Abstract
The Puf family of RNA-binding proteins regulates mRNA translation and decay via interactions with 3′ untranslated regions (3′ UTRs) of target mRNAs. In yeast, Puf3p binds the 3′ UTR of COX17 mRNA and promotes rapid deadenylation and decay. We have investigated the sequences required for Puf3p recruitment to this 3′ UTR and have identified two separate binding sites. These sites are specific for Puf3p, as they cannot bind another Puf protein, Puf5p. Both sites use a conserved UGUANAUA sequence, whereas one site contains additional sequences that enhance binding affinity. In vivo, presence of either site partially stimulates COX17 mRNA decay, but full decay regulation requires the presence of both sites. No other sequences outside the 3′ UTR are required to mediate this decay regulation. The Puf repeat domain of Puf3p is sufficient not only for in vitro binding to the 3′ UTR, but also in vivo stimulation of COX17 mRNA decay. These experiments indicate that the essential residues involved in mRNA decay regulation are wholly contained within this RNA-binding domain.
Keywords: decay, turnover, mRNA, Puf, yeast, 3′ UTR
INTRODUCTION
Post-trantranscriptional control is essential for proper regulation of gene expression. In particular, regulation of translation and mRNA degradation play important roles in somatic cells, in germline sex determination, and during early development (for reviews, see Gray and Wickens 1998; Guhaniyogi and Brewer 2001). This control is often mediated by regulatory elements located in the 3′ untranslated regions (UTRs) of mRNAs (for reviews, see Grzybowska et al. 2001; Mazumder et al. 2003). Proteins that sequence-specifically bind these regulatory elements can either enhance or inhibit translation and/or decay of the bound mRNAs (for review, see Derrigo et al. 2000). However, many questions still remain regarding the specific mechanisms by which such RNA-binding proteins regulate mRNA metabolism.
Members of the Puf family of RNA-binding proteins regulate both translation and decay in diverse eukaryotes (for review, see Wickens et al. 2002). Pumilio from Drosophila melanogaster promotes abdominal segmentation in the early embryo by binding the 3′ UTR of hunchback mRNA and subsequently repressing its translation (Murata and Wharton 1995) and promoting its deadenylation (Wreden et al. 1997). Pumilio also represses translation of cyclin B mRNA for regulation of germline stem cell development (Forbes and Lehmann 1998; Asaoka-Taguchi et al. 1999; Parisi and Lin 1999), and plays a role in anterior patterning (Gamberi et al. 2002). The FBF proteins of Caenorhabditis elegans promote the sperm/oocyte switch by binding the 3′ UTR of fem-3 mRNA and repressing its expression (Zhang et al. 1997), and control germline stem cell maintenance by repressing gld-1 mRNA expression (Crittenden et al. 2002). In Dictyostelium, PufA inhibits development of fruiting bodies by repressing pkaC mRNA expression (Souza et al. 1999), and Puf1 is specifically expressed in undifferentiated spores of Saprolegnia parasitica (Andersson and Cerenius 2002). In Saccharomyces cerevisiae, Puf proteins appear to have a significant role in regulation of mRNA decay. Puf3p binds the 3′ UTR of COX17 mRNA and promotes its de-adenylation and subsequent decay (Olivas and Parker 2000), whereas Puf5p binds the 3′ UTR of the HO mRNA and stimulates its decay (Tadauchi et al. 2001). Regulation of RNA expression by Pumilio and FBF requires recruitment of additional proteins to the RNA (Kraemer et al. 1999; Sonoda and Wharton 1999, 2001), but Puf protein partners have yet to be identified in unicellular eukaryotes.
The Puf family is characterized by a conserved RNA-binding domain consisting of eight imperfect repeats of a 36-amino-acid sequence plus short flanking regions. Structural analyses of the Puf repeat domain from both Pumilio (Edwards et al. 2001) and human PUM1 (Wang et al. 2001) show that each repeat sequence folds into three α-helices, with the repeats aligning to form an extended rainbow shape. A crystal structure of the human PUM1 repeat domain bound to a minimal Drosophila Nanos response element (NRE) sequence revealed a modular binding method, where conserved amino acids within each repeat contact and stack with successive bases along the RNA (Wang et al. 2002). Sequences within the Pumilio repeat domain are also sufficient for translational repression in vivo (Wharton et al. 1998).
All Puf proteins studied bind to RNA targets with a conserved UGU sequence, with specificity conferred by flanking sequences. For example, human PUM1 (Zamore et al. 1997; Wang et al. 2002), murine PUM2 (White et al. 2001), and Xenopus Pum (Nakahata et al. 2001) proteins bind to sequences containing UGUANAUA, which is also found in box B of the bipartite NRE target of DmPUM (Murata and Wharton 1995). In contrast, C. elegans FBF binds a target sequence containing UCUUGUGU (Zhang et al. 1997), whereas yeast Puf5 binds a target sequence containing AGUUGUGU (Tadauchi et al. 2001), where the underlined nucleotides have been shown to be important for binding.
In the major pathway of mRNA degradation in yeast, transcripts are first deadenylated, which alters the mRNP structure to allow decapping, then the body of the mRNA is exposed for rapid 5′–3′ exonucleolytic degradation (Decker and Parker 1993; Hsu and Stevens 1993; Muhlrad et al. 1994, 1995). Different mRNAs can exhibit highly variable rates of deadenylation and decapping, often due to specific 3′-UTR elements that modulate these rates (for reviews, see Jacobson and Peltz 1996; Tucker and Parker 2000). In this work we have identified the 3′-UTR sequences that specifically recruit Puf3p to the COX17 mRNA and show that two separate binding sites can individually bind Puf3p. The two sites contain similar but distinct sequences that yield different binding affinities toward Puf3p in vitro. In contrast, each individual site promotes equal stimulation of COX17 mRNA decay in vivo, and both sites are required for wild-type stimulation of deadenylation and decay rates. This indicates that multiple Puf3p signals can combine to increase activity more than individual Puf3p signals alone. We also demonstrate that the repeat domain of Puf3p is sufficient for both binding to the COX17 mRNA and signaling to the decay machinery, supporting a conserved role of the Puf repeat domain as an independent regulator of mRNA metabolism.
RESULTS
The repeat domain of Puf3p is sufficient and specific for binding COX17 mRNA
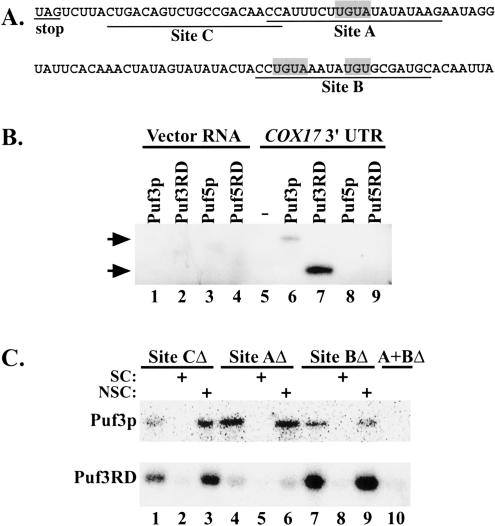
Our previous work identified yeast COX17 mRNA as a target of Puf protein regulation. We found that Puf3p directly binds the 3′ UTR of COX17 mRNA and promotes rapid deadenylation and decay of this transcript, whereas deletions of the other four Puf genes in yeast have no effect on COX17 mRNA decay in vivo (Olivas and Parker 2000). In this work, we wished to determine how the specificity of binding and regulation of COX17 mRNA is attained by Puf3p. First, we wanted to verify that binding of the COX17 3′ UTR by Puf3p is mediated by its Puf repeat domain, as has been shown in other organisms (Zamore et al. 1997; Zhang et al. 1997). For this experiment, we assayed binding in vitro using glutathione S-transferase (GST)-tagged proteins purified from Escherichia coli encompassing either the full-length Puf3 sequence or only the Puf3 repeat domain (Puf3RD). These purified proteins were then incubated with in vitro transcribed, uniformly radiolabeled RNA of the COX17 3′-UTR sequence (Fig. 1A ▶) or a nonspecific vector RNA sequence. The reactions were UV cross-linked to attach the radiolabel of any bound RNA to the protein, then treated with RNase T1 to degrade unbound RNA. As shown in Figure 1B ▶, both full-length Puf3 protein and Puf3RD become radiolabeled when incubated with the COX17 3′ UTR (lanes 6,7) but not with the nonspecific RNA (lanes 1,2). Puf3RD is therefore sufficient for specific binding to COX17 mRNA. The difference in band intensities between the bound Puf3 and Puf3RD lanes reflects different amounts of protein used in the assay, as we had difficulty purifying stable, full-length protein.
FIGURE 1.
Puf3p and Puf3RD bind to specific sites in the COX17 3′UTR. (A) The COX17 3′-UTR sequence used in binding reactions is shown. The shaded boxes highlight three UGU(A) sequences in the 3′ UTR. DNA oligos complementary to the underlined Sites A, B, and/or C were used in RNase H reactions to delete the corresponding sites from the 3′ UTR. (B) In vitro binding reactions of uniformly radio-labeled transcripts (Vector or COX17 3′ UTR) in the presence or absence of GST-tagged proteins (Puf3p, Puf3RD, Puf5p, or Puf5RD) were UV cross-linked and digested of unbound RNA. Radiolabel that remains bound to the protein represents a direct interaction between the RNA and the Puf protein. Shown is an SDS-polyacrylamide gel of radiolabeled Puf proteins in digested binding reactions. Positions of full-length Puf3p (124 kDa) and Puf3RD (74 kDa) based on Western analysis are shown by the arrows. Sizes of Puf5p (121 kDa) and Puf5RD (81.5 kDa) have been verified by Western analysis. (C) In vitro binding reactions of radiolabeled COX17 3′ UTR deleted of Site C (lanes 1–3), Site A (lanes 4–6), Site B (lanes 7–9), or Site A and B (lane 10) in the presence of Puf3p (top panel) or Puf3RD (bottom panel) were UV cross-linked, digested of unbound RNA, and electrophoresed as described in A. Excess unlabeled full-length COX17 3′ UTR or Vector RNA were used as specific (SC, lanes 2,5,8) or non-specific (NSC, lanes 3,6,9) competitors, respectively.
We next tested whether other yeast Puf proteins can also bind the COX17 3′ UTR. Because we have previously shown that deletions of PUF1, PUF2, PUF4, and PUF5 do not affect COX17 mRNA decay (Olivas and Parker 2000), the simplest model is that only Puf3p is able to bind and subsequently regulate the mRNA. Alternatively, other Puf proteins might also bind to the target sequence, with only Puf3p correctly interacting or signaling to stimulate deadenylation and decay. To test these alternatives, we purified GST-Puf5p and GST-Puf5 repeat domain (Puf5RD) proteins from E. coli and repeated the binding assays with the COX17 3′ UTR. As shown in Figure 1B ▶, neither Puf5p nor Puf5RD is able to bind the COX17 3′-UTR sequence (lanes 8,9). Our Puf5p and Puf5RD protein preparations were able to bind HO 3′-UTR sequences in binding assays, as has been shown previously (Tadauchi et al. 2001), verifying that our proteins were active (data not shown). Thus, the inability of Puf5p to bind the COX17 3′ UTR demonstrates that not all Puf proteins can bind to the same target sequence, and suggests that specificity of mRNA regulation can derive from preferential Puf binding.
Puf3p binds to either of two UGU-containing regions of the COX17 3′ UTR
The COX17 3′ UTR contains three UGU triplets, two of which are part of UGUA sequences (shaded boxes in Fig. 1A ▶). To determine whether one or more of these UGU triplets is involved in binding Puf3p, we incubated Puf3p or Puf3RD in vitro with uniformly radiolabeled RNA sequences encompassing different regions of the COX17 3′ UTR. We created the various RNA sequence regions by hybridizing in vitro transcribed RNA of the complete COX17 3′ UTR to oligos complementary to Sites A, B, or C, then treated with RNaseH to delete the respective RNA sequences from the 3′ UTR (Fig. 1A ▶). The remaining undigested RNA fragments were used in binding reactions as described above.
Interestingly, both Puf3p and Puf3RD are able to bind to the COX17 3′-UTR RNA and become radiolabeled even when any single site is deleted (Fig. 1C ▶, lanes 1,4,7). This binding is specific, because the interactions can be competed with excess unlabeled COX17 3′-UTR RNA (Fig. 1C ▶, lanes 2,5,8), but not with excess unlabeled nonspecific vector RNA (lanes 3,6,9). Because Site A contains one UGUA sequence, whereas Site B contains both a UGUA and a UGU sequence, this demonstrates that no single UGU(A) sequence is absolutely required for Puf3p binding. However, neither Puf3p nor Puf3RD is able to bind RNA that is missing both Sites A and B (Fig. 1C ▶, lane 10). Together, these results suggest that Puf3p binding requires at least one UGU-containing region, but either one is sufficient.
Puf3p binding requires UGUA as well as specific surrounding sequences
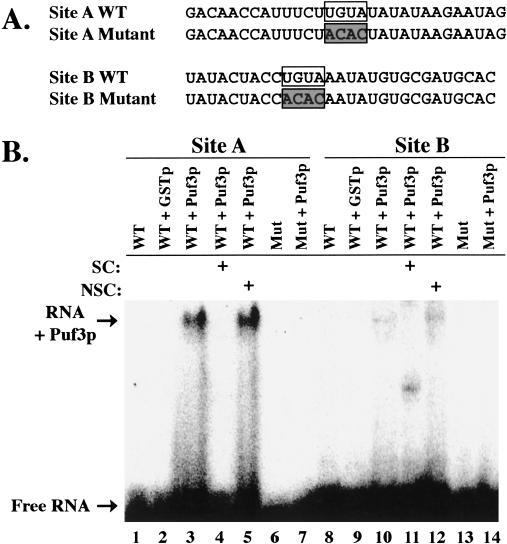
We next wished to verify that the binding of Puf3p to the COX17 3′ UTR required the UGUA sequences located within each of the two sites, A and B. Therefore, we transcribed and labeled small (29–30 nt) RNAs encompassing the wild-type sequences of either site or mutant sequences in which each UGUA had been changed to ACAC (Fig. 2A ▶). After in vitro binding of purified full-length Puf3p to these RNAs, the resulting complexes were analyzed by gel mobility shift experiments. As shown in Figure 2B ▶, Puf3p is able to bind wild-type RNA sequences of both Sites A and B (lanes 3 and 10, respectively). Binding occurs through Puf3p and not simply the GST tag, as there is no binding to GST protein alone (Fig. 2B ▶, lanes 2,9). Moreover, interactions with wild-type sequences are specific, as the complexes can be competed with excess unlabeled wild-type COX17 3′-UTR RNA (Fig. 2B ▶, lanes 4,11), but not with excess unlabeled nonspecific vector RNA (Fig. 2B ▶, lanes 5,12). These minimal RNA sequences are thus sufficient for specific complex formation. In contrast, binding of Puf3p to either mutant sequence lacking UGUA is undetectable (Fig. 2B ▶, lanes 7,14). Therefore, a UGUA target sequence is required for Puf3p binding to the COX17 3′ UTR.
FIGURE 2.
Puf3p binding to minimal Sites A and B requires UGUA. (A) Sequences of the 29–30-nt transcripts of wild-type (WT) and mutant Site A and Site B used in binding reactions are shown. UGUA regions are boxed. Mutant transcripts contain ACAC (shaded boxes) in place of UGUA. (B) In vitro binding reactions of radiolabeled RNA (Site A WT, lanes 1–5; Site A mutant, lanes 6,7; Site B WT, lanes 8–12; Site B mutant, lanes 13,14) in the presence or absence of 20 μM GST (lanes 2,9) or 0.14 μM GST-Puf3p (lanes 3–5,7,10–12,14) were separated on a native polyacrylamide gel. Excess unlabeled full-length COX17 3′ UTR or Vector RNA were used as specific (SC, lanes 4,11) or nonspecific (NSC, lanes 5,12) competitors, respectively. Positions of free radiolabeled RNA (Free RNA) and RNA bound to Puf3p (RNA + Puf3p) are indicated.
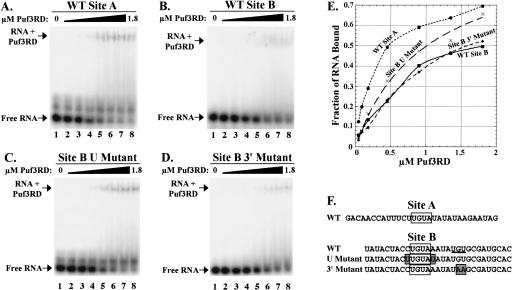
Although Puf3p binds specifically to either Site A or Site B in the COX17 3′ UTR, it is apparent from Figure 2 ▶ that Puf3p binds more strongly to Site A than to Site B (cf. lanes 3 and 10). To get an estimate of the affinity of Puf3p for these target sites, we incubated increasing concentrations of protein (45 nM to 1.8 μM) with 200 pM of radiolabeled Site A or Site B RNA, then analyzed the complexes by gel mobility experiments. For these and subsequent binding experiments, we used our purified Puf3RD protein, because we had already shown it was sufficient for specific binding and because it was much easier to purify in a stable and concentrated form than the full-length protein. As shown in Figure 3A ▶, at 0.45 μM concentration of protein added (lane 5), half of the Site A RNA is shifted from free form into a complex, giving an estimated equilibrium dissociation constant (KD) of 0.45 μM (Fig. 3E ▶). In contrast, Site B shows a fourfold decrease in binding affinity, with an estimated KD of 1.8 μM (Fig. 3B,E ▶). Our KD value for the higher affinity Site A is at least fivefold weaker than that found for a GST-fusion protein of the Drosophila Pum-Homology Domain (DmPUM-HD) binding a 181-nt target RNA (25–100 nM; Zamore et al. 1997). Part of the reason for this difference in affinity is likely the low specific activity of our protein. By performing a binding experiment using increasing concentrations of RNA with a fixed amount of protein, we found that binding was saturated at a concentration equal to 5%–10% of the total protein concentration (data not shown). This percent of active protein is also consistent with data we obtained from a thermodynamic simulation program, which calculates the fraction of RNA that should be bound to protein at any given [RNA], [active protein], and KD. From this simulation, an [active protein] equal to 5% of our [total protein], and a KD equal to 10% of our experimental KD replicated the fractions of RNA bound in the saturation experiment. Therefore, we estimate that our apparent KD of 0.45 μM is ~10-fold weaker than the actual KD of Puf3RD with Site A. Another issue that would affect our affinity is the small, 30-nt size of our Site A RNA target. Other studies with the DmPUM-HD have shown a fourfold decrease in affinity to a short 30-nt RNA target versus a 181-nt target, with no change in specificity (Zamore et al. 1999). Nevertheless, although our determination of Puf3RD binding affinity may be significantly underestimated from in vivo concentrations, the results of our studies comparing the relative affinities of Puf3RD toward Site A and Site B targets remain valid and suggest that different UGUA flanking sequences contribute to the binding energy.
FIGURE 3.
Differential binding affinities of Puf3RD to Sites A and B. In vitro binding reactions of radiolabeled wild-type (WT) Site A (A), WT Site B (B), Site B U Mutant (C), or Site B 3′ Mutant (D) transcripts in the absence or presence of increasing concentrations of Puf3RD were separated on native polyacrylamide gels. Concentrations of Puf3RD used in binding reactions were 0, 0.045, 0.09, 0.18, 0.45, 0.9, 1.35, and 1.8 μM in lanes 1–8, respectively. Positions of free radiolabeled RNA (Free RNA) and RNA bound to Puf3RD (RNA + Puf3RD) are indicated. (E) Data from the gel mobility shifts in A–D are plotted, with the μmolar concentration of Puf3RD used in the binding reaction on the x-axis and the fraction of RNA shifted from free form to bound form on the y-axis. Best-fit binding curves are shown for WT Site A (circle), WT Site B (square), Site B U Mutant (X), and Site B 3′ Mutant (diamond). Data points are averages of multiple experiments. (F) Sequences of wild-type (WT) and mutant Site A and Site B transcripts used in binding reactions are shown. UGUA regions are boxed. Sequences altered in the mutant transcripts are indicated by shaded boxes. The second UGU sequence in wild-type Site B is underlined.
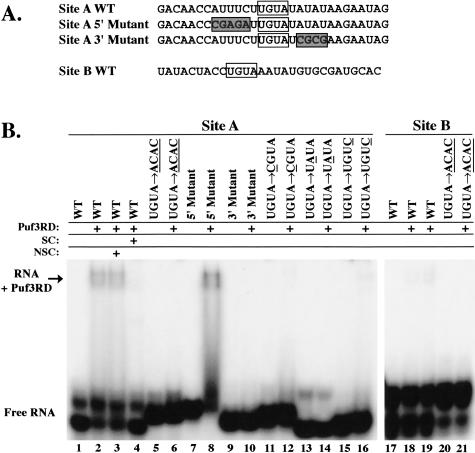
To further examine the importance of individual nucleotides within the UGUA sequence as well as the role of flanking sequences, we tested binding of Puf3RD to a series of mutant target sequences (Fig. 4A ▶). In our initial experiments to detect differences in binding affinity to these mutants, we used a single 0.45-μM concentration of Puf3RD, the concentration equal to its experimentally determined KD value with the wild-type Site A target. As expected from our analyses with the full-length Puf3p, the Puf3RD was unable to bind the Site A target sequence when the UGUA was mutated to ACAC (Fig. 4B ▶, lane 2 vs. lane 6). The interaction of Puf3RD with the wild-type target is specific, as it cannot be competed with excess unlabeled RNA of the UGUA → ACAC Site A mutant, but can be competed with excess unlabeled wild-type Site A RNA (Fig. 4B ▶, lanes 3 and 4, respectively). We also created mutant RNAs with CGUA, UAUA, and UGUC sequences in place of the wild-type UGUA of Site A (where the bold, underlined nucleotide is mutant). Puf3RD was unable to bind RNA with the UAUA mutation (Fig. 4B ▶, lane 14), whereas binding to RNAs with the CGUA and UGUC mutations was very weak, with KD values > 1.8 μM (Fig. 4B ▶, lanes 12,16; data not shown). This clearly demonstrates the significant role that each of these nucleotides plays in binding to Puf3RD.
FIGURE 4.
Puf3RD binding requires additional sequences flanking the conserved UGUA regions. (A) Sequences of wild-type (WT) and mutant Site A and Site B transcripts used in binding reactions are shown. UGUA regions are boxed. Sequences altered in the mutant transcripts are indicated by shaded boxes. (B) In vitro binding reactions of radiolabeled transcripts in the absence or presence of 0.45 μM Puf3RD were separated on a native polyacrylamide gel. Base substitutions in UGUA mutants of Site A and Site B transcripts are given, with altered bases underlined. Excess unlabeled wild-type Site A RNA was used as specific competitor (SC, lane 4), and excess unlabeled mutant Site A RNA containing a UGUA → ACAC alteration was used as nonspecific competitor (NSC, lane 3). Lane 19 contains 0.9 μM Puf3RD. Similar results were also obtained with full-length Puf3p (data not shown). Positions of free radiolabeled RNA (Free RNA) and RNA bound to Puf3RD (RNA + Puf3RD) are indicated.
To determine whether specific sequences upstream of and downstream from the UGUA were important for binding, we mutated 5 nt 5′ of the UGUA, or 4 nt 3′ of the UGUA (Fig. 4A ▶, Site A 5′ and 3′ Mutants, respectively). The 5′ mutation had no detrimental effect on binding (Fig. 4B ▶, lane 8), but replacing the 3′ AUAU with CGCG completely inhibited binding (Fig. 4B ▶, lane 10). A downstream AUA region is also involved in binding human PUM-HD in crystal structures (Wang et al. 2002), murine PUM2 in RNA selection experiments (White et al. 2001), and is present downstream from the box B region of the Drosophila NRE (Murata and Wharton 1995). Thus, an expanded recognition sequence of UGUANAUA is conserved across several Puf proteins.
Both Site A and Site B of COX17 contain the conserved UGUANAUA sequence. However, as was shown in Figure 3 ▶, Puf3RD binds to Site B with fourfold lower affinity than to Site A. Binding to Site B is nevertheless specific, as mutation of the UGUA of Site B to ACAC eliminates binding (Fig. 4B ▶, lane 21). The difference in Site A and Site B binding affinities suggests that other flanking sequences surrounding the UGUANAUA of Site A are important for efficient binding. Because our Site A 5′ Mutant had no effect on Puf3RD binding, we reasoned that such upstream sequences of Site B were also likely not critical for binding affinity. However, the nucleotides immediately flanking the UGUA in Site A (UUGUAU) are different than those in Site B (CUGUAA). To test the importance of these nucleotides, we mutated Site B to contain flanking uracil bases (Fig. 3F ▶, Site B U Mutant). Incubation of Site B U mutant RNA with increasing concentrations of Puf3RD provides a KD estimation of 1 μM (Fig. 3C,E ▶). This approximately twofold increase in binding affinity versus the wild-type Site B brings the affinity to within half of that of the wild-type Site A target, thus demonstrating that the uracils flanking the UGUA are important for achieving high affinity binding to Puf3RD. It is possible that only one of the flanking uracils is required. However, the crystal structure of human PUM-HD complexed with RNA of the Drosophila NRE box B shows that all nucleotides of a UGUAUAUA sequence interact with the protein, whereas the upstream flanking uracil forms a turn in the RNA to allow internal base stacking interactions (Wang et al. 2002). Furthermore, an upstream flanking uracil is conserved in several Puf targets, including both boxes of the Drosophila NRE (Murata and Wharton 1995), the fem-3 PME of C. elegans (Zhang et al. 1997), and the HO target of yeast Puf5p (Tadauchi et al. 2001), and a downstream flanking uracil is also found in box B of the Drosophila NRE (Murata and Wharton 1995).
COX17 Site B contains both the conserved UGUANAUA as well as a second downstream UGU sequence (underlined in Fig. 3F ▶). To test whether this second UGU is required for Puf3RD binding to Site B, we created a mutant in which the second UGU was mutated to UAA (Fig. 3F ▶, Site B 3′ Mutant). As shown in Figure 3 ▶, D and E, the Site B 3′ mutant RNA binds to Puf3RD with a KD of 1.6 μM, which is an affinity very similar to wild-type Site B RNA. This suggests that although the second UGU is not required for binding of Puf3RD to Site B, the alteration of the sequence to UAA, which mimics the downstream sequence of Site A, does not significantly enhance binding.
Puf3p regulation of COX17 mRNA in vivo requires both UGUA elements
Our results have established that the COX17 3′ UTR contains two sites that individually bind Puf3p via a UGUAN AUA sequence in vitro, and additional flanking sequences make Site A a higher affinity target than Site B. To determine whether one or both of these sites are required for Puf3p regulation of COX17 mRNA decay in vivo, we examined the decay of either wild-type or mutant versions of the mRNA in a transcriptional pulse-chase. In this experiment, COX17 is expressed from a plasmid under the control of the regulatable GAL10 promoter, such that transcription of COX17 mRNA is induced by addition of galactose to the growth medium, then rapidly repressed by addition of glucose (Decker and Parker 1993). This creates a pulse of newly synthesized transcripts whose deadenylation and subsequent decay can be monitored over time. The COX17 gene was mutagenized at Site A, Site B, or both sites by changing the UGUA sequences to ACAC.
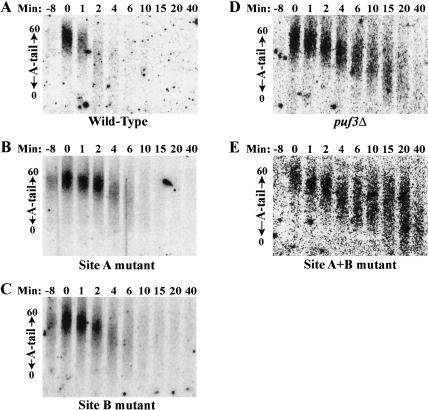
As we have reported in previous work (Olivas and Parker 2000), a pulse of wild-type COX17 transcripts expressed in a wild-type yeast strain are synthesized with a poly(A) tail of 45–60 residues (Fig. 5A ▶, lane 0). The poly(A) tails then deadenylate heterogeneously, with transcripts reaching a fully deadenylated state within 2–4 min, and all transcripts nearly completely degraded within 6 min. (Fig. 5A ▶). In contrast, a pulse of wild-type COX17 transcripts expressed in a puf3Δ strain begin with approximately the same poly(A) tail length of 35–60 residues (Fig. 5D ▶, lane 0), but then deadenylate at a much slower rate such that transcripts do not become fully deadenylated until 15 min. Moreover, transcripts with short poly(A) tails persist to 40 min, indicative of a role of Puf3p in stimulating terminal deadenylation and/or decapping (Fig. 5D ▶).
FIGURE 5.
Both UGUA regions are required for in vivo regulation of COX17 mRNA deadenylation and decay by Puf3p. Shown are Northern blot analyses of transcriptional pulse-chase experiments examining decay of the COX17 transcript from wild-type PUF3 (A); wild-type PUF3, Site A mutant (B); wild-type PUF3, Site B mutant (C); puf3Δ (D), and wild-type PUF3, Site A + B mutant (E) strains. Minutes after transcriptional repression are indicated above each blot. The -8 lane in each blot corresponds to background levels of RNA expression prior to galactose induction of the COX17 transcript. The distribution of poly(A) tail lengths are indicated by arrows on the left of each blot, from a maximum length of 60 A’s (top) to a fully deadenylated length of 0 A’s (bottom).
We next analyzed our mutant COX17 mRNAs in the wild-type PUF3 yeast strain. It was predicted that mutation of Site A, with its higher affinity for Puf3RD, might have a larger effect on decay than mutation of Site B. To our surprise, mutation of either Site A or Site B had an equal, yet intermediate effect on decay (Fig. 5B,C ▶). For each individual mutant, the pulse of COX17 transcripts did not become fully deadenylated until 6 min, and persisted to 10 min. By comparison, decay of COX17 mRNA with both sites mutated (Fig. 5E ▶) looked identical to the decay of wild-type COX17 mRNA in the puf3Δ strain (Fig. 5D ▶), with fully deadenylated species attained only after 15 min, and transcripts with short poly(A) tails persisting to 40 min. It appears that decay of each COX17 single mutant in the first 2 min progresses at a slow rate of deadenylation, similar to that seen in the COX17 double mutant and the puf3Δ. Then, in contrast to the COX17 double mutant and puf3Δ, deadenylation in the single mutants proceeds more rapidly between 2 and 6 min, and there is no buildup of transcripts with short poly(A) tails. This suggests that binding of a single Puf3p to the COX17 3′ UTR is sufficient to promote rapid terminal deadenylation and/or decapping, but may be less efficient in the initial phase of deadenylation than the binding of two Puf3 proteins.
Together, these results demonstrate that regulation of COX17 mRNA decay by Puf3p requires binding of the protein to the UGUA sequences in the transcript’s 3′ UTR. Moreover, binding of Puf3p to either site alone equally enhances deadenylation and decay of the COX17 transcript versus no Puf3p recruitment to the 3′ UTR, but full regulation of COX17 decay requires binding to both sites. This is similar to what is seen in Drosophila, where the hunchback mRNA contains two Pumilio binding sites that are both required for full translational repression (Wharton and Struhl 1991; Curtis et al. 1997).
The COX17 3′ UTR is sufficient for Puf3p regulation
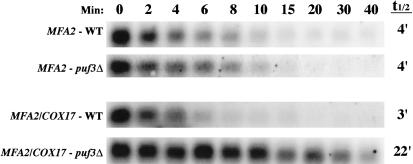
We have shown that Puf3p binds to two UGUA elements in the 3′ UTR of COX17 in vitro, and both elements are required for Puf3p regulation of mRNA decay in vivo. The simplest interpretation is that these 3′-UTR elements are sufficient for directing Puf3p-mediated decay regulation of the COX17 mRNA. However, Puf3p regulation of decay could require additional COX17 sequences outside the 3′ UTR. To test these models, we examined whether the COX17 3′ UTR could direct Puf3p decay regulation when attached to another RNA that is not normally regulated by Puf proteins. We replaced the 3′ UTR of the plasmid-expressed MFA2 gene with the 3′ UTR of COX17, creating an MFA2/COX17 hybrid construct. The wild-type MFA2 construct and hybrid MFA2/COX17 construct were then transformed into a cox17Δ strain containing a temperature-sensitive lesion in RNA polymerase II (rpb1-1; Herrick et al. 1990) and either wild-type PUF3 or a puf3Δ. A transcriptional shutoff was performed by expressing the MFA2 or MFA2/COX17 mRNAs to steady-state levels under the control of the regulatable GAL upstream activating sequence; then transcription was rapidly repressed by simultaneous addition of glucose and a shift to high temperature. As expected, wild-type MFA2 mRNA is not regulated by Puf3p, decaying with a half-life of 4 min in both the wild-type PUF3 and puf3Δ strains (Fig. 6 ▶). In contrast, the MFA2/ COX17 mRNA is under the control of Puf3p regulation, decaying with a short half-life of 3 min in the wild-type PUF3 strain, but stabilized approximately sevenfold in the puf3Δ strain to a half-life of 22 min (Fig. 6 ▶). These half-lives of the MFA2/COX17 hybrid mRNA in the wild-type and puf3Δ strains are identical to those seen for endogenous COX17 mRNA in the same strains. Therefore, these observations indicate that the COX17 3′ UTR is sufficient to direct full regulation of decay by Puf3p.
FIGURE 6.
The COX17 3′ UTR is sufficient to promote Puf3p regulation of mRNA decay. Shown are Northern blot analyses of the decay of MFA2 mRNA or the hybrid MFA2/COX17 mRNA expressed from a wild-type (WT) strain or a puf3Δ strain. Minutes following transcriptional repression are indicated above the set of blots, with the half-lives (t1/2) as determined from multiple experiments.
The Puf3 repeat domain is sufficient for COX17 decay regulation
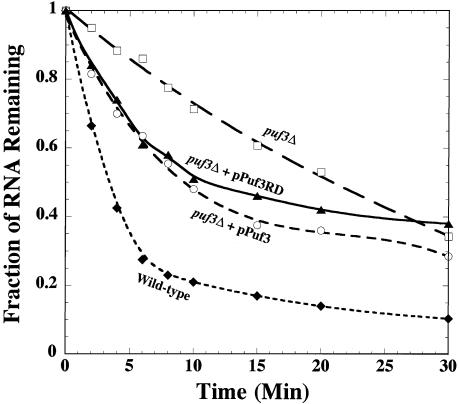
The repeat domains of Puf proteins typically compose less than one-half of the total protein. For example, the repeat domain of Puf3p occupies only one-third of the protein, and the repeat domain of Pumilio occupies only one-fourth of the protein. Yet expression of just the repeat domain of Pumilio is sufficient for nearly complete rescue of Pumilio’s activity of translational repression of hunchback mRNA in vivo in a pum deletion background (Wharton et al. 1998). It is possible that Puf3p is similar to Pumilio in acting solely through its repeat domain, or it may stimulate decay by a different mechanism that requires additional protein sequences outside of its repeat domain. To test these possibilities, we transformed yeast lacking endogenous PUF3 with plasmids expressing either full-length Puf3p or just the Puf3RD. The decay rates of COX17 mRNA in the wild-type, puf3Δ, and transformed puf3Δ strains were then compared by using the rpb1-1 lesion to shut off transcription following a shift to high temperature. As shown in Figure 7 ▶ and in previous work (Olivas and Parker 2000), COX17 mRNA in wild-type PUF3 cells decays with a half-life of ~3 min, whereas in the puf3Δ strain the half-life is dramatically increased to 22 min. In comparison, expression of either the full-length Puf3p or the Puf3RD in the puf3Δ strain rescues the rapid decay of COX17, with half-lives of 9 min and 11 min, respectively (Fig. 7 ▶). It is unclear why even full-length Puf3p does not completely rescue the COX17 decay rate to wild-type levels, but it might be due to the different level of expression achieved from the high-copy 2μm plasmid versus endogenous Puf3p levels. On the plasmid, PUF3 expression is under the control of the high-level, constitutive GPD promoter. Nonetheless, the important finding is that expression of Puf3RD rescues decay of the COX17 mRNA to nearly the same level as full-length Puf3p, indicating that the repeat domain contains the minimal elements necessary for both binding to the mRNA as well as signaling for rapid decay.
FIGURE 7.
The Puf3RD rescues decay of COX17 mRNA in a puf3Δ strain. Data from Northern blot analyses of COX17 decay are plotted, with minutes following transcriptional repression on the x-axis and the fraction of RNA remaining as compared to the steady-state RNA level at time 0 on the y-axis. Decay was monitored in the following strains: wild-type (closed diamond), puf3Δ (open square), puf3Δ transformed with a plasmid expressing Puf3p (open circle), and puf3Δ transformed with plasmid expressing Puf3RD (closed triangle). Data points are averages of multiple experiments.
DISCUSSION
Two target sequences in the COX17 3′ UTR specifically recruit Puf3p for rapid mRNA decay
Puf proteins play important roles in regulating mRNA metabolism in eukaryotes. In yeast, Puf3p promotes deadenylation and degradation of COX17 mRNA. In this report we provide several lines of evidence that the COX17 3′ UTR contains two distinct binding sites for recruiting Puf3p and accelerating mRNA decay. First, deletion of both sites from the COX17 3′ UTR abolishes binding of Puf3p in vitro, whereas deletion of either single site retains 3′-UTR binding activity (Fig. 1 ▶). Second, short RNAs encompassing either individual site are sufficient for binding Puf3p in vitro (Fig. 2 ▶). Third, mutation of either single site slows deadenylation and decay of the COX17 mRNA in vivo, and mutation of both sites further slows deadenylation and decay to the same extent as a puf3Δ (Fig. 5 ▶). Finally, no other regions outside the COX17 3′ UTR are required for Puf-mediated mRNA decay, and attachment of the COX17 3′ UTR to another mRNA causes decay to occur with the same Puf3p-dependent rates as COX17 mRNA itself (Fig. 6 ▶).
We hypothesize that each binding site within the COX17 3′ UTR recruits a Puf3 protein, which can individually stimulate decay. The two sites in the COX17 3′ UTR thus would allow two Puf3 proteins to be recruited for increased stimulation of decay. A similar situation is seen in Drosophila, where the hunchback mRNA contains two NRE-binding sequences that are both required for full translational repression, although one site contributes more activity than the other site (Wharton and Struhl 1991; Curtis et al. 1997). In addition, a single Puf repeat domain of Pumilio is known to bind a single NRE target sequence (Zamore et al. 1999). Interestingly, the distance between the two NRE sequences in the hunchback mRNA, 45 nt from center to center, is very similar to the 41 nt between the two COX17 binding sites, although the significance of this distance is unclear. RNA footprinting studies of Pumilio binding the NRE show protection of ~30 nt over the NRE (Wharton et al. 1998), suggesting that two Puf proteins could bind side by side to target sequences located ~40–45 nt apart. Other studies have shown that the in vitro binding affinity of Pumilio to RNAs containing two NREs is no greater than binding to a single NRE, indicating independent binding to each site (Zamore et al. 1999), although cooperativity of binding and/or Puf function in vivo is not known. Nonetheless, these results suggest that the mechanism(s) by which Puf proteins both repress translation and promote decay allows for individual Pufs to function independently, or for multiple Pufs to act in combination.
Several proposals have been suggested as to how Puf proteins might function (Wickens et al. 2002). In one model, Puf proteins may directly, or through intermediates, interact with the decay machinery to enhance its activity on the target mRNA. Because Puf proteins can repress translation of nonpolyadenylated mRNAs (Chagnovich and Lehmann 2001), this model would have to include a mechanism for interacting with the translation machinery as well. In a second model, Puf proteins may perturb the mRNP structure, perhaps by disrupting other RNA-binding proteins, so as to produce global changes in the mRNA that would affect both translation and decay. Though there is no direct evidence for either model, it is easier to imagine how multiple Puf proteins bound to an mRNA could combine to disrupt mRNP structure than for multiple Puf proteins to all interact with the decay and translation machinery.
We also provide evidence that the COX17 3′-UTR binding sites preferentially bind the Puf3 protein versus another yeast Puf protein, Puf5p (Fig. 1 ▶). This observation supports a model in which the Puf proteins in yeast target different mRNAs through distinct binding specificities of each Puf. The two Puf3p binding sites in COX17 both contain a UGUANAUA sequence, which is also conserved in the binding sites of Pumilio, human PUM1, murine PUM2, and Xenopus Pum. This conservation of binding sequences correlates to the similarity of the amino acid sequences between the repeat domains of these Puf proteins, which all group to the same branch of an unrooted phylogenetic tree (Wickens et al. 2002). In contrast, the other yeast Pufs fall on other branches of the tree, which suggests that they might indeed have altered target specificities versus Puf3p. Puf5p has already been shown to bind an alternative sequence containing UUGUGU (Tadauchi et al. 2001). Additional support for this hypothesis comes from a recent microarray analysis that identified mRNAs associated with each of the five yeast Puf proteins (Gerber et al. 2004). Specifically, each Puf protein was found to interact with a discrete set of mRNAs, and similar yet distinct conserved sequence motifs were identified in the 3′ UTRs of the mRNAs targeted by Puf3p, Puf4p, and Puf5p. All the sequence motifs contain UGUR followed by UA located 2, 3, or 4 nt downstream. Because of the similarity in these sequence motifs, it is possible that the yeast Puf proteins could have overlapping target specificities. In fact, 12% of the mRNAs identified in the microarray screen bound to more than one Puf protein. In such cases, functional specificity could still occur through variations in protein partners or signals. Closer examination of the mRNA targets of the five yeast Puf proteins will address these issues in the future.
The UGUA(U/A)AUA binding sequence contained within both COX17-binding sites matches the conserved Puf3p target sequence motif identified in the microarray analysis: (U/C)(A/C/U)UGUA(U/A)AUA (Gerber et al. 2004). Yet our results show that the UGUA flanking regions provide additional target specificity. In particular, Puf3p binding to Site A is greater than binding to Site B, in part due to uracil bases flanking the UGUA sequence (Fig. 3 ▶). Thus, our data indicate a preferred Puf3-binding site of UUGUAUAUA. We also found that nucleotides upstream of this sequence had no effect on binding, including the first (U/C) nucleotide position of the microarray-derived conserved sequence. Surprisingly, both sites appear to be equally important for Puf-mediated regulation of COX17 mRNA decay in vivo (Fig. 5 ▶). These results are opposite to those found in Drosophila, where the two NRE sites of hunchback mRNA have equal in vitro binding affinities (Zamore et al. 1999), yet contribute unequally to translational repression (Curtis et al. 1997). The most likely explanation for these results is that surrounding 3′-UTR sequences and/or protein factors influence the functional activity of Puf proteins in vivo. Together, our characterization of the Puf3p binding site will allow us to better evaluate novel mRNA targets of Puf protein binding and regulation.
We have already begun to analyze other mRNAs that contain the consensus Puf3 binding site. Specifically, a search of 500 nt of 3′-UTR sequence of all known and hypothetical ORFs identified eight genes in yeast (including COX17) that contain the full UUGUAUAUAUAA sequence, while 142 genes contain the shortened UU GUAUAUA sequence, and 528 genes contain at least UGUAUAUA. Several of these genes were also found in our previous microarray experiment that identified mRNAs differentially expressed in the absence of Puf proteins (Olivas and Parker 2000). Furthermore, preliminary RNA analyses of several of these candidates show that many are indeed under Puf protein regulation and are currently under study. Thus, Puf proteins appear to be widespread regulators of mRNA metabolism in yeast.
The repeat domain of Puf3 is an independent regulator of mRNA decay
Expression of just the repeat domain of Puf3p rescues rapid COX17 mRNA decay in a puf3Δ strain (Fig. 7 ▶). Thus, sequences necessary for both mRNA binding and decay regulation are contained within this region. The repeat domain of Pumilio is also sufficient to regulate translation in Drosophila (Wharton et al. 1998). Pumilio function requires interactions with Nanos and Brat, and their sites of interaction have been mapped to the outer surface of the rainbow-shaped Puf repeat domain (Edwards et al. 2001). Although no Puf3p interacting partners required for mRNA decay have yet been identified in yeast, our results would argue that any such interactions would also map to the repeat domain, and efforts are underway to study this possibility. Furthermore, the ability of the conserved repeat domain to regulate both translation and decay further supports a model in which the Puf-mediated signal affects both processes through a similar mechanism, such as mRNP rearrangements.
The regions outside the Puf repeat domain have no known function, even though they usually comprise well over half of the total protein. Although these regions may enhance the activity of the repeat domain (Wharton et al. 1998), the conservation of such large N-terminal regions among Puf proteins suggests that these regions are important for other unknown activities. Systematic screens to identify protein–protein interactions have identified eight candidate Puf3p interactors (Ito et al. 2001; Gavin et al. 2002; Ho et al. 2002). However, none of these candidates have known functions in mRNA metabolism, none bind directly to the Puf3RD in yeast two-hybrid experiments (data not shown), and deletions of several of the candidates have no effect on COX17 mRNA decay (data not shown). Therefore, if these candidate proteins do function with Puf3p, they may be acting through the N-terminal domain, and may be involved in processes other than mRNA metabolism. Further studies are needed to unveil the roles the Puf N-terminal domains might play.
MATERIALS AND METHODS
Yeast strains
The genotypes of the S. cerevisiae strains used are given in Table 1 ▶.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
| yWO5 | MATa, his4-539, leu2-3,112, trp1-1, ura3-52, cup1::LEU2/PM | Hatfield et al. 1996, yRP840 |
| yWO7 | MATα, leu2-3,112, ura3-52, rpb1-1 | Olivas and Parker 2000, yRP693 |
| yWO43 | MATα, his4-539, leu2-3,112, trp1-1, ura3-52, cup1::LEU2/PM, rpb1-1, puf3::Neor | Olivas and Parker 2000 yRP1360 |
| yWO50 | MATa, his3-1,15, his4-539, leu2-3,112, trp1-1, ura3, rpb1-1, cox17::TRP1 | Olivas and Parker 2000, yRP1546 |
| yWO51 | MATa, his4-539, leu2-3,112, trp1-1, ura3, rpb1-1, cox17::TRP1, puf3::Neor | Olivas and Parker 2000, yRP1547 |
Protein expression and purification
The GST-PUF3 fusion construct pWO3 was previously created (pRP1020; Olivas and Parker 2000) in pGEX-6P-1 (Amersham Biosciences). To create the GST-PUF3 Repeat Domain fusion construct, a fragment containing the PUF3 Repeat Domain (amino acids 465–879) was isolated from pWO3 by digesting with XbaI (filled in by Klenow Fragment) and NotI, then inserted into a derivative of pGEX-3X (Amersham Biosciences) to yield pWO12. The GST-PUF5 fusion construct was created by PCR amplifying the complete PUF5 ORF from yWO5 and inserting it into pBlue-script (Stratagene) between XmaI and XhoI to yield pWO17. Because PUF5 contains an intron from nucleotides 4–643 of its ORF, we digested pWO17 with ClaI (filled in with Klenow Fragment) and XhoI to isolate the sequence encoding amino acids 15–859 and cloned it into pGEX-6P-3 (Amersham Biosciences) to create the GST-Puf5p expression vector pWO18. To create the GST-PUF5 Repeat Domain fusion construct, pWO17 was digested with EcoRI and HindIII to isolate the sequence encoding amino acids 92–592 and this fragment was ligated into pBluescript, producing pWO19. An EcoRI-SalI fragment of pWO19 was then cloned into pGEX-6P-3 to make the GST-Puf5RD expression vector pWO20. All constructs were verified by sequence analysis. The GST fusion constructs pWO3, pWO12, pWO18, and pWO20 were transformed into the protease-deficient E. coli strain BL-21, and GST fusion proteins were purified as recommended by the manufacturer. Protein eluates were dialyzed into 50 mM Tris-HCL (pH 8.0), and expression products were verified by Western analysis with anti-GST antibodies.
In vitro binding analyses
In vitro transcribed RNA containing the COX17 3′-UTR sequence was made from pWO6 (pRP1019; Olivas and Parker 2000). After digestion of pBS or pWO6 with MseI, RNA was transcribed using T7 RNA polymerase in the presence or absence of α-32P UTP to produce 145- and 147-nt transcripts, respectively. Transcription reactions were treated with DNase I. Radiolabeled transcripts were purified by separation on denaturing polyacrylamide gels, elution from gel slices, and ethanol precipitation. To produce COX17 3′-UTR transcripts deleted of Sites A, B, or C, the 147-nt transcript was annealed to complementary oligos oWO8, oWO7, or oWO2, respectively, then cleaved with RNase H prior to gel purification of the remaining RNA (see Fig. 1A ▶).
Binding reactions with RNA transcribed from pWO6 or pBS included radiolabeled RNA (500,000 cpm) and 1× binding buffer (10 mM HEPES at pH 7.5, 50 mM KCl, 1 mM EDTA, 2 mM DTT, 200 U/ml RNasin, 0.1 mg/ml bovine serum albumin, 0.01% Tween-20, 0.1 mg/ml poly(rU) and 10 μg/ml yeast tRNA) in the presence or absence of GST-Puf3p (0.2 μM), GST-Puf3RD (0.5 μM), GST-Puf5p (0.2 μM), or GST-Puf5RD (0.3 μM), and in the presence or absence of ~10-fold excess unlabeled transcript in a total of 15 μl. Reactions were incubated for 30 min at 24°C, then subjected to UV cross-linking (energy mode 8000 × 100 μJ/cm2). Cross-linked reactions were treated with 100 U of RNase T1 for 30 min prior to loading on SDS-7.5% polyacrylamide (29:1 acrylamide:bis-acrylamide) gels.
Short RNAs of Sites A and B (29–30 nt) were transcribed from single-stranded oligonucleotide templates containing the 18-nt T7 RNA polymerase promoter annealed to a complementary primer. RNAs were transcribed using the T7-MEGAshortscript kit (Ambion) as recommended by the manufacturer with the following changes: The reaction contained 500 μM each of ATP, CTP, and GTP, 50 μM of UTP, 40 μCi of α-32P UTP (800 Ci/mmole) for labeled reactions, and 20 U RNasin. Radiolabeled transcripts were purified by separation on denaturing polyacrylamide gels, elution from gel slices, and ethanol precipitation. Unlabeled transcripts were purified using a Nucleotide Removal Kit (Qiagen).
Binding reactions with short RNAs included radiolabeled RNA (20,000 cpm) and 1× binding buffer in the presence or absence of GST-Puf3p or GST-Puf3RD, and in the presence or absence of ~10-fold excess unlabeled transcript in a total of 30 μl. Reactions were incubated for 30 min at 24°C, 5 μg of heparin was added, and reactions incubated for a further 10 min at 24°C, then reactions were electrophoresed on 8% nondenaturing polyacrylamide gels for 2.5 h at 200 V at 4°C. To estimate KD values, the fraction of RNA bound for each gel lane was calculated using the definition Fraction bound = RNA Shifted/(RNA Shifted + Free RNA), where RNA Shifted and Free RNA represent the storage phosphor signals of all shifted complexes or the free RNA species, respectively. Best-fit curves were obtained for the binding data using Kaleidagraph software.
In vivo COX17 mRNA decay analysis
Transcriptional pulse-chase experiments were performed essentially as described (Decker and Parker 1993) on strains yWO50 (wild-type) and yWO51 (puf3Δ). These strains contain the temperature-sensitive rbp1-1 allele for RNA polymerase II and are deleted for the endogenous COX17 gene. Regulated expression of wild-type or mutant COX17 mRNA was accomplished by transformation of the above yeast strains with pWO5 (pG74/ST30; Beers et al. 1997), in which the COX17 gene is under the control of the GAL10 promoter, or mutant versions of the plasmid. Site A (pWO7), Site B (pWO8), and Site A + B (pWO9) mutants were generated by site-directed mutagenesis of pWO5 using the Quick Change XL kit (Stratagene). All mutations were verified by sequencing. To monitor poly(A) tail lengths, COX17 mRNA was cleaved just upstream of the stop codon using RNase H reactions with oWO1 as described (oCOX17-C; Olivas and Parker 2000), and RNA was separated on 6% polyacrylamide/7.7 M urea gels at 300 V for 4 h and transferred to nylon membrane for probing with oWO2 (oCOX17-P; Olivas and Parker 2000).
Steady-state transcriptional shutoff experiments were performed essentially as described (Caponigro et al. 1993) on strains yWO7 (wild-type) and yWO43 (puf3Δ) that contain the rpb1-1 allele. yWO43 was also analyzed after transformation with plasmids expressing full-length Puf3p (pWO13) or the Puf3RD (pWO14) under the control of the constitutive GPD promoter. pWO13 was created by insertion of the PUF3 ORF into a derivative of pG-1 as previously described (pRP1021; Olivas and Parker 2000). pWO14 was created by digestion of pWO12 with BamHI and NotI (filled in with Klenow fragment) to isolate the PUF3RD, and insertion of this fragment into a derivative of pG-1 between BamHI and NcoI (filled in with Klenow fragment) to place the PUF3RD ORF just downstream of an inserted FLAG tag sequence and the GPD promoter. Steady-state transcriptional shutoff experiments were also performed on strains yWO50 and yWO51, each transformed with plasmids expressing MFA2 RNA (pRP485) or the hybrid MFA2/COX17 3′-UTR RNA (pWO25). pRP485 was created as described (Decker and Parker 1993), with the MFA2 RNA expressed under the control of the GAL1 UAS. The MFA2 3′ UTR of pRP485 was replaced by the COX17 3′ UTR to produce pWO25 by insertion of a BamHI-HindIII fragment containing the COX17 3′ UTR from pWO23 into the BglII-HindIII sites of pRP485. A 514-bp PCR product containing 490 bp of COX17 sequence 3′ of the stop codon was inserted into pUC18 between XbaI and PstI to create pWO23. Northern blots were normalized for loading to the stable scRI RNA, an RNA polymerase III transcript (Felici et al. 1989).
Acknowledgments
We gratefully thank Roy Parker, Marc Spingola, Elizabeth Kellogg, Cynthia Dupureur, and members of the Olivas laboratory for helpful discussions and review of the manuscript. We also thank Fred Scherrer for assistance with Figure 6 ▶ and Alex Tzagoloff and Roy Parker for providing reagents. This work was supported by a grant to W.M.O. from the National Institutes of Health (GM63759).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7270204.
REFERENCES
- Andersson, M.G. and Cerenius, L. 2002. Pumilio homologue from Saprolegnia parasitica specifically expressed in undifferentiated spore cysts. Eukaryot. Cell 1: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka-Taguchi, M., Yamada, M., Nakamura, A., Hanyu, K., and Kobayashi, S. 1999. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1: 431–437. [DOI] [PubMed] [Google Scholar]
- Beers, J., Glerum, D.M., and Tzagoloff, A. 1997. Purification, characterization and localization of yeast COX17p, a mitochondrial copper shuttle. J. Biol. Chem. 272: 33191–33196. [DOI] [PubMed] [Google Scholar]
- Caponigro, G., Muhlrad, D., and Parker, R. 1993. A small segment of the MATa1 transcript promotes mRNA decay in Saccharomyces cerevisiae: A stimulatory role for rare codons. Mol. Cell. Biol. 13: 5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnovich, D. and Lehmann, R. 2001. Poly (A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc. Natl. Acad. Sci. 98: 11359–11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden, S.L., Bernstein, D.S., Bachorik, J.L., Thompson, B.E., Gallegos, M., Petcherski, A.G., Moulder, G., Garstead, R., Wickens, M., and Kimble, J. 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663. [DOI] [PubMed] [Google Scholar]
- Curtis, D., Treiber, D.K., Tao, F., Zamore, P.D., Williamson, J.R., and Lehmann, R. 1997. A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J. 16: 834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J. and Parker, R. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 7: 1632–1643. [DOI] [PubMed] [Google Scholar]
- Derrigo, M., Cestelli, A., Savettieri, G., and Di Liegro, I. 2000. RNA–protein interactions in the control of stability and localization of messenger RNA. Int. J. Mol. Med. 5: 111–123. [PubMed] [Google Scholar]
- Edwards, T.A., Pyle, S.E., Wharton, R.P., and Aggarwal, A.K. 2001. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105: 281–289. [DOI] [PubMed] [Google Scholar]
- Felici, F., Cesareni, G., and Hughes, J.M.X. 1989. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol. Cell. Biol. 9: 3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes, A. and Lehmann, R. 1998. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125: 679–690. [DOI] [PubMed] [Google Scholar]
- Gamberi, C., Peterson, D.S., He, L., and Gottlieb, E. 2002. An anterior function for the Drosophila posterior determinant Pumilio. Development 129: 2699–2710. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J. Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Gerber, A.P., Herschlag, D., and Brown, P.O. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLOS Biol. 2: 342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, N.K. and Wickens, M. 1998. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 14: 399–458. [DOI] [PubMed] [Google Scholar]
- Grzybowska, E.A., Wilczynska, A., and Siedlecki, J.A. 2001. Regulatory functions of 3′UTRs. Biochem. Biophys. Res. Commun. 288: 291–295. [DOI] [PubMed] [Google Scholar]
- Guhaniyogi, J. and Brewer, G. 2001. Regulation of mRNA stability in mammalian cells. Gene 265: 11–23. [DOI] [PubMed] [Google Scholar]
- Hatfield, L., Beelman, C.A., Stevens, A., and Parker, R. 1996. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick, D., Parker, R., and Jacobson, A. 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Hsu, C.L. and Stevens, A. 1993. Yeast cells lacking 5′ → 3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13: 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, T., Yoshida, M., Hattori, M., and Sakaki, Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, A. and Peltz, S.W. 1996. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 65: 693–739. [DOI] [PubMed] [Google Scholar]
- Kraemer, B., Crittenden, S., Gallegos, M., Moulder, G., Barstead, R., Kimble, J., and Wickens, M. 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9: 1009–1018. [DOI] [PubMed] [Google Scholar]
- Mazumder, B., Seshadri, V., and Fox, P.L. 2003. Translational control by the 3′-UTR: The ends specify the means. Trends Biochem. Sci. 28: 91–98. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C.J., and Parker, R. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ → 3′ digestion of the transcript. Genes & Dev. 8: 855–866. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15: 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, Y. and Wharton, R. 1995. Binding of Pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80: 747–756. [DOI] [PubMed] [Google Scholar]
- Nakahata, S., Katsu, Y., Mita, K., Inoue, K., Nagahama, Y., and Yamashita, M. 2001. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 276: 20945–20953. [DOI] [PubMed] [Google Scholar]
- Olivas, W. and Parker, R. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19: 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M. and Lin, H. 1999. The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics 153: 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonada, J. and Wharton, R.P. 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes & Dev. 13: 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Drosophila Brain Tumor is a translational repressor. Genes & Dev. 15: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, G.M., da Silva, A.M., and Kuspa, A. 1999. Starvation promotes Dictyostelium development by relieving PufA inhubition of PKA translation through the YakA kinase pathway. Development 126: 3263–3274. [DOI] [PubMed] [Google Scholar]
- Tadauchi, T., Matsumoto, K., Herskowitz, I., and Irie, K. 2001. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M. and Parker, R. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69: 571–595. [DOI] [PubMed] [Google Scholar]
- Wang, X., Zamore, P.D., and Hall, T.M. 2001. Crystal structure of a Pumilio homology domain. Mol. Cell 7: 855–865. [DOI] [PubMed] [Google Scholar]
- Wang, X., McLachlan, J., Zamore, P.D., and Hall, T.M.T. 2002. Modular recognition of RNA by a human Pumilio-homology domain. Cell 110: 501–512. [DOI] [PubMed] [Google Scholar]
- Wharton, R.P. and Struhl, G. 1991. RNA Regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67: 955–967. [DOI] [PubMed] [Google Scholar]
- Wharton, R.P., Sonoda, J., Lee, T., Patterson, M., and Murata, Y. 1998. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1: 863–872. [DOI] [PubMed] [Google Scholar]
- White, E.K., Moore-Jarrett, T., and Ruley, H.E. 2001. PUM2, a novel murine Puf protein, and its consensus RNA-binding site. RNA 7: 1855–1866. [PMC free article] [PubMed] [Google Scholar]
- Wickens, M., Bernstein, D.S., Kimble, J., and Parker, R. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18: 150–157. [DOI] [PubMed] [Google Scholar]
- Wreden, C., Verrotti, A.C., Schisa, J.A., Lieberfarb, M.E., and Strickland, S. 1997. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development 124: 3015–3023. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Williamson, J.R., and Lehmann, R. 1997. The pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3: 1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Bartel, D.P., Lehmann, R., and Williamson, J.R. 1999. The PUMILIO–RNA interaction: A single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry 38: 596–604. [DOI] [PubMed] [Google Scholar]
- Zhang, B., Gallegos, M., Puoti, A., Durkin, E., Fields, S., Kimble, J., and Wickens, M.P. 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484. [DOI] [PubMed] [Google Scholar]