32,2,7GpppG.
Keywords: decapping; in vitro RNA decay; scavenger; DcpS; m7GpppG, m32,2,7GpppG; trimethylguanosine (TMG) cap; Dcp1/Dcp2; nematode; Ascaris; C. elegans
INTRODUCTION
Spliced leader (SL) RNA trans-splicing contributes a 5′ terminal exon to pre-mRNAs to form the mature 5′ end of the mRNA. SL trans-splicing is present in a diverse spectrum of eukaryotes including sarcomastigophoran protozoa (euglenoids and trypanosomes and their relatives), Cnidaria (Hydra vulgaris), flatworms (free-living and parasitic: polyclads, flukes, and tapeworms), nematodes (e.g., Caenorhabditis elegans and Ascaris), and chordates (tunicate, Ciona intestinalis; Nilsen 2001). Addition of the 5′ terminal exon from trans-splicing SL RNAs generates cellular mRNAs with a short, conserved sequence in the mRNA 5′ untranslated region (UTR) and a distinct hypermethylated cap (in metazoa) derived from the donated spliced leader, an N2,2,7-trimethyl-GpppN cap (m32,2,7GpppN or TMG cap). Non-trans-spliced mRNAs have an N7-methyl-GpppN RNA cap (m7GpppN) with diverse 5′ UTR sequences. Therefore, two populations of mRNAs coexist in metazoan cells that exhibit spliced leader addition: trans-spliced and non-trans-spliced mRNAs.
Both mRNA cap and 5′ UTR sequence and structure are known to play important roles in translation, and cap structure is also an important determinant in transcript stability. The mRNA cap is recognized and bound directly or indirectly by a number of different cap-binding proteins. Cap-binding or -interacting proteins include the translation initiation factor eIF4E (Gingras et al. 1999; Von Der Haar et al. 2004), nuclear cap-binding complex (consisting of cap-binding proteins CBP 20 and 80; Izaurralde et al. 1994, 1995), and decapping proteins (Dcp1, Dcp2, and DcpS; for review, see Decker and Parker 2002). Proteins that interact with the metazoan hypermethylated cap (m32,2,7GpppN) and/or spliced leader sequence of a trans-spliced mRNA are likely to play an important role in mRNA metabolism, and may mediate the use and/or cellular discrimination of trans-spliced versus non-trans-spliced mRNAs.
Decapping proteins are involved in cleaving the unusual 5′-ppp-5′phosphodiester linkage in the RNA cap (G-5′ppp5′-N). The presence of the RNA cap triphosphate bridge increases mRNA stability presumably as it prevents 5′to 3′ exoribonuclease degradation of the mRNA. mRNA decay pathways have been extensively characterized in yeast and more recently in higher eukaryotes. A major mechanism of mRNA decay in yeast proceeds from 5′ to 3′ following removal of the cap. Decay is initiated by deadenylation of the mRNA (Tucker and Parker 2000; Parker and Song 2004). This typically leads to decapping of the intact RNA by a protein complex known as Dcp1/Dcp2 (Beelman et al. 1996; Dunckley and Parker 1999; Parker and Song 2004). Removal of the cap exposes the 5′ end of the RNA to exoribonucleolytic decay by Xrn1. A second general pathway, often called the 3′ to 5′ pathway, involves deadenylation of the mRNA followed by 3′ to 5′ exoribonuclease degradation of the mRNA body by the exosome (Tucker and Parker 2000; Parker and Song 2004). Processive 3′ to 5′ degradation of the mRNA body finally leads to the production of a 5′ cap dinucleoside triphosphate (m7GpppN or short m7GpppN capped oligonucleotide). The m7GpppN or oligonucleotide cap is subsequently hydrolyzed by what has been named a “scavenger” decapping activity (DcpS; Nuss et al. 1975; Wang and Kiledjian 2001; Liu et al. 2002). The 3′to 5′pathway is typically considered a less active pathway for mRNA turnover in yeast. However, a recent genome-wide analysis of mRNA decay using expression profiling and various mutant yeast strains suggests the possibility that the 3′ to 5′ decay may be more important than previously realized (He et al. 2003). Yeast cells blocked in both the 5′ to 3′ and 3′ to 5′ pathways are not viable, illustrating the importance of mRNA turnover in gene expression (Anderson and Parker 1998).
The predominant general pathway of mRNA decay in higher eukaryotes is currently unclear. Experiments using mammalian cells, and particularly in vitro studies, suggest that an important mechanism is through the 3′ to 5′ decay pathway (Chen et al. 2001; Wang and Kiledjian 2001; Mukherjee et al. 2002) leading to cap dinucleoside triphosphate, m7GpppN, which is cleaved by the scavenger activity (DcpS; Wang and Kiledjian 2001). Both types of decapping activity, DcpS and Dcp1/Dcp2, have been described in mammalian cells (Gao et al. 2001; Wang and Kiledjian 2001; Wang et al. 2002; Lejeune et al. 2003). A recent RNAi study demonstrated that the C. elegans 5′ to 3′ exoribonuclease is required in the latter stages of embryogenesis when epithelial cell movements are required for ventral closure (Newbury and Woollard 2004). Overall, however, the nature and contribution of different mRNA decay pathways and decapping enzymes to mRNA turnover and their importance in development or viability in multicellular organisms have not been characterized.
SL1 trans-splicing is responsible for the maturation of ~50 to 90% of mRNAs in the nematodes C. elegans and Ascaris, respectively (Zorio et al. 1994; Maroney et al. 1995). The function(s) of SL1 trans-splicing in nematode gene expression remains unknown. Among the possible post-transcriptional functions that have been postulated for addition of the TMG-capped-SL sequence to mRNAs are roles in mRNA translation, stability, processing, localization, and/or transport. SL1 trans-splicing is required for early C. elegans development (Ferguson et al. 1996), and the SL1 sequence with its TMG cap have been shown to functionally collaborate to enhance translational efficiency of a trans-spliced mRNA in an in vitro system (Maroney et al. 1995). However, our in vitro and in vivo translation studies in Ascaris embryos indicate that the TMG-capped-SL sequence does not confer a measurable translational advantage on recipient mRNAs (L.S. Cohen, M. Mikhli, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, unpubl.; S. Lall, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, in prep.). A second SL sequence (SL2) with a hypermethylated cap and sequence divergent from SL1 (~50% sequence similarity) is also present in C. elegans (Huang and Hirsh 1989). SL2 is added to internal splice acceptor sites between mRNAs in polycistronic primary trancripts contributing to the maturation of these operon RNAs into monocistronic mRNAs (Spieth et al. 1993; Blumenthal and Gleason 2003). Although SL1 trans-splicing is capable of resolving polycistronic transcripts under some conditions, it is unclear whether this is a primary function of SL1 trans-splicing and the overall function of SL1 remains unclear (Williams et al. 1999; Liu et al. 2001).
Several decapping enzymes that hydrolyze the m7GpppN cap on mRNAs or free cap dinucleoside triphosphates have recently been characterized in yeast and mammalian cells (Beelman et al. 1996; LaGrandeur and Parker 1998; Dunckley and Parker 1999; Wang and Kiledjian 2001; Liu et al. 2002; Lykke-Andersen 2002; Salehi et al. 2002; van Dijk et al. 2002, 2003; Wang et al. 2002; Piccirillo et al. 2003). mRNA decay, deadenylation, and decapping activities have also recently been described in trypanosomes (Milone et al. 2002, 2004). However, activities in other species have not been well characterized, and neither mRNA decay pathways nor decapping activities have been examined directly in trans-splicing metazoa. In trans-splicing metazoa, the two distinctly capped mRNA populations, and particularly the presence of a distinct cap and the SL sequence on some mRNAs may have a profound influence on mRNA translation and stability. Analysis of the decay pathways and associated proteins may provide insight into the function of trans-splicing, and could lead to potential targets for rational drug design against a number of divergent parasitic worms. In this paper, we describe mRNA transcript decay that yields mRNA caps as well as the m7GpppG and m32,2,7GpppG decapping activities present in nematodes using Ascaris embryo extracts. We have also produced and characterized a recombinant C. elegans dinucleoside triphosphate scavenger type decapping protein (DcpS) that can hydrolyze m7GpppG and m32,2,7GpppG.
RESULTS
Decay and decapping activity in Ascaris embryo whole cell extracts
Two distinct mRNA cap-cleaving activities have been described in eukaryotes. One activity, Dcp1/Dcp2, decaps m7GpppN-mRNA substrates producing m7Gpp + pN-RNA products (Beelman et al. 1996; Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002; Steiger et al. 2003). A second activity described as a scavenger activity, DcpS, hydrolyzes N7–methyl-guanosine-dinucleoside triphosphate (m7GpppN) or a short oligonucleotide cap (m7GpppNn) producing m7Gp and ppN products (Nuss et al. 1975; Nuss and Furuichi 1977; Wang and Kiledjian 2001; Liu et al. 2002). These two cap cleaving activities can be distinguished by their reaction products, m7Gp for the scavenger/dinucleoside triphosphate activity (DcpS) and m7Gpp for the Dcp1/Dcp2 RNA decapping activity.
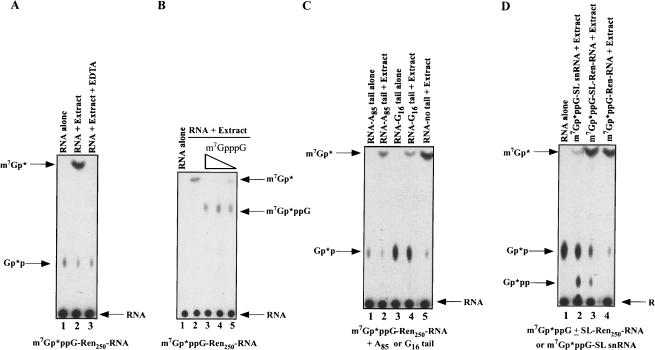
We prepared Ascaris embryo extracts competent for cap-dependent translation that demonstrate cap and poly(A)-tail translation synergism (S. Lall, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, in prep.). These extracts were considered to be physiologically relevant for analysis of decapping activities and mRNA turnover. Cap-labeled m7Gp*ppG-RNA (where the * follows the 32P-labeled phosphate) was incubated in 32–64 cell Ascaris embryo extracts for 30–60 min and the decay and decapping reaction products characterized by thin layer chromatography and PAGE. Analysis of the reaction products derived from incubation of a 250 nucleotide cap-labeled m7Gp*ppG-Ren250-RNA (derived from pRLnull; see Supplementary Fig. 1 ▶) indicated that the predominant RNA decapping product observed was m7Gp* (Fig. 1A ▶, lane 2). The decay and decapping reactions are linear for at least 30 min, and dependent on extract concentration (data not shown). Similar results were obtained using an ~1 kb RNA with a complete open reading frame and poly(A)-tail and an actin mRNA (data not shown).
FIGURE 1.
RNA decay and decapping activity in Ascaris embryo extracts illustrates contribution of 3′ to 5′ exoribonuclease and scavenger activities. (A) m7Gp*ppG 32P-cap-labeled RNA undergoes decay and decapping in Ascaris extracts producing m7Gp*. A 250 nucleotide cap-labeled RNA substrate (the * follows the 32P-labeled phosphate) derived from the 5′ end of pRLnull was incubated in Ascaris embryo extract for 50 min at 30°C, aliquots of the reactions were then applied to PEI-cellulose thin layer chromatography (TLC) plates, the plates developed in 0.45 M ammonium sulfate, and labeled substrate and products detected by autoradiography. Comigration of appropriate markers followed by UV shadowing, enzymatic conversion assays, and inhibition studies were used to identify and confirm reaction products. Note that the Gp*p spot is derived from the cap-labeling reaction and is not present in gel-purified RNA substrates that were used in other experiments with similar results. (B) Inhibition of scavenger activity during RNA decay leads to accumulation of m7GpppG in Ascaris extracts. Reactions were carried out with or without cap analogs. Concentrations of competitive cap inhibitors are 200, 50, and 20 μM. (C) Addition of a 3′ poly(A)-tail or G16 to the RNA substrate leads to reduction in m7Gp* product. (D) Inherent SL snRNA structure and SL RNP formation in Ascaris extracts leads to a reduction in decay and scavenger product. Note that the SL snRNA undergoes reduced decay and cap hydrolysis (cf. lane 2 and lanes 3,4), while an mRNA with only the m7Gp*ppG-capped 22 nucleotide SL sequence added to its 5′ end is degraded and hydrolyzed normally (cf. lanes 3 and 4). RNA alone = m7Gp*ppG-SL snRNA. Reactions in (B)–(D) were carried out and analyzed as described in (A).
Several lines of evidence suggest that the m7Gp* product observed in the whole cell extract decay reactions results from (1) 3′ to 5′ exoribonucleolytic decay of the RNA producing a short m7Gp*ppG substrate that is (2) rapidly cleaved by a dinucleoside triphosphate hydrolase (“scavenger” or DcpS-like) activity. The exosome and 3′ to 5′ exoribonucleolytic RNA decay in other systems are known to require Mg++ and are inhibited by EDTA. Notably, m7Gp* derived from RNA substrates in the Ascaris embryo extracts is greatly reduced in the presence of EDTA (Fig. 1A ▶, cf. lanes 2 and 3), suggesting that the RNA decay or scavenger activity is inhibited. Under our RNA decay assay conditions, the m7Gp*ppG product derived from 3′ to 5′ exoribonucleolytic decay of the RNA—and the substrate of the scavenger activity—does not accumulate in the reactions. m7Gp*ppG substrate (or short cap oligonucleotides) accumulates in RNA decay reactions when DcpS activity is specifically inhibited by m7GpppG cap or m7Gpp nucleotides or in extract fractions with reduced scavenger dinucleoside triphosphate hydrolase activity (Fig. 1B ▶, cf. lane 2 and lanes 3–5; e.g., S130 pellet; see below). Because our analysis of recombinant nematode DcpS scavenger activity indicates that it does not require Mg++ or Mn++ (data not shown), these data support the existence of an EDTA inhibited 3′to 5′ exoribonucleolytic pathway that generates substrate for the scavenger-like enzyme.
RNA elements and/or RNA binding proteins that decrease 3′to 5′ exoribonucleolytic decay of an RNA should lead to less m7Gp*ppG product generated by the exosome, reducing substrate available for the scavenger decapping activity, and a consequent reduction in m7Gp* product. Addition of either an 85 nucleotide poly(A)-tail or G16 to the 3′end of the RNA, sequences known to reduce 3′ to 5′exonucleolytic decay of RNA substrates, did lead to a decrease in m7Gp* product (Fig. 1C ▶, cf. lane 5 and lanes 2,4). The most striking example of decreased m7Gp* product as a result of decreased 3′to 5′ exoribonucleolytic decay is observed when the spliced leader RNA (SL snRNA) is the substrate. This small 110 nucleotide RNA is predicted to form a three-stem-loop structure, and is known to assemble into a complex SL RNP in Ascaris embryo extracts (Maroney et al. 1990a; Denker et al. 1996, 2002). We predict this SL RNP would be relatively resistant to 3′ to 5′ exoribonucleolytic decay, leading to minimal m7Gp*ppG substrate formation. As predicted, low levels of m7Gp* were generated in the extracts from the SL snRNA substrate (Fig. 1D ▶, cf. lanes 2 and 4). Addition of the spliced leader sequence alone to the 5′ end of a truncated mRNA is not predicted to lead to the formation of a complex structure and RNP, and does not similarly reduce m7Gp* formation (Fig. 1D ▶, cf. lane 3 and lanes 2,4). We interpret the sum of these observations (EDTA inhibition of the 3′ to 5′ decay, m7GpppG accumulation following inhibition of scavenger activity, and 3′ poly(A) or (G) and SL snRNP structure leading to reduction in m7Gp* formation) to indicate that the rate-limiting step in the complete decay of the RNA in these reactions is the 3′ to 5′ exoribonucleolytic decay of the RNA. In support of this interpretation, incubation of a labeled RNA containing a 3′ subterminal G16 or G30 tract in the extract leads to the predicted 3′–5′ decay intermediate stalled at the G-tracts as shown in other systems (data not shown; Anderson and Parker 1998; Milone et al. 2002)
The combined activity of 3′ to 5′ exoribonucleolytic decay and scavenger cleavage of the resulting cap dinucleoside triphosphate product were observed on a number of different RNA substrates. In addition, similar decay, RNA decapping, and dinucleoside triphosphate hydrolase activities were also observed in C. elegans whole cell embryo extracts (Supplementary Fig. 2F; data not shown).
Decay and decapping of m32,2,7GpppG-capped RNAs
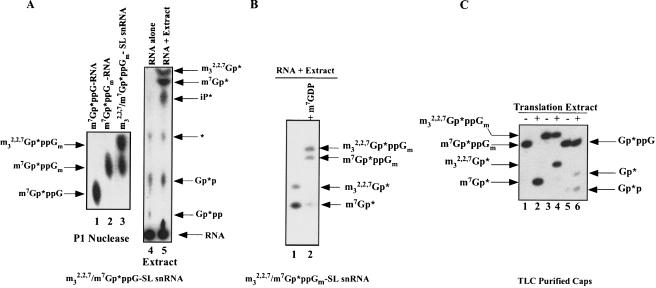
Both m32,2,7GpppG- and m7GpppN-capped mRNAs are substrates for RNA decay in nematodes. To determine whether the scavenger activity in the nematode extracts could cleave an m32,2,7Gp*ppG cap substrate, a cap-labeled trimethylguanosine (m32,2,7Gp*ppG) RNA was prepared by hypermethylation of an m7Gp*ppG cap-labeled SL snRNA in Ascaris embryo extracts as previously described (Maroney et al. 1990a). SL snRNA hypermethylation is not complete in these reactions, leading to a mixed substrate containing m7Gp*ppGm and m32,2,7Gp*ppGm capped SL snRNAs as deduced from nuclease P1 and T1 digestion and TLC and PAGE analysis of the products (Fig. 2A ▶, lane 3; Supplementary Fig. 2B,C; data not shown). Incubation of the mono-and tri-methyl-capped-SL snRNAs in whole cell Ascaris embryo extracts resulted in the accumulation of both m7Gp* and m32,2,7Gp* products (Fig. 2A ▶, lane 5; Fig. 2B ▶, lane 1). 3′ to 5′ exoribonucleolytic decay of these RNAs should lead to the formation of both m32,2,7GpppG and m7GpppG di-nucleoside triphosphates. When nematode scavenger activity is inhibited in the extract with 100 μM m7GDP, both m32,2,7GpppG and m7GpppG dinucleoside triphosphates accumulate from m32,2,7GpppG and m7GpppG capped RNAs (Fig. 2B ▶, lane 2). Overall, these experiments indicate that RNA decay in Ascaris extracts can lead to m7Gp*ppG and m32,2,7Gp*ppG products. Both products are then substrates for a scavenger dinucleoside triphosphate hydrolase(s) that cleaves both cap structures.
FIGURE 2.
Decay and decapping of an m32,2,7Gp*ppG/m7Gp*ppG 32P-cap-labeled RNA in Ascaris extracts leads to both m7Gp* and m32,2,7Gp* products. (A) Hypermethylation and decay of the SL snRNA in Ascaris extracts. Hypermethylation of the Ascaris SL snRNA in extracts leads to a mixed SL snRNA consisting of ~50% m7Gp*ppGm SL snRNA and 50% m32,2,7Gp*ppGm SL snRNA as illustrated by P1 nuclease digestion and TLC analysis of the released cap (see lane 3 and compare to lanes 1 and 2; see also Supplementary Fig. 2A–C). Incubation of the mixed m7Gp*ppGm-/m32,2,7Gp*ppGm-SL snRNA in extracts leads to both m7Gp* and m32,2,7Gp* cap-derived products (see lane 5) as illustrated by TLC and autoradiography. Although m7GDP can comigrate with inorganic phosphate (iP) in this system, the product in lane 5 was shown to be iP based on the absence of its accumulation when phosphatase inhibitors were included in the reaction and the lack of conversion of this product to a triphosphate when incubated with nucleoside diphosphokinase (data not shown; Wang and Kiledjian 2001). Note that the Gp*p and Gp*pp spots are derived from the cap-labeling reaction and are not present in gel-purified RNA substrates that were used in other experiments with similar results. The reaction in lane 5 was carried with an approximately fivefold higher extract protein and longer incubation times than the reactions illustrated in Figure 1 ▶. * = unknown. (B) Inhibition of scavenger activity during decay reactions in Ascaris extracts leads to accumulation of methylated cap guanosine dinucleoside triphosphates. A mixed substrate, cap-labeled SL snRNA (m32,2,7/m7Gp*ppGm-SL snRNA) was incubated in Ascaris extract with or without 100 μM m7GDP, the reactions phenol:chloroform extracted, resolved by denaturing 25% PAGE, and detected by autoradiography. Decay and decapping of the substrate leads to accumulation of both m7Gp* and m32,2,7Gp* (see lane 1). Inhibition of scavenger activity with 100 μM m7GDP leads to accumulation of the substrates for the scavenger enzymes, m7Gp*ppGm and m32,2,7Gp*ppGm (lane 2). Accumulation of a small amount of m7Gp* results from incomplete inhibition of the scavenger activity in lane 2. Note that PAGE assay is more sensitive than the TLC assay. (C) Scavenger activity in Ascaris translation extracts hydrolyzes m32,2,7Gp*ppGm. TLC purified cap-labeled substrates (see A; Supplementary Fig. 2A–D) were incubated with Ascaris extract, the reactions phenol:chloroform extracted, the reaction products resolved by 25% denaturing PAGE, and detected by autoradiography. The single, 30-min time-point assays were carried out within the linear range of extract concentration and incubation time using saturating substrate.
Scavenger activity on mono- and trimethylated caps in Ascaris extracts
The scavenger decapping activity on m7Gp*ppGm and m32,2,7Gp*ppGm was further directly examined in the Ascaris embryo extracts using dinucleoside triphosphate substrates generated by P1 nuclease cleavage of cap-labeled RNAs. The nematode scavenger activity was shown to cleave both m7Gp*ppGm and m32,2,7Gp*ppGm substrates (Fig. 2C ▶, lanes 2,4). The scavenger hydrolase activity in Ascaris embryo extracts is robust, 5.3 × 10−12 mol/min/mg protein using 0.1 fmole of m7Gp*ppG substrate. Extract scavenger activity is at least 50% less active on a m32,2,7Gp*ppGm substrate compared to that observed with a m7Gp*ppGm substrate (Fig. 2C ▶, cf. lanes 2 and 4). Reduced activity of the scavenger enzyme on the trimethyl-compared to the monomethyl-cap substrate is supported by inhibition studies that indicate m7GpppG is a stronger competitive inhibitor of the nematode scavenger activity than m32,2,7GpppG (Table 1 ▶; data not shown). Scavenger hydrolysis of Gp*ppG in the extracts is low, demonstrating the activity has a preference for N7-methylation of the cap (Fig. 2C ▶, cf. lanes 2, 4, and 6). In addition, similar decay and decapping activities were also observed in C. elegans embryo extracts (Supplementary Fig. 2F; data not shown).
TABLE 1.
Competitive inhibitor analysis of recombinant nematode DcpS
| Approximate μM for 50% inhibition | ||
| Dinucleotide and nucleotide competitor | C. elegans rDcpS | Human rDcpS |
| m7GpppG | 2.75 | 0.38 |
| m7GpppC | 4 | ND |
| m7GpppU | 4.5 | ND |
| m7GpppA | 4 | ND |
| m7GpppGm | 8 | ND |
| m7GppppG | 3 | ND |
| m7GTP | 0.3 | ND |
| m7GDP | 0.12 | ND |
| m7GMP | 2.5 | ND |
| m32,2,7GpppG | 64 | 8.5 |
| m32,2,7GpppA | 76 | ND |
| m32,2,7GDP | >500 | ND |
| GpppG | >500 | ND |
| GDP | >500 | ND |
| ApppG | >500 | ND |
Cap cleavage reactions were conducted at 30°C for 30 min with m7Gp*ppG substrate and 10 ng of recombinant DcpS protein with different concentrations of dinucleoside triphosphate or nucleotide competitors (0.01–500 μM). Reaction products were resolved by TLC and detected by autoradiography. Assays were carried out within the linear range of protein, time, and using saturating substrate. Substrate to product conversion was analyzed with a Molecular Dynamics Storm 860 and ImageQuant software.
ND = Not determined.
Direct RNA decapping and relative levels of scavenger and RNA decapping activities in Ascaris extracts
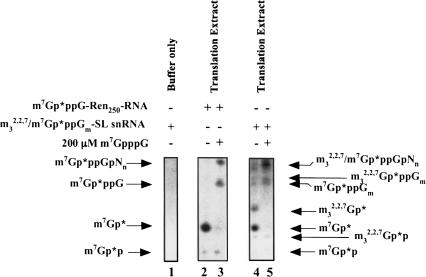
Direct RNA decapping of m7Gp*ppG-RNA and m32,2,7Gp*ppGm-SL snRNA by a Dcp1/Dcp2 type activity producing m7Gp*p and m32,2,7Gp*p was also observed in Ascaris whole embryo extracts with the addition of Mn++ and the use of a more sensitive denaturing PAGE assay (Fig. 3 ▶, lanes 2,4). These assays were also carried out with either cold m7GpppG or m7GDP competitor to attempt to enhance and facilitate Dcp1/Dcp2 product formation as previously described (Gao et al. 2001). These cap analogs can inhibit scavenger decapping activity and sequester cap-binding proteins, which may facilitate better access of Dcp1/Dcp2 to the RNA cap. In general, use of cap analogs to sequester cap-binding proteins did not significantly enhance (≤twofold) our ability to detect potential Dcp1/Dcp2 activity and guanosine diphosphate product formation in the nematode extracts (Fig. 3 ▶, cf. lanes 2–5). Mn++ enhances the nematode extract Dcp1/Dcp2 activity (L.S. Cohen, M. Mikhli, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, unpubl.) as described for the yeast and human enzymes (Wang et al. 2002; Steiger et al. 2003). However, even in the presence of Mn++ (Fig. 3 ▶), we observed only relatively low levels of Dcp1/Dcp2 activity using a variety of extract preparations and different assay conditions. Although Dcp1/2 type activity and its products (m7Gp*p and m32,2,7Gp*p) are observed in the Ascaris extracts, the predominant decapping products observed are monophosphates consistent with a dinucleoside triphosphate or oligonucleotide “scavenger” decapping activity (DcpS). We estimate that the combined activity of 3′ to 5′ exoribonucleolytic decay of the RNA substrate and scavenger dinucleoside triphosphate hydrolysis of the resulting cap dinucleotides by the scavenger is at least ~15-fold greater (and ~60-fold in the absence of Mn++) than the activity of Dcp1/Dcp2 RNA decapping activity in the extract (Fig. 3 ▶, lane 2; data not shown).
FIGURE 3.
Dcp1/Dcp2 activity in Ascaris translation extracts. Cap-labeled RNAs were incubated in Ascaris translation extracts with and without 200 μM m7GpppG, the reactions phenol:chloroform extracted, resolved by 25% denaturing PAGE, and products detected by autoradiography. Note the relative ratios of monophosphate (3′ to 5′ decay and scavenger activity) and diphosphate (Dcp1/Dcp2-like activity) nucleotide decay products in lanes 2 and 4. The diphosphate products do not increase dramatically when scavenger activity is inhibited with m7GpppG. Dcp1/Dcp2 products (m7Gp*p and m32,2,7Gp*p), although limited, are highly reproducible.
Overall, we have observed two types of decapping activities (scavenger- and Dcp1/Dcp2-like) in Ascaris extracts, each of which can cleave mono- and trimethylated caps. We have further shown that a recombinant C. elegans Dcp2 can directly decap m32,2,7Gp*ppGm-SL snRNA producing m32,2,7Gp*p (Supplementary Fig. 1E, lanes 8,16).
Cap dinucleoside triphosphate hydrolase and RNA decapping activity in cellular fractions
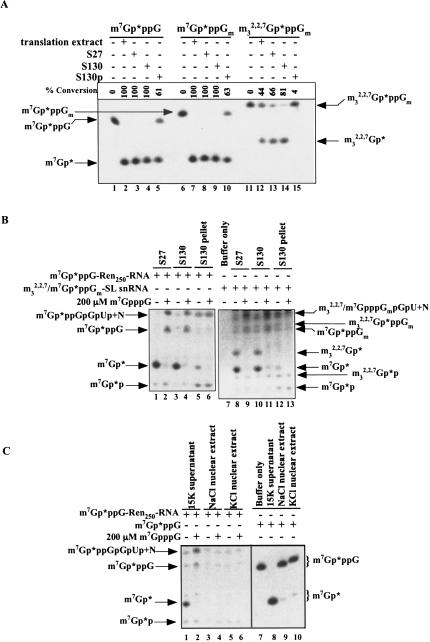
To assess the distribution of the decapping enzyme activity in several cellular fractions, whole-cell embryo extracts were subjected to some initial fractionation as described in the Materials and Methods, and the fractions assayed for both DcpS and Dcp1/Dcp2 type activities. DcpS activity, assessed by dinucleoside cap cleavage leading to m7Gp* or m32,2,7Gp*, was observed in the 27,000 × g (S27) and 130,000 × g (S130) supernatant fractions, but reduced in the 130,000 × g pellet fractions and nuclear extracts (Fig. 4A, C ▶). 3′ to 5′ exoribonuclease activity (producing cap dinucleoside triphosphate or combined with scavenger activity, m7Gp* or m32,2,7Gp*) was observed in all of the fractions examined, although lower levels were observed in nuclear extracts and the 130,000 × g pellet (Fig. 4B, C ▶). Dcp1/Dcp2 RNA decapping activity was observed in all fractions including the S27 supernatant, S130 pellet, S130 pellet salt wash, and nuclear extracts (see Fig. 4B, C ▶; data not shown) with the highest levels observed in the S130 pellet and S130 pellet salt wash. Thus, the two decapping activities exhibit differences in their cellular fractionation.
FIGURE 4.
Distribution of scavenger, 3′ to 5′ exoribonuclease, and Dcp1/Dcp2 activites in Ascaris embryo extract fractions. (A) Distribution of scavenger activity in fractionated Ascaris extracts. TLC purified cap substrates (derived from nuclease P1-treated RNA illustrated in Fig. 2A ▶) were incubated in crudely fractionated Ascaris extract prepared as described in Materials and Methods and analyzed as described in Figure 3 ▶. S27 = 27,000 × g Ascaris embryo extract supernatant; S130 = 130,000 × g supernatant; S130 pellet = 130,000 × g pellet. (B) Dcp1/Dcp2-like and 3′ to 5′ decay/DcpS activity in fractionated Ascaris extracts. m7Gp*ppG-Ren250-RNA or mixed m7Gp*ppGm-/m32,2,7Gp*ppGm-SL snRNA was incubated in Ascaris embryo extract and analyzed as described in Figure 3 ▶. Exosome/DcpS activity is defined by the formation of m7Gp* and m32,2,7Gp*. Dcp1/Dcp2-like activity is defined as the formation of m7Gp*p and m32,2,7Gp*p. Note that both the DcpS and Dcp1/Dcp2 products are produced from the two capped RNA substrates. Hypermethylated SL snRNA is 50% monomethylated and 50% tri-methylated as illustrated in Figure 2A ▶, nuclease P1, lane 3. The m7Gp*p and m32,2,7Gp*p products of Dcp1/Dcp2, although limited, are highly reproducible. It should be noted that Dcp2 has reduced activity on RNA substrates with the spliced leader sequence (L.S. Cohen, M. Mikhli, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, unpubl.), the substrate used to generate the hypermethylated cap. (C) 3′ to 5′ decay/DcpS, scavenger, and Dcp1/Dcp2 activity in nuclear extracts. m7Gp*ppG-cap-labeled RNA (lanes 1–6) or m7Gp*ppG dinucleoside triphosphate (lanes 7–10) was incubated in Ascaris embryo extract fractions and analyzed as described in Figure 3 ▶. Nuclear extracts were prepared as described in Materials and Methods.
A C. elegans protein with scavenger DcpS activity
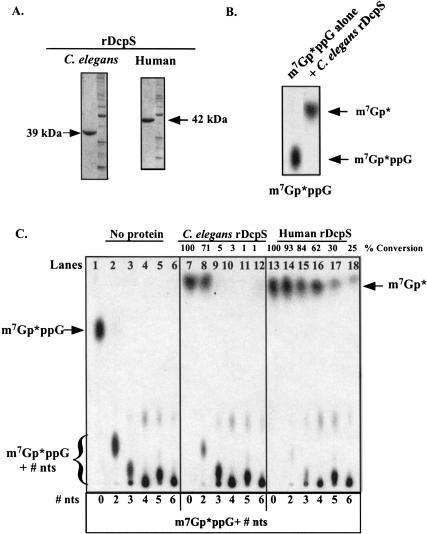
Because a DcpS ortholog likely carries out the scavenger activity observed in nematode extracts, we searched for and identified a putative C. elegans ortholog of the human DcpS in the C. elegans′ database. The C. elegans DcpS open reading frame was amplified by RT-PCR, cloned into a bacterial expression vector, and the bacterially expressed 6×-His-tagged protein purified by Ni2+-NTA agarose and then ion exchange chromatography. The purified protein (>90% pure; Fig. 5A ▶) is ribonuclease-free and readily cleaved m7Gp*ppG dinucleoside triphosphate to m7Gp* demonstrating that the identified C. elegans protein exhibited scavenger dinucleoside triphosphate hydrolase activity (Fig. 5B ▶). The specific activity of the recombinant C. elegans DcpS for m7Gp*ppG is ~2 × 10−11 moles/min/mg protein. A similarly purified recombinant human DcpS protein was also prepared and assayed for a comparison of activities and substrate specificity (see below; Fig. 5A ▶).
FIGURE 5.
Recombinant nematode DcpS hydrolyzes free cap and maximum substrate length of recombinant nematode and human DcpS. (A) SDS-PAGE of recombinant DcpS proteins. Bacterially expressed, histidine tagged C. elegans and human DcpS were purified using nickel affinity and then Q-Sepharose Fast Flow column chromatography. Purified protein preparations were >90% pure based on Coomassie Blue staining of SDS-PAGE gels. (B) Recombinant C. elegans DcpS hydrolyzes m7Gp*ppG to m7Gp*. m7Gp*ppG in buffer alone or with 100 ng of recombinant DcpS protein was incubated for 30 min at 30°C and aliquots of the reactions applied to PEI-cellulose TLC plates, the plates developed in 0.45 M ammonium sulfate, and the substrate and product detected by autoradiography. (C) Maximum cap substrate length of recombinant nematode DcpS. Cap-labeled dinucleotide and oligonucleotide substrates were generated as described in the Materials and Methods. Cap cleavage reactions were carried out at 30°C for 30 min using 100 ng of recombinant protein and aliquots of the reactions applied to PEI-cellulose TLC plates, the plates developed in 0.45 M ammonium sulfate, and the substrate and product detected by autoradiography. Substrate to product conversion was determined from phosphoimager analysis. The substrate sequences are as follows: 0 = m7GpppGOH, +2 = m7GpppGpGpUOH, +3 = m7GpppGpGpGpCOH, +4 = m7GpppGpGpGpGpUOH, +5 = m7GpppGpGpGpApApCOH, and +6 = m7GpppGpGpApGpApCOH.
C. elegans DcpS Hydrolyzes up to m7Gp*ppG + 2 nucleotide substrates, but not longer substrates
To examine the substrate length requirement of the nematode DcpS, cap-labeled m7Gp*ppGNn oligonucleotide substrates of different lengths were prepared using nuclease P1 or RNase A digestion of different cap-labeled RNAs (see Materials and Methods). As illustrated in Figure 5C ▶ (lanes 7,8), m7Gp*ppG and m7Gp*ppG + 2 nucleotide substrates were readily cleaved to m7Gp* by C. elegans DcpS, but dinucleoside triphosphate cap substrates with 3–6 additional 3′ nucleotides (lanes 9–12) were poor substrates for the dinucleoside triphosphate hydrolase activity. The recombinant human enzyme, by comparison, readily cleaves cap dinucleosides triphosphates with up to four additional nucleotides and retains some activity with m7Gp*ppG + 6 oligonucleotide (Fig. 5C ▶, lanes 13–18). These data on the human enzyme are in agreement with those described by Wang et al. (2002), who demonstrated that the human enzyme retains some activity on substrates as long as 10 nucleotides. Thus, nematode DcpS requires a shorter substrate than the human enzyme and exhibits minimal hydrolase activity on m7Gp*ppG + 3 nucleotides or greater.
2′-O-Ribose is methylated in nematode caps
Capped RNAs from higher eukaryotes typically contain a 2′-O-methyl in the ribose of the first and second template encoded bases of the RNA (known as cap 2). Lower eukaryote mRNAs such as yeast do not have 2′-O-methyl added to the mRNA cap (Cap 0; Mager et al. 1976; Sripati et al. 1976). The Ascaris SL snRNA has an m32,2,7GpppGm cap (cap 1) as shown by analysis of an in vivo 32P-labeled SL snRNA (Maroney et al. 1990a,b). Transfer of the m32,2,7GpppGm cap 1 with the 22 nucleotide spliced leader to mRNAs by trans-splicing (Liou and Blumenthal 1990; Van Doren and Hirsh 1990) would thus produce trans-spliced mRNAs with an m32,2,7GpppGm (cap 1). As the m32,2,7GpppGm cap is derived from a m7GpppG-SL snRNA in vivo, it is likely that non-trans-spliced mRNAs in nematodes would have a m7GpppNm cap (cap 1). In fact, we have observed 2′-O-ribose methylation of m7GpppG-capped RNA in Ascaris extracts (data not shown).
C. elegans DcpS hydrolyzes both m7GpppG and m32,2,7GpppG (cap 0 and cap 1) dinucleoside triphosphate substrates
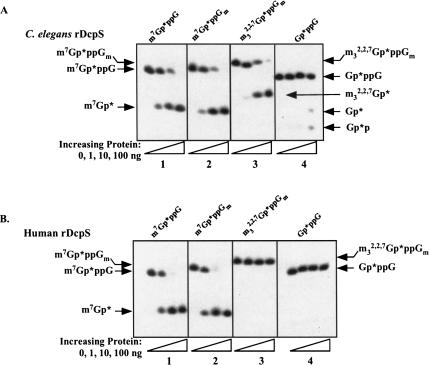
To further examine the substrate specificity of nematode DcpS, we prepared and TLC purified labeled m7Gp*ppG, m7Gp*ppGm, m32,2,7Gp*ppG, and m32,2,7Gp*ppGm caps from cap-labeled RNAs treated with nuclease P1 (see Supplementary Fig. 2A–D). As we observed hydrolysis of both methylated and hypermethylated dinucleoside cap substrates in the Ascaris embryo extracts, we hypothesized that the C. elegans DcpS would cleave both m7Gp*ppG and m32,2,7Gp*ppG caps. As hypothesized, both substrates were in fact converted to their respective Gp* products (Fig. 6A ▶, panels 1–3).
FIGURE 6.
Comparison of cap substrate hydrolysis for recombinant C. elegans and human DcpS. Cap cleavage reactions were carried out at 30°C with recombinant C. elegans DcpS (A) or 37°C with recombinant human DcpS (B) for 30 min using TLC purified and characterized substrates. Reactions were carried out and analyzed as described in Figure 2C ▶ using 25% denaturing PAGE and detected by autoradiography. Control reactions using a similarly purified recombinant protein (β-galactosidase) demonstrated no degradation of RNA or cleavage of cap dinucleotides.
We then compared the substrate specificity of C. elegans and human DcpS for cap 0 and cap 1 m7GpppG and m32,2,7GpppG. As illustrated, both the nematode and human enzymes are active on either m7GpppG cap 0 or cap 1 (Fig. 6A, B ▶, panels 1,2). The C. elegans DcpS is also active on both cap 0 or cap 1 m32,2,7GpppG (Fig. 6B ▶, panel 3; data not shown). However, under similar assay conditions, the human DcpS enzyme did not hydrolyze either m32,2,7GpppG cap 0 or cap 1 (Fig. 6 ▶, cf. A and B, panel 3; data not shown). Thus, the recombinant nematode DcpS has significant m32,2,7GpppG hydrolase activity compared to the human enzyme. Furthermore, the 2′-O-methyl ribose does not significantly affect the activity of either the nematode or human DcpS.
C. elegans DcpS has minimal activity on a guanine dinucleoside triphosphate substrate that is not methylated (Gp*ppG) compared to methylated caps (Fig. 6A ▶, panel 4). However, at ~100 ng recombinant protein a small, but detectable amount of Gp*ppG hydrolysis is observed. Very high levels of recombinant C. elegans DcpS protein (~500 ng) in an extended incubation lead to more efficient hydrolysis (see Supplementary Fig. 2D, lanes 5,8). Notably, the specificity for which phosphodiester bond is hydrolyzed appears reduced with the Gp*ppG substrate as both Gp* and Gp*p products are observed. Only monophosphate products are typically observed with methylated caps even with excess DcpS protein. In comparison, up to 100 ng of human DcpS under the same assay conditions had almost no activity on a Gp*ppG substrate (Fig. 6B ▶, cf. panel 4).
Competition experiments were also carried out using dinucleoside triphosphates and nucleotide competitors to gain further insight into the substrate specificity of the recombinant nematode DcpS enzyme. The concentration of competitor required to produce 50% inhibition of the recombinant enzyme is shown in Table 1 ▶. The most effective inhibitors were m7GTP (0.3 μM) and m7GDP (0.12 μM) nucleotides followed by m7GMP (2.5 μM), then m7GpppG/C/A/U (2.75, 4, 4, and 4.5 μM, respectively), m7GppppG (3 μM), and m7GpppGm (8 μM). Trimethylated guanine dinucleoside triphosphate substrates (m32,2,7GpppG and m32,2,7GpppA) were significantly less active as inhibitors (64 or 74 μM required for 50% inhibition). The 3′ nucleotide in a methylated guanine dinucleoside triphosphate does not lead to significant differences in level of inhibition. This is consistent with the likely variation in the +1 nucleotide of different non-trans-spliced mRNA transcripts. Neither ApppG nor nonmethylated guanine dinucleoside triphosphate (GpppG) or guanine nucleotides inhibited the enzyme. Interestingly, although trimethylated dinucleoside triphosphate substrates (m32,2,7GpppG) can inhibit the enzyme, the trimethylated nucleotide diphosphate (m32,2,7GDP) demonstrated minimal inhibition. In addition, nucleotide concentrations required for 50% inhibition of the nematode recombinant enzyme were in general approximately five- to eightfold higher than those observed for the human DcpS enzyme.
C. elegans DcpS binds to both m7GTP- and m32,2,7GTP-sepharose
The previous experiments indicated that C. elegans recombinant DcpS hydrolyzes m32,2,7GpppG, whereas the human enzyme has minimal activity on this hypermethylated cap. To independently examine cap binding to mono- and trimethylguanosine nucleotides, cap-binding affinity experiments (Table 2 ▶) were carried out using the recombinant nematode and human DcpS enzymes and m7GTP- and m32,2,7GTP-Sepharose as previously described (Jankowska-Anyszka et al. 1998; Keiper et al. 2000; Miyoshi et al. 2002). The recombinant nematode DcpS protein bound with equivalent efficiency to the m7GTP- and m32,2,7GTP-Sepharose. In contrast, the human DcpS protein bound very poorly to m32,2,7GTP-Sepharose and bound more tightly than C. elegans DcpS to the m7GTP-Sepharose. As a positive control we used another nematode cap-binding protein, Ascaris eIF4E, which binds equally to both m7GTP- and m32,2,7GTP-Sepharose (S. Lall, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, in prep.). As negative controls that do not bind cap alone, we used C. elegans Dcp2 (L.S. Cohen, M. Mikhli, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, unpubl.) and nuclear cap-binding protein 80 (CBP80; R.E. Davis, unpubl.). These data are consistent with and provide additional experimental support to the cleavage studies (Fig. 6A, B ▶, panel 3) that indicate the nematode DcpS has a higher affinity for trimethylated guanine nucleotides than does the human DcpS.
TABLE 2.
Recombinant nematode DcpS binds to both m7GTP- and m32,2,7GTP-Sepharose
| Recombinant protein | Cap analog affinity matrix | Relative binding affinity |
| C. elegans DcpS | m7GTP-Sepharose | ++ |
| m32,2,7GTP-Sepharose | ++ | |
| Human DcpS | m7GTP-Sepharose | ++++ |
| m32,2,7GTP-Sepharose | − | |
| Ascaris eIF4E | m7GTP-Sepharose | +++ |
| m32,2,7GTP-Sepharose | +++ | |
| C. elegans Dcp2 | m7GTP-Sepharose | − |
| or C. elegans CBP80 | m32,2,7GTP-Sepharose | − |
N7-methyl guanosine diphosphate (m7GDP) is a relatively poor substrate for C. elegans DcpS
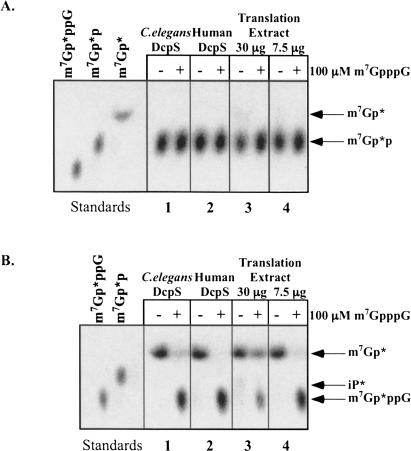
Recently, experiments illustrating conversion of m7GDP to m7GMP by recombinant human DcpS were described (van Dijk et al. 2003). These studies suggested care should be taken in interpreting the contribution of different cellular decapping activities simply based on the accumulation of m7GMP or m7GDP products in cellular extracts because DcpS activity could be hydrolyzing the product of Dcp1/Dcp2 activity (e.g., m7GDP) to m7GMP. To determine whether the nematode extract and/or recombinant DcpS enzyme was active in the conversion of m7GDP to m7GMP, we generated and TLC purified m7Gp*p substrate from m7Gp*ppG-RNA using recombinant nematode Dcp2 (L.S. Cohen, M. Mikhli, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, unpubl.). Using similar assay conditions as described by van Dijk et al. (2003), we have not been able to observe significant conversion of m7GDP to m7GMP with either doubly purified, recombinant C. elegans or human DcpS preparations (Fig. 7A ▶, panels 1 and 2). m7Gp*ppG cap TLC purified during the same experiment is readily cleaved and mixing experiments indicate that the TLC purified m7Gp*p added to an m7Gp*ppG cleavage reaction does not contain TLC extracted components that inhibit the enzyme activity (Fig. 7B ▶, panels 1,2; data not shown). Liu et al. (2002), in their characterization of the human protein, also noted that the human protein did not hydrolyze m7GDP. Furthermore, in Ascaris whole-cell embryo extract assays, very little labeled m7GDP is converted to m7GMP (Fig. 7A ▶, panels 3,4). The activity responsible for this conversion could be one of several different enzymes. Importantly, (1) the percentage of m7GMP derived from m7GDP in the extracts would contribute only a very small percentage of total m7GMP to our in vitro decapping reactions, and (2) the levels of m7GDP produced do not increase more than twofold in extract assays when DcpS is inhibited with m7GpppG. This suggests it is unlikely the major m7Gp* products observed in our whole embryo extract assays represent significant levels of DcpS conversion of m7Gp*p derived from Dcp1/Dcp2 de-capping of RNA, and that our estimates of the relative amounts of DcpS compared to Dcp1/Dcp2 activity are not subject to this limitation. Our interpretation of the data is that the predominant decay/decapping pathway in nematode embryos utilizes 3′ to 5′ exonuclease decay of the RNA followed by scavenger cleavage of the produced m7GpppN and m32,2,7GpppG dinucleoside triphosphates.
FIGURE 7.
Recombinant nematode DcpS and Ascaris embryo extract activity on m7GDP. Doubly purified recombinant C. elegans and human DcpS (150 ng) or Ascaris extract (30 and 7.5 μg) was incubated with TLC purified (A) m7Gp*p or (B) m7Gp*ppG and the reaction products analyzed by TLC and detected by autroradiography. Recombinant DcpS assays were carried out in a buffer as described (van Dijk et al. 2003) for 1 h with the human enzyme at 37°C and the nematode enzyme and extracts at 30°C. Reactions carried out with the decapping buffer used in the current study produced identical results. (B) Illustrates that a similarly purified substrate from the same experiment is a substrate for DcpS. Mixing experiment indicate that the TLC purified m7Gp*p is not inhibitory to DcpS hydrolysis of m7Gp*ppG in B (data not shown).
DISCUSSION
Summary
Analysis of decapping activities in nematode embryo extracts indicates the predominant activity is a scavenger de-capping activity (DcpS-like) that hydrolyzes m7GpppG and m32,2,7GpppG products of 3′ to 5′ exoribonucleolytic RNA decay. Dcp1/Dcp2-like RNA decapping activity is detectable in nematode embryo extracts but present at significantly lower levels. Scavenger activity is very robust, and the combined activities of 3′ to 5′ exoribonucleolytic decay of RNA followed by N7-guanosine nucleoside triphosphate hydrolase activities greatly exceeds (~15-fold) that observed for direct decapping of RNA by Dcp1/Dcp2-like activity. These data suggest that 3′ to 5′ decay appears to be the predominant general pathway of decay in nematode embryo extracts and may be the default pathway for mRNA degradation in vitro, at least for the RNA substrates examined.
Our characterization of C. elegans recombinant DcpS activity indicates the enzyme has limited activity on guanosine dinucleoside triphosphates (GpppG) unless the first base is methylated at the N7 position. Nematode scavenger enzyme readily hydrolyzes m7GpppG, m7GpppGm, m7GpppGp, m7GpppGmp, and m7GpppGmpGp (some data not shown). Capped-oligonucleotides up to m7GpppG + 2 nucleotides are actively hydrolyzed, but longer capped oligo-nucleotides are poor substrates. Nematode DcpS hydrolyzes tri-methyl-guanosine cap dinucleoside tri-phosphates including m32,2,7GpppG, m32,2,7GpppGm, m32,2,7GpppGp, and m32,2,7GpppGmpGp. Nematode DcpS is more active on m7GpppG versus m32,2,7GpppG, as illustrated by both direct hydrolysis of cap-labeled substrates (at least 2:1) and inhibition assays. Notably, in comparison, human DcpS has minimal activity on the trimethylated cap (m32,2,7GpppG) and is active on longer oligonucleotide substrates. The data suggest there are differences between the human and nematode DcpS enzymes in the substrate binding pocket and/or its flexibility.
Structural basis for DcpS substrate requirements
Recently, the structure of human DcpS bound to m7GpppG or m7GpppA and insight into the mechanism of DcpS were reported (Gu et al. 2004). The DcpS structure indicates the protein is an asymmetric dimer that contains both an open nonproductive and a closed productive DcpS–cap complex. Many residues interact with the cap and/or each other to create a pocket that is favorable for m7GpppG binding, as well as the structural changes associated with the closing of the pocket and subsequent cleavage of the cap. Differences in substrate and substrate length specificity observed among Schizosaccharmyces pombe (substrate can be a full-length capped RNA), nematodes (both m7G-and m32,2,7GpppG cap and Cap + 2 nucleotides), and humans (Cap + 10 nucleotides) suggests that the binding pocket of DcpS enzymes exhibit unusual flexibility or distinct structural features associated with the open and closed conformations for cap binding and cleavage. It remains to be determined what overall structural determinants contribute to these differences.
We have identified a variety of additional DcpS orthologs in databases ranging from early eukaryotes (Tetrahymena, Entamoeba, Chlamydomonas, and a diatom) and metazoa including DcpS sequences from organisms with trans-splicing (tunicate: Ciona; flatworm: Schistosoma; C. elegans, and two additional parasitic nematodes: Brugia malayi and Heterodera glycines; data not shown). Comparison of these sequences (particularly DcpS orthologs from trans-splicing organisms) with the mechanistic and structural information derived from analysis of human DcpS will enable us to carry out nematode DcpS structure/function studies that should provide insight into the difference in cap-binding specificity observed in nematodes, and if these differences may be exploitable for rational drug design in the development of new anti-nematode drugs.
Nematode scavenger activity and decapping of m32,2,7GpppG-capped RNAs
During the course of our studies the characterization a C. elegans operon containing a histidine triad protein, dcs-1 was reported (Kwasnicka et al. 2003). This is the same protein we have characterized as C. elegans DcpS. Our data are in general agreement with their kinetic analysis and inhibition data indicating that the protein is relatively specific for N7-methyl-guanine nucleotides. Their Ki values for m7GpppG, m7GTP, and m7GDP were in the 2.2–3.5-μM range, and the Ki value for trimethylated cap was 28 μM, approximately eightfold higher than the monomethylated nucleotides. From their Ki data, Kwasnicka et al. (2003) concluded that the scavenger enzyme was not likely to function on m32,2,7GpppG-capped snRNAs. Our data indicate that the protein can function on an m32,2,7GpppG-capped snRNA, the SL snRNA, as well as a U1 snRNA (data not shown). In addition, of seminal importance in our view is whether the scavenger activity in nematodes is sufficient to act on m32,2,7GpppG derived from trans-spliced mRNAs representing ~70% of the nematode mRNA population. Although the substrate affinity of nematode DcpS is clearly significantly lower for m32,2,7GpppG than m7GpppG, the levels of scavenger activity in nematode extracts are very high. DcpS activity associated with the exosome complex (or subset of exosome complex proteins) actively engaged in 3′ to 5′ RNA decay could also have altered kinetic properties, and might have access to higher local concentrations of substrate. Human DcpS activity was previously shown to be present both in free form and a complex, likely the exosome (Wang and Kiledjian 2001). We have made similar observations in the Ascaris extracts (data not shown). We have rarely observed significant accumulation of m32,2,7GpppG or m7GpppG caps in our embryo extract decay reactions (or in vivo following biolistic introduction of labeled RNAs; L.S. Cohen, M. Mikhli, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, unpubl.) where endogenous mRNAs in the extracts are simultaneously undergoing decay. Taken together, the high levels of nematode embryo DcpS-like activity, the in vitro decay data, and the likely association of DcpS with the exosome complex (data not shown) suggests to us that the activity in vivo is likely to be sufficient to hydrolyze both m32,2,7GpppG and m7GpppG caps derived from 3′ to 5′ decay. Alternatively, it remains possible that additional scavenger activities are present in nematodes that can hydrolyze trimethylguanosine caps. Recently, a U8 snoRNA binding protein was identified as a nuclear decapping enzyme active on RNA substrates producing GDP (and its methylated derivatives) and p-RNA products (Ghosh et al. 2004). Although this protein can cleave m32,2,7GpppG-capped RNA, and is conserved in vertebrates, it does not appear to be present in the genomes of lower eukaryotes including C. elegans.
Cellular distribution of decapping activities
Our initial extract studies suggest that the DcpS activity is primarily cytoplasmic in 32–64 cell Ascaris embryos. Studies by the Kiledjian laboratory (Liu et al. 2002) also found that the human DcpS protein was primarily found in the cytoplasmic fraction, with little protein observed in the nuclear fraction based on immunoprecipitation experiments. However, analysis of the S. pombe, COS cell, and human DcpS proteins using fluorescence and immunocytochemistry indicate that they localize primarily to the nucleus (Salehi et al. 2002; Kwasnicka et al. 2003; Cougot et al. 2004; M. Kiledjian, pers. comm.). The predominantly nuclear localization of the COS cell DcpS led Kwasnicka et al. (2003) to propose that the enzyme might function to decap aberrant transcripts in the nucleus that might result under stress conditions. Studies are currently underway in our laboratory to examine the localization of DcpS protein using immunocytochemistry in developing nematode embryos and to examine the interaction of nematode DcpS with other cap-binding proteins and exosome components.
Conclusion
The nematode-decapping enzymes, DcpS and Dcp1/Dcp2, have the ability to hydrolyze the typical eukaryotic m7GpppG cap as well as the m32,2,7GpppG cap added to mRNAs through trans-splicing. An in vitro decay system is described that indicates RNA decay by a 3′ to 5′ pathway followed by DcpS hydrolysis of the resulting cap appears to be the predominant nematode decay pathway in embryo extracts. These studies provide the foundation for further studies to characterize the contribution and interplay of translation and decay in the expression of trans-spliced and non-trans-spliced nematode mRNAs.
MATERIALS AND METHODS
Preparation of whole-cell embryo and nuclear extracts
Whole-cell extracts were prepared from 32–64 cell Ascaris suum embryos using methods similar to those described by Nilsen and collaborators (Hannon et al. 1990a,b; Maroney et al. 1995) with a metal dounce, 10 mM KCl, and protease inhibitors. These Ascaris embryo extracts are competent for cap-dependent translation and exhibit mRNA cap and poly(A)-tail synergism (S. Lall, C. Fried-man, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, in prep.). In some experiments, extracts were prepared with a modified buffer system (10 mM Tris-HCl, pH 7.5, 1 mM KoAc, 1.5 mM MgoAc, 1 mM DTT, and protease inhibitors) and crudely fractionated as described (Brewer and Ross 1990; Ross 1999; Wang and Kiledjian 2000). Ascaris embryo nuclear extracts were prepared as described (Seidl and Moritz 1998), the nuclear pellets extracted with either 400 mM KCl or 350 mM NaCl, and the resulting nuclear extracts dialyzed. C. elegans whole-cell embryo extracts were generously provided by Margaret MacMorris and Tom Blumenthal.
Transcription template and RNA preparation
PCR templates (see Supplementary Fig. 1) for in vitro transcription reactions were prepared from Promega plasmid pRLnull or pAscSL_snRNA using primers provided in the Supplementary Figure 1 ▶. These PCR reactions generate templates with a T7 promoter, different 5′ UTRs with or without the spliced leader sequence, and 3′ ends either with or without a poly(A) tail or a homopolymeric 3′ G16 tract. Transcription reactions produced from either a Riboprobe or MegaScript kit as described by the manufacturer (Promega and Ambion) were DNase I-treated, extracted with TRIzol Reagent (Invitrogen), and the RNAs precipitated twice, once with isopropanol and then with ammonium acetate/ethanol. Precipitated RNAs were further washed with 70% ethanol, dissolved in water, quantitated spectrophotometrically, and examined by aga-rose-formaldehyde, denaturing gel electrophoresis.
Cap-labeled substrate preparation
Cap-labeled RNAs were prepared from uncapped RNA substrates using 32P-α-GTP (Perkin-Elmer), and recombinant vaccinia RNA guanylyltransferase and (guanine- N7)-methyltransferase (generously provided by Stewart Shuman) or human capping enzyme (generously provided by Aaron Shatkin). In some experiments, the cap was further methylated at the 2′-O-ribose by inclusion of mRNA cap-specific 2′-O-methyltransferase (generously provided by Paul Gershon). For many experiments, cap-labeled RNAs were gel purified by denaturing PAGE prior to use.
Cap-labeled oligonucleotides of defined length (see Fig. 5C ▶) were derived by treating cap-labeled RNA transcripts (derived from pRLnull, pBlueScript II SK, pGL3 Basic, or the Megascript positive control template) with RNase A and characterized prior to use by thin layer chromatography (TLC) and PAGE.
Cap dinucleoside triphosphates (Gp*ppG, m7Gp*ppG or m32,2,7Gp*ppG, where the * follows the 32P-labeled phosphate) were typically generated from cap-labeled RNAs by Nuclease P1 (Calbiochem and Sigma) digestion of the RNA (see Supplementary Fig. 2A,B). In some cases, caps were produced from cap-labeled RNAs by T1 digestion (Ambion), which produces caps or capped oligonucleotides with a 3′ phosphate. To generate pure cap-labeled substrates, labeled RNAs and caps were either gel purified or purified by TLC and elution of individual substrates in 1× decapping buffer.
Ascaris m32,2,7Gp*ppGm-SL snRNA was produced by hypermethylation of an m7Gp*ppGm-SL snRNA in Ascaris whole-cell embryo extracts and purified as previously described (Maroney et al. 1990b).
Whole cell extract decay analysis
Ascaris embryo extract mRNA decay and decapping assays were carried out using the decapping buffer of Zhang et al. (1999) (50 mM Tris, pH 7.9, 30 mM (NH4)2SO4, 1 mM MgCl2, 1 mM DTT) with the addition of 2.5 mM MnCl2, which increases Dcp2 activity. RNA substrates in the reactions ranged from ~0.04–13 ng (0.5–163 fmole) per reaction, corresponding to 5000–160,000 dpm. Cap dinucleoside triphosphate substrates (e.g., m7Gp*ppG, m7Gp*ppGm, or their nonmethylated or hypermethylated derivatives) ranged from ~500–64,000 dpm/reaction (0.05–0.64 fmole). Reactions typically contained 7.5 μg of protein (protein range in reactions was ~2–30 μg), the reactions were incubated at 30°C for 30–60 min, and then extracted with 1:1 phenol:chloroform/isoamyl alcohol. For specific activity determinations, reactions were carried out within the linear range of protein and time using a set substrate concentration described in the text. Reactions were spotted onto TLC plates or loaded on denaturing PAGE gels, resolved as described below, and detected by autoradiography.
Cloning, expression, and purification of C. elegans DcpS
Total C. elegans RNA was isolated from mixed stage C. elegans cultures using Trizol®. First-strand cDNA was generated using SuperScript II reverse transcriptase and oligo dT primers (Invitrogen). The C. elegans DcpS open reading frame was amplified from the cDNA using specific primers (see Supplementary Fig. 1 ▶) and Expand High Fidelity Polymerase (Roche). The DcpS coding region PCR product was digested with NdeI and BamHI (Promega) and cloned into pET16b (Novagen) using DH5α as a host. Recombinants were identified and confirmed by DNA sequencing. Clones were then transformed into Rosetta DE3 (Novagen) for protein expression. Protein expression was induced with 0.4 mM isopropyl β-D-thiogalactoside overnight at room temperature. Frozen bacterial pellets were resuspended in ice-cold lysis buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 300 mM urea, 10% glycerol, 1% Triton X-100, 10 mM imidazole), lysozyme was added to a final concentration of 1 mg/mL, the suspension incubated on ice for 30 min, and then sonicated. The 6×-His-tagged DcpS was bound to Ni2+-nitrilotriacetic acid (NTA)-agarose (Qiagen Inc.) for 60 min at 4°C, unbound proteins removed with a washing buffer (20 mM HEPES, pH 7.5, 300 mM NaCl), and the bound proteins eluted with wash buffer containing increasing concentrations of imidazole (20–300 mM imidazole). Fractions containing DcpS were dialyzed against 20 mM Tris pH 7.5, 150 mM NaCl, loaded onto a HiTrap Q FF anion exchange column (Pharmacia) equilibrated in the same buffer, and bound protein eluted with a gradient from 0.15 to 1 M NaCl. Those fractions containing DcpS activity were dialyzed against 0.2 mM EDTA, 20 mM Tris, pH 7.5, 50 mM KCl, 20% glycerol, 1 mM DTT, 0.5 mM PMSF and stored at–80°C. Human DcpS in pET28, generously provided by Kiledjian et al. (Liu et al. 2002), was expressed and purified in a manner identical to the nematode protein.
Recombinant DcpS assay
Recombinant DcpS cap cleavage assays were carried out as described for the embryo extracts at 30°C for 30 min with ~1–150 ng of two step purified (His-Tag and anion exchange) recombinant protein. Inhibition assays contained unlabeled dinucleoside tri-phosphates and guanosine nucleoside phosphates that were pre-mixed in reaction buffer before the addition of the protein. DcpS reactions were stopped on ice, extracted with phenol/chloroform, the reaction products resolved on TLC plates or PAGE gels, and detected by autoradiography. Substrate to product conversion was determined by phosphoimager analysis using Molecular Dynamics STORM 860 and ImageQuant software. For the inhibition experiments, 50% inhibition was determined by graphical analysis of percent conversion of substrate to product and determination of the amount of inhibitor needed for 50% inhibition.
Substrate characterization and dinucleoside triphosphate (cap) cleavage and RNA decapping product analysis
Capped RNA and cap substrates and their reaction products were characterized and identified using a variety of methods including comigration with known standards using TLC or denaturing PAGE analysis and several enzyme shift strategies (treatment with alkaline phosphatase [Roche], phosphatase inhibitors [Sigma], diphosphonucleotide kinase [Sigma], tobacco acid pyrophosphatase [Epicentre], nucleotide pyrophosphatase [Sigma], Nuclease P1 [Calbiochem], Nuclease T1 [Ambion], and RNase A [Ambion]) (see Supplementary Fig. 2; data not shown). TLC was carried out using PEI-cellulose as the stationary phase and ammonium sulfate (0.45 M) or lithium chloride (0.75 M) as the mobile phase. PAGE analysis was done using 20% or 25% denaturing gels as described (Bergman et al. 2002). Reaction substrates and products were then quantified by phosphoimager analysis of the TLC or PAGE separations using Molecular Dynamics STORM 860 and ImageQuant software.
m7GTP- and m32,2,7GTP-sepharose binding assays
Recombinant C. elegans DcpS, human DcpS, C. elegans Dcp2 (L.S. Cohen, M. Mikhli, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, unpubl.), Ascaris eIF4E (S. Lall, C. Friedman, M. Jankowska-Anyszka, J. Stepinski, E. Darzynkiewicz, and R.E. Davis, unpubl.), and C. elegans CBP80 (R.E. Davis, unpubl.) were mixed with m7GTP-Sepharose or m32,2,7GTP-Sepharose beads (Jankowska et al. 1993) in binding buffer (20 mM Tris, pH 7.5, 25 mM KCl, 0.2 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 20% glycerol), the samples incubated for 45 min at 4°C with gentle shaking, the sepharose beads washed with binding buffer, and the bound proteins eluted with binding buffer containing 500 mM KCl. Proteins that remained bound to the sepharose beads were removed from the beads by denaturation using heat and Laemmli SDS PAGE loading buffer. Aliquots of the proteins (starting material, flow-through, wash, and eluate) were analyzed by SDS PAGE, stained with SYPRO Ruby (Molecular Probes), and analyzed with a Molecular Dynamics Storm 860 and ImageQuant software.
Other reagents
The following dinucleoside cap analogs were synthesized as previously described: m7GpppG, GpppG, m7GppppG, and m32,2,7GpppG, according to Stepinski et al. (1995), and m7GpppC, m7GpppU, m7GpppA, and m7GpppGm, according to Jankowska et al. (1996). ApppG and m32,2,7GpppA were prepared by a new recently developed strategy (Stepinski et al. 2001) using adenosine 5′-phoshorimidazolide and GDP and m32,2,7GDP, respectively, as substrates and carrying out the coupling reaction in dimethylformamide in the presence of zinc chloride. Nucleotide competitors m7GMP, m7GDP, m7GTP, and m32,2,7GDP were prepared from GMP, GDP, GTP, and m22,2GDP, respectively, by the methylation method described earlier (Darzynkiewicz et al. 1985). The synthesis of m7GTP- and m32,2,7GTP-Sepharose 4B were also carried out by the known method (Jankowska et al. 1993).
ELECTRONIC SUPPLEMENTARY DATA
Supplementary material can be found online at http://www.uchsc.edu/sm/bbgn/Davislab/RNA_DcpS.html.
Acknowledgments
Human DcpS coding region in pET-28a (pET28-hDcpS) was generously provided by Mike Kiledjian, recombinant vaccinia RNA guanylyltransferase and (guanine-N7)-methyltransferase by Stew-art Shuman, human RNA guanylyltransferase by Aaron Shatkin, mRNA cap-specific 2′-O-methyltransferase by Paul Gershon, and C. elegans whole embryo extract by Margaret MacMorris and Tom Blumenthal. We thank Mike Kiledjian and his laboratory for exceptionally helpful discussions and sharing reagents and information. Thanks to Carol J. Wilusz and Jeffrey Wilusz for generously providing reagents, information, and encouragement. Thanks to Paul Copeland and Scott Kinzy for helping us to carry out gel filtration experiments in their laboratory, and to Chris Lima and Mike Kiledjian for access to data prior to publication. We thank members of the Davis lab for their comments on the manuscript. This work was supported by National Institutes of Health (NIH) Grant AI49558 and CUNY-CSI startup funds to R.E.D., and Grants #PBZ-KBN-059/T09/10 and KBN 3 P04A 021 25 from the Polish Committee for Scientific Research to E.D.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7690504.
REFERENCES
- Anderson, J.S.J. and Parker, R.P. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., and Parker, R. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646. [DOI] [PubMed] [Google Scholar]
- Bergman, N., Opyrchal, M., Bates, E.J., and Wilusz, J. 2002. Analysis of the products of mRNA decapping and 3′-to-5′ decay by denaturing gel electrophoresis. RNA 8: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal, T. and Gleason, K.S. 2003. Caenorhabditis elegans operons: Form and function. Nat. Rev. Genet. 4: 110–118. [DOI] [PubMed] [Google Scholar]
- Brewer, G. and Ross, J. 1990. Messenger RNA turnover in cell-free extracts. Methods Enzymol. 181: 202–209. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464. [DOI] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., and Seraphin, B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Ekiel, I., Tahara, S.M., Seliger, L.S., and Shatkin, A.J. 1985. Chemical synthesis and characterization of 7-methyl-guanosine cap analogs. Biochemistry 24: 1701–1707. [Google Scholar]
- Decker, C.J. and Parker, R. 2002. mRNA decay enzymes: Decappers conserved between yeast and mammals. Proc. Natl. Acad. Sci. 99: 12512–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker, J.A., Maroney, P.A., Yu, Y.T., Kanost, R.A., and Nilsen, T.W. 1996. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA 2: 746–755. [PMC free article] [PubMed] [Google Scholar]
- Denker, J.A., Zuckerman, D.M., Maroney, P.A., and Nilsen, T.W. 2002. New components of the spliced leader RNP required for nematode trans-splicing. Nature 417: 667–670. [DOI] [PubMed] [Google Scholar]
- Dunckley, T. and Parker, R. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18: 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, K.C., Heid, P.J., and Rothman, J.H. 1996. The SL1 trans-spliced leader RNA performs an essential embryonic function in Caenorhabditis elegans that can also be supplied by SL2 RNA. Genes & Dev. 10: 1543–1556. [DOI] [PubMed] [Google Scholar]
- Gao, M., Wilusz, C.J., Peltz, S.W., and Wilusz, J. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20: 1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, T., Peterson, B., Tomasevic, N., and Peculis, B.A. 2004. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol. Cell 13: 817–828. [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Gu, M., Fabrega, C., Liu, S.-W., Liu, H., Kiledjian, M., and Lima, C.D. 2004. Insights into the structure, mechanism and regulation of scavenger decapping activity. Mol. Cell 14: 67–80. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J., Maroney, P.A., Ayers, D.G., Shambaugh, J.D., and Nilsen, T.W. 1990a. Transcription of a nematode trans-spliced leader RNA requires internal elements for both initiation and 3′ end-formation. EMBO J. 9: 1915–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon, G.J., Maroney, P.A., Denker, J.A., and Nilsen, T.W. 1990b. Trans splicing of nematode pre-messenger RNA in vitro. Cell 61: 1247–1255. [DOI] [PubMed] [Google Scholar]
- He, F., Li, X., Spatrick, P., Casillo, R., Dong, S., and Jacobson, A. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12: 1439–1452. [DOI] [PubMed] [Google Scholar]
- Huang, X.Y. and Hirsh, D. 1989. A second trans-spliced RNA leader sequence in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. 86: 8640–8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde, E., Lewis, J., McGuigan, C., Jankowska, M., Darzynkiewicz, E., and Mattaj, I.W. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E., Lewis, J., Gamberi, C., Jarmolowski, A., McGuigan, C., and Mattaj, I.W. 1995. A cap-binding protein complex mediating U snRNA export. Nature 376: 709–712. [DOI] [PubMed] [Google Scholar]
- Jankowska, M., Temeriusz, A., Stolarski, R., and Darzynkiewicz, E. 1993. Synthesis of m2,7GTP- and m2,2,7GTP-Sepharose 4B: New affinity resins for isolation of cap binding proteins. Collect. Czech. Chem. Commun. 58: 132–137. [Google Scholar]
- Jankowska, M., Stepinski, J., Stolarski, R., Wieczorek, Z., Temeriusz, A., Haber, D., and Darzynkiewicz, E. 1996. 1H NMR and fluorescence studies of new mRNA 5′-cap analogues. Collect. Czech. Chem. Commun. 61: S197–SS202. [Google Scholar]
- Jankowska-Anyszka, M., Lamphear, B.J., Aamodt, E.J., Harrington, T., Darzynkiewicz, E., Stolarski, R., and Rhoads, R.E. 1998. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and tri-methylated mRNA cap structures. J. Biol. Chem. 273: 10538–10542. [DOI] [PubMed] [Google Scholar]
- Keiper, B.D., Lamphear, B.J., Deshpande, A.M., Jankowska-Anyszka, M., Aamodt, E.J., Blumenthal, T., and Rhoads, R.E. 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 275: 10590–10596. [DOI] [PubMed] [Google Scholar]
- Kwasnicka, D.A., Krakowiak, A., Thacker, C., Brenner, C., and Vincent, S.R. 2003. Coordinate expression of NADPH-dependent flavin reductase, Fre-1, and Hint-related 7meGMP-directed hydrolase, DCS-1. J. Biol. Chem. 278: 39051–39058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur, T.E. and Parker, R. 1998. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, F., Li, X., and Maquat, L.E. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12: 675–687. [DOI] [PubMed] [Google Scholar]
- Liou, R.F. and Blumenthal, T. 1990. Trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol. Cell. Biol. 10: 1764–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Huang, T., MacMorris, M., and Blumenthal, T. 2001. Interplay between AAUAAA and the trans-splice site in processing of a Caenorhabditis elegans operon pre-mRNA. RNA 7: 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Rodgers, N.D., Jiao, X., and Kiledjian, M. 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 21: 4699–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager, W.H., Klootwijk, J., and Klein, I. 1976. Minimal methylation of yeast messenger RNA. Mol. Biol. Rep. 3: 9–17. [DOI] [PubMed] [Google Scholar]
- Maroney, P.A., Hannon, G.J., Denker, J.A., and Nilsen, T.W. 1990a. The nematode spliced leader RNA participates in trans-splicing as an Sm snRNP. EMBO J. 9: 3667–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney, P.A., Hannon, G.J., and Nilsen, T.W. 1990b. Transcription and cap trimethylation of a nematode spliced leader RNA in a cell-free system. Proc. Natl. Acad. Sci. 87: 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney, P.A., Denker, J.A., Darzynkiewicz, E., Laneve, R., and Nilsen, T.W. 1995. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: A role for spliced leader addition in translational efficiency. RNA 1: 714–723. [PMC free article] [PubMed] [Google Scholar]
- Milone, J., Wilusz, J., and Bellofatto, V. 2002. Identification of mRNA decapping activities and an ARE-regulated 3′ to 5′ exonuclease activity in trypanosome extracts. Nucleic Acids Res. 30: 4040–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2004. Characterization of deadenylation in trypanosome extracts and its inhibition by poly(A)-binding protein Pab1p. RNA 10: 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, H., Dwyer, D.S., Keiper, B.D., Jankowska-Anyszka, M., Darzynkiewicz, E., and Rhoads, R.E. 2002. Discrimination between mono- and trimethylated cap structures by two isoforms of Caenorhabditis elegans eIF4E. EMBO J. 21: 4680–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, D., Gao, M., O’Connor, J.P., Raijmakers, R., Pruijn, G., Lutz, C.S., and Wilusz, J. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury, S. and Woollard, A. 2004. The 5′-3′ exoribonuclease xrn-1 is essential for ventral epithelial enclosure during C. elegans embryogenesis. RNA 10: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen, T.W. 2001. Evolutionary origin of SL-addition trans-splicing: Still an enigma. Trends Genet. 17: 678–680. [DOI] [PubMed] [Google Scholar]
- Nuss, D.L. and Furuichi, Y. 1977. Characterization of the m7G(5′)pppN-pyrophosphatase activity from HeLa cells. J. Biol. Chem. 252: 2815–2821. [PubMed] [Google Scholar]
- Nuss, D.L., Furuichi, Y., Koch, G., and Shatkin, A.J. 1975. Detection in HeLa cell extracts of a 7-methyl guanosine specific enzyme activity that cleaves m7GpppNm. Cell 6: 21–27. [DOI] [PubMed] [Google Scholar]
- Parker, R. and Song, H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11: 121–127. [DOI] [PubMed] [Google Scholar]
- Piccirillo, C., Khanna, R. and Kiledjian, M. 2003. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA 9: 1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J. 1999. Assays for analyzing exonucleases in vitro. Methods 17: 52–59. [DOI] [PubMed] [Google Scholar]
- Salehi, Z., Geffers, L., Vilela, C., Birkenhager, R., Ptushkina, M., Berthelot, K., Ferro, M., Gaskell, S., Hagan, I., Stapley, B., et al. 2002. A nuclear protein in Schizosaccharomyces pombe with homology to the human tumour suppressor Fhit has decapping activity. Mol. Microbiol. 46: 49–62. [DOI] [PubMed] [Google Scholar]
- Seidl, C. and Moritz, K.B. 1998. A novel UV-damaged DNA binding protein emerges during the chromatin-eliminating cleavage period in Ascaris suum. Nucleic Acids Res. 26: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth, J., Brooke, G., Kuersten, S., Lea, K., and Blumenthal, T. 1993. Operons in C. elegans: Polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell 73: 521–532. [DOI] [PubMed] [Google Scholar]
- Sripati, C.E., Groner, Y., and Warner, J.R. 1976. Methylated, blocked 5′ termini of yeast mRNA. J. Biol. Chem. 251: 2898–2904. [PubMed] [Google Scholar]
- Steiger, M., Carr-Schmid, A., Schwartz, D.C., Kiledjian, M., and Parker, R. 2003. Analysis of recombinant yeast decapping enzyme. RNA 9: 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepinski, J., Bretner, M., Jankowska, M., Felczak, K., Stolarski, R., Wieczorek, Z., Cai, A.-L., Rhoads, R.E., Temeriusz, A., Haber, D., et al. 1995. Synthesis and properties of P1,P2-, P1,P3- and P1,P4-dinucleoside di-, tri- and tetraphosphate mRNA 5′-cap analogues. Nucleosides Nucleotides 14: 717–721. [Google Scholar]
- Stepinski, J., Waddell, C., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 2001. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA 7: 1486–1495. [PMC free article] [PubMed] [Google Scholar]
- Tucker, M. and Parker, R. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69: 571–595. [DOI] [PubMed] [Google Scholar]
- van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., and Seraphin, B. 2002. Human Dcp2: A catalytically active mRNA de-capping enzyme located in specific cytoplasmic structures. EMBO J. 21: 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk, E., Le Hir, H., and Seraphin, B. 2003. DcpS can act in the 5′-3′ mRNA decay pathway in addition to the 3′-5′ pathway. Proc. Natl. Acad. Sci. 100: 12081–12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren, K. and Hirsh, D. 1990. mRNAs that mature through trans-splicing in Caenorhabditis elegans have a trimethylguanosine cap at their 5′ termini. Mol. Cell. Biol. 10: 1769–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Der Haar, T., Gross, J.D., Wagner, G., and McCarthy, J.E. 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 11: 503–511. [DOI] [PubMed] [Google Scholar]
- Wang, Z. and Kiledjian, M. 2000. Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. EMBO J. 19: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751–762. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Jiao, X., Carr-Schmid, A., and Kiledjian, M. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. 99: 12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C., Xu, L., and Blumenthal, T. 1999. SL1 trans splicing and 3′-end formation in a novel class of Caenorhabditis elegans operon. Mol. Cell. Biol. 19: 376–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Williams, C.J., Wormington, M., Stevens, A., and Peltz, S.W. 1999. Monitoring mRNA decapping activity. Methods 17: 46–51. [DOI] [PubMed] [Google Scholar]
- Zorio, D.A., Cheng, N.N., Blumenthal, T., and Spieth, J. 1994. Operons as a common form of chromosomal organization in C. elegans. Nature 372: 270–272. [DOI] [PubMed] [Google Scholar]