Abstract
Evidence is presented that morpholino, 2′-O-methyl, phosphorothioate, and RNA antisense oligonucleotides can direct site-specific -1 translational frameshifting when annealed to mRNA downstream from sequences where the P- and A-site tRNAs are both capable of repairing with -1 frame codons. The efficiency of ribosomes shifting into the new frame can be as high as 40%, determined by the sequence of the frameshift site, as well as the location, sequence composition, and modification of the antisense oligonucleotide. These results demonstrate that a perfect duplex formed by complementary oligonucleotides is sufficient to induce high level -1 frameshifting. The implications for the mechanism of action of natural programmed translational frameshift stimulators are discussed.
Keywords: frameshifting, antisense, morpholino, phosphorothioate, 2′-O-methyl, recoding
INTRODUCTION
An increasing number of examples of highly efficient programmed frameshifting sites have been described (for reviews, see Farabaugh 1996; Baranov et al. 2002; Namy et al. 2004). Programmed frameshifting occurs at shift prone sites in response to stimulatory signals within the mRNA, which are often downstream RNA structural elements. The frame-shift site and stimulators direct the ribosome into a new frame at a specific location within an overlap of coding sequences in two different frames. The trans-frame products produced are generally functional proteins, and in some cases the efficiency of frameshifting is up to 50%. This extension of the genetic code (Gesteland et al. 1992; Gesteland and Atkins 1996) governs the expression of a subset of genes in probably all organisms.
A high proportion of eukaryotic −1 programmed frame-shift sites are a heptanucleotide sequence of the form X XXY YYZ, where the P- and A-site tRNAs can repair with the mRNA by shifting 1 nt toward the 5′ end of the gene. High levels of frameshifting are achieved by cis-acting stimulatory sequences located downstream from the shift site. H-type pseudoknots (Pleij et al. 1985) are the most widespread of 3′ stimulators in eukaryotes (for reviews, see Giedroc et al. 2000; Brierley and Pennell 2001). Mutagenic and structural data for several of the frameshift stimulators suggest key features that differ between frameshift stimulators. For example, the Mouse Mammary Tumor Virus (MMTV) pseudoknot requires a wedged adenosine at the interface between the two stacked stems, creating a bend in the pseudoknot (Shen and Tinoco 1995) that is critical for frameshift stimulation (Chen et al. 1995, 1996; Kang et al. 1996; Kang and Tinoco 1997). However, for several other frameshift stimulating pseudoknots, neither the wedge adenine nor the bent structure are present (Michiels et al. 2001; Wang et al. 2002). The crystal structure of the Beet Western Yellow Virus (BWYV) frameshift pseudoknot (Su et al. 1999; Egli et al. 2002) reveals several significant structural features. Notably, there is a triplex formed between the adenosine-rich loop 2 and stem 1. Mutagenic studies demonstrated that this interaction is critical for frameshifting efficiency (Kim et al. 1999). Although three-dimensional structural data are lacking for the frameshift pseudoknot of the Infectious Bronchitis Virus (IBV), extensive mutational analysis and chemical probing (Liphardt et al. 1999; Napthine et al. 1999) show that this pseudoknot has an extended stem I and large loop 2 relative to the MMTV or BWYV pseudoknots described above. The sequence and size of loop 2 are unimportant in this case, leading to the conclusion that interactions with this loop are not required for stimulatory function.
Several experiments demonstrate that simple blocks to ribosome progression in the form of RNA secondary structures (Brierley et al. 1991; Somogyi et al. 1993; Chen et al. 1995; Liphardt et al. 1999; Napthine et al. 1999) are not sufficient to induce mammalian ribosome frameshifting. Nevertheless, several naturally occurring simple stem loop frameshift stimulators are known. Escherichia coli dnaX mRNA (Flower and McHenry 1990; Tsuchihashi and Kornberg 1990) has a well-characterized stem loop stimulator, and the stability of the stem is the critical feature for frame-shift stimulation (Larsen et al. 1997). For many years it was thought that the human immunodeficiency virus (HIV) gag-pol programmed frameshift stimulator was a simple stem loop (Parkin et al. 1992), and increases in the stability of this stem were reported to increase frameshifting (Bidou et al. 1997). Later work showed that a more complex structure, the nature of which is controversial, is involved in HIV decoding (Dinman et al. 2002; Dulude et al. 2002).
Although the mechanism of pseudoknot stimulation is unknown, the IBV (Somogyi et al. 1993; Kontos et al. 2001) and Saccharomyces cerevisiae L-A virus (Tu et al. 1992; Lopinski et al. 2000) pseudoknots have been shown to pause the ribosome. Pausing is very likely the mechanism of frameshift stimulation in examples where frameshifting is stimulated by a stop codon (Weiss et al. 1987, 1990; Gramstat et al. 1994) or a “hungry” codon in the A-site due to limitation in the abundance of aminoacyl tRNA (for review, see Gallant and Lindsley 1998). However pausing, as measured in these experiments, does not appear to be sufficient for pseudoknot frameshift stimulation, as stem loops and pseudoknots of similar thermodynamic stability that cause ribosome pausing are not necessarily effective frameshift stimulators (Tu et al. 1992; Somogyi et al. 1993; Kontos et al. 2001). In addition, variations of the IBV pseudoknot have demonstrated a lack of correlation between the extent of pausing and the efficiency of frameshifting (Kontos et al. 2001). Nevertheless, it cannot be ruled out that a specific kinetic barrier to ribosome translocation during unwinding of the structure, not detected in the above experiments, is achieved only by certain stimulatory elements (Bidou et al. 1997; Brierley and Pennell 2001).
To further address the role of secondary structure in frameshift stimulation, we tested simple secondary structures derived from modified antisense oligonucleotides annealed downstream from a shift-prone site. For this purpose, several shift-prone sites located between two overlapping reading frames were translated in vitro in the presence of morpholino, 2′-O-Me, phosphorothioate, and RNA antisense oligonucleotides.
RESULTS
Morpholino oligonucleotide-induced frameshifting on the UUUUUUA shift site
The ability of morpholino antisense oligonucleotides to induce ribosomal frameshifting was determined by in vitro transcription and translation of the dual luciferase reporter vector, p2Luc, in the presence or absence of morpholino oligonucleotides. p2Luc contains the Renilla and firefly luciferase genes on either side of a multiple cloning site, and can be transcribed using the T7 promoter located upstream of the Renilla luciferase gene (Grentzmann et al. 1998). Sequences containing a shift-prone site were cloned between the two reporter genes such that the downstream firefly luciferase gene is in the −1 reading frame and the production of full-length protein requires a −1 shift in reading frame. The resulting constructs were transcribed and translated in vitro with complementary morpholinos, using rabbit reticulocyte lysates in the presence of 35S methionine and analyzed by electrophoresis on SDS polyacrylamide gels as described in Materials and Methods.
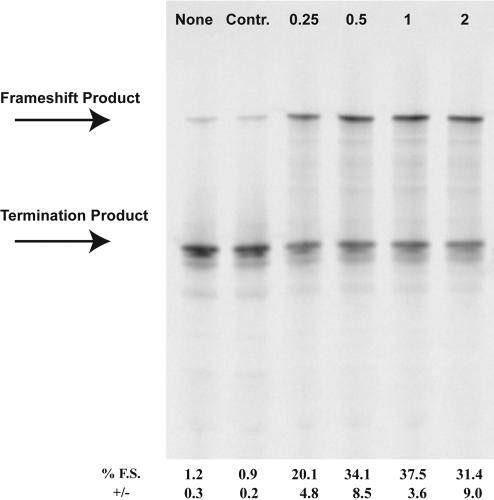
The ability of a morpholino antisense oligonucleotide to induce translational frameshifting at the highly shifty UUUUUUA (U6A) sequence was initially examined. A single morpholino oligonucleotide, MOAB, was designed to anneal 3 nt downstream from the shift site within the vector p2LucU6A. Titration of the morpholino oligonucleotide into coupled transcription/translation reactions revealed a maximal frameshifting level of ~40% with 1 μM morpholino oligonucleotide, as compared to 1% in the absence of morpholino oligonucleotide or with a control sense morpholino oligonucleotide (Fig. 1 ▶). To verify that the morpholino oligonucleotide was activating ribosomal frame-shifting and not transcription slippage, RNA was transcribed in the absence of morpholino oligonucleotide and added to reticulocyte lysate translations in the presence of 1 μM MOAB morpholino oligonucleotide. Frameshifting levels were increased to nearly the same level, ~30-fold, as observed in coupled reactions, demonstrating that the morpholino oligonucleotide acts to induce frameshifting during translation (data not shown; see Materials and Methods for experimental details).
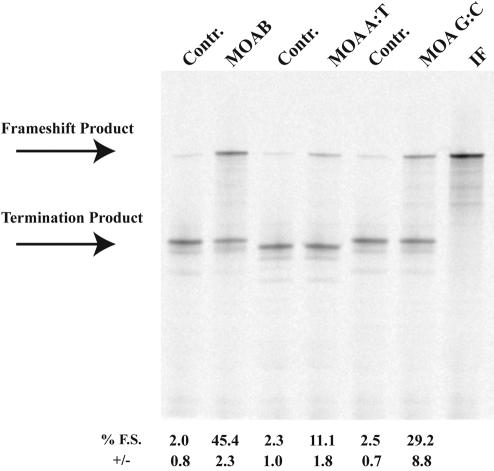
FIGURE 1.
Morpholino-induced frameshifting on the UUUUUUA frameshift site. p2LucU6A was transcribed and translated in rabbit reticulocyte lysate in the absence or presence of increasing amounts of antisense MOAB morpholino. Control (Contr.) morpholino is a morpholino oligonucleotide corresponding to the sense sequence of MOAB. SDS PAGE (4%–12% Bistris polyacrylamide gel) of 35S-methionine-labeled protein products from transcription and translation reactions is shown. Concentrations are shown in micromolar and the location of the full-length frameshift product and nonframeshift termination product are indicated. Average percent frameshifting (% F.S.) and standard deviations (+/−) from the mean are shown below each lane of the gel.
Spacer effects
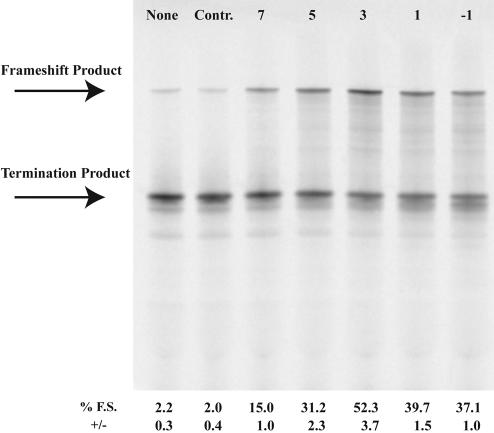
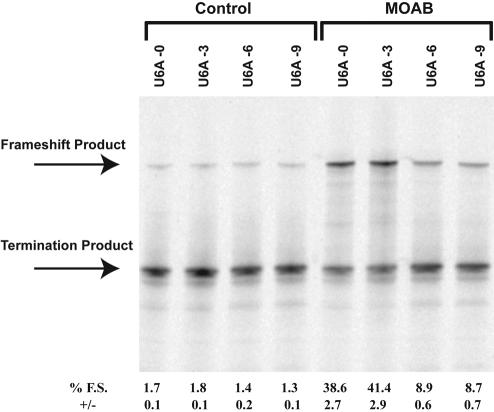
The spacer length between frameshift sites and downstream stimulators in programmed −1 frameshifting is important for optimal frameshift stimulation (Brierley et al. 1989; Kollmus et al. 1994). To determine the optimal spacer length between the shift site and the morpholino antisense oligonucleotide:RNA hybrid, morpholino antisense oligos MOA−1, MOAA, MOAB, MOAC, and MOAD were designed to hybridize downstream from the UUUUUUA site in p2LucU6A such that either −1, 3, 5, 7, or 9 nt separate the last A of the shift site from the 3′ end of the morpholino oligonucleotide (“−1” hybridizes with the A of the UUUUUUA sequence). The greatest level of frameshifting was observed with a 3-nt spacer (Fig. 2 ▶). However, using this approach, the morpholino oligonucleotide sequence is, by necessity, altered at each location. To address the spacer effect without altering the morpholino sequence, three additional p2Luc constructs were produced p2LucU6A-0, p2LucU6A-6, p2LucU6A-9 such that either 0, 6, or 9 nt separate the last A of the UUUUUUA shift site from the first complementary base (3′ end) of the MOAB morpholino oligonucleotide. Spacer distances of 0 or 3 nt stimulated frameshifting to ~40% whereas 6- and 9-nt spacers reduced frameshifting to just below 10% (Fig. 3 ▶). We conclude that the optimal spacer length is between 0 and 5 nt downstream from the shift site.
FIGURE 2.
Spacer length effect on morpholino-induced frameshifting. p2LucU6A was transcribed and translated in rabbit reticulocyte lysate in the absence or presence of increasing amounts of antisense morpholinos MOAD, MOAC, MOAB, MOAA, or MOA-1 that anneal 7, 5, 3, 1, or −1 nt downstream from the frameshift site, respectively. Control (Contr.) morpholino is a morpholino oligonucleotide corresponding to the sense sequence of MOAB. SDS PAGE (4%–12% Bistris polyacrylamide gel) of 35S-methionine-labeled protein products from transcription and translation reactions is shown. Each morpholino was included at a concentration of 1 μM. The location of the full-length frameshift product and nonframeshift termination product are indicated. Average percent frameshifting (% F.S.) and standard deviations (+/−) from the mean are shown below each lane of the gel.
FIGURE 3.
Spacer length effect on morpholino-induced frameshifting. Plasmids p2LucU6A-0, p2LucU6A, p2LucU6A-6, and p2LucU6A-9 were transcribed and translated in rabbit reticulocyte lysate in the absence or presence of increasing amounts of antisense morpholino MOAB that anneals 0, 3, 6, and 9 nt downstream from the frameshift site for each construct, respectively. SDS PAGE (4%–12% Bistris polyacrylamide gel) of 35S-methionine-labeled protein products from transcription and translation reactions is shown. MOAB was included at a concentration of 1 μM. The location of the full-length frameshift product and nonframeshift termination product are indicated. Average percent frameshifting (% F.S.) and standard deviations (+/−) from the mean are shown below each lane of the gel.
Morpholino oligonucleotide specificity
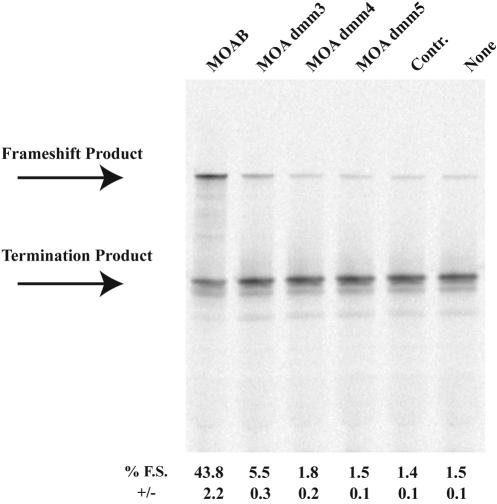
To test the sequence specificity for the action of morpholino oligonucleotides to induce frameshifting, we tested the effect of three morpholino oligonucleotides (MOA dmm3, MOA dmm4, and MOA dmm5) corresponding to the MOAB sequence but containing 3, 4, or 5 mismatched nucleotides, respectively (see Materials and Methods). Three mismatches reduced frameshifting levels on the UUUUUUA shift site in in vitro transcription/translation reactions from ~40% to 5%, and additional mismatches reduced frameshifting levels to background levels (Fig. 4 ▶). These results demonstrate sequence specificity and suggest that morpholino oligonucleotides are unlikely to affect ribosome frame maintenance at nontarget sites.
FIGURE 4.
Specificity of morpholino-induced frameshifting. p2LucU6A was transcribed and translated in rabbit reticulocyte lysate in the absence or presence of antisense morpholinos, MOAB, MOA dmm3, MOA dmm4, or MOA dmm5 that contain 0, 3, 4, or 5 mismatches, respectively. Each morpholino is added at a concentration of 1 μM. Control (Contr.) morpholino is a morpholino oligonucleotide corresponding to the sense sequence of MOAB. SDS PAGE (4%–12% Bistris polyacrylamide gel) of 35S-methionine-labeled protein products from transcription and translation reactions is shown. The location of the full-length frameshift product and nonframeshift termination product are indicated. Average percent frameshifting (% F.S.) and standard deviations (+/−) from the mean are shown below each lane of the gel.
Sequence effects
The sequence composition of the morpholino oligonucleotide:RNA hybrid may influence ribosome frameshifting due to increases in the thermodynamic stability of G:C-rich sequences relative to those which are A:U rich. Two additional p2Luc constructs were made with the first 6 nt after the 3-nt spacer changed to be entirely A and U (p2LucU6A A:T) or G and C (p2LucU6A G:C) nucleotides. Corresponding complementary morpholino oligonucleotides, MOA A:T and MOA G:C, were synthesized and tested in in vitro transcription and translation reactions for their ability to induce frameshifting on the UUUUUUA frameshift site. Interestingly, MOAB with 50% G:C composition gave higher frameshifting efficiencies (~45% frameshifting) than MOA A:T (~10% frameshifting) or MOA G:C (~30% frameshifting; Fig. 5 ▶). These results demonstrate a morpholino oligonucleotide:RNA hybrid sequence effect on ribosomal frameshifting and suggest that an intermediate thermodynamic stability may result in maximal frameshift induction.
FIGURE 5.
The effect of sequence composition on morpholino induced frameshifting. p2LucU6A, p2LucU6A A:T, and p2LucU6A G:C were transcribed and translated in rabbit reticulocyte lysate in the absence or presence of the complementary antisense morpholinos, MOAB, MOA A:T, and MOA G:C, respectively. Control (Contr.) morpholino is a morpholino oligonucleotide corresponding to the sense sequence of MOAB. Each morpholino is added at a concentration of 1 μM. (IF) p2LucU6A in which the six Us have been deleted. SDS PAGE (4%–12% Bistris polyacrylamide gel) of 35S-methionine-labeled protein products from transcription and translation reactions is shown. The location of the full-length frameshift product and non-frameshift termination product are indicated. Average percent frame-shifting (% F.S.) and standard deviations (+/−) from the mean are shown below each lane of the gel.
Heptanucleotide shift sites
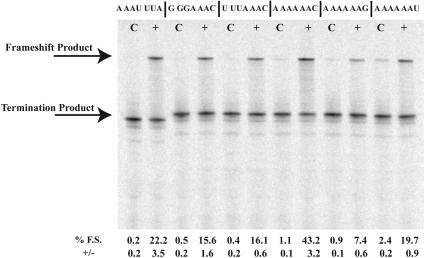
Morpholino oligonucleotide-induced frameshifting was examined at six additional heptanucleotide frameshift motifs, AAAUUUA, GGGAAAC, UUUAAAC, AAAAAAC, AAAA AAG, and AAAAAAU. The complementary MOAB morpholino oligonucleotide was added to 1 μM in in vitro transcriptions/translations of p2LucAAAUUUA, p2LucGG GAAAC, p2LucUUUAAC, p2LucA6C, p2LucA6G, and p2LucA6U (Fig. 6 ▶). Frameshifting on the A AAA AAC shift site was equivalent, ~40%, to that observed at the UUUUUUA sequence, whereas changing the C to either a U or a G reduced frameshift levels to 20% and 7%, respectively. Frameshifting at the AAAUUUA, GGGAAAC, and UUUAAAC sites ranged between 20% and 15%. Thus, frameshift stimulation is not unique to the UUUUUUA site and varies in efficiency depending upon the P- and A-site codons.
FIGURE 6.
The effect of the heptanucleotide motif on morpholino-induced frameshifting. p2LucAAAUUUA, p2LucGGGAAAC, p2LucUUUAAAC, p2LucAAAAAAC, p2LucAAAAAAG, and p2LucAAAAAAU were transcribed and translated in rabbit reticulocyte lysate in the presence of the complementary antisense morpholino, MOAB. Control © morpholino is a morpholino oligonucleotide corresponding to the sense sequence of MOAB. Each morpholino is added at a concentration of 1 μM. SDS PAGE (4%–12% Bistris polyacrylamide gel) of 35S-methionine-labeled protein products from transcription and translation reactions is shown. The location of the full-length frameshift product and nonframeshift termination product are indicated. Average percent frameshifting (% F.S.) and standard deviations (+/−) from the mean are shown below each lane of the gel.
2′-O-methyl-, phosphorothioate-, and RNA antisense oligonucleotide-induced frameshifting
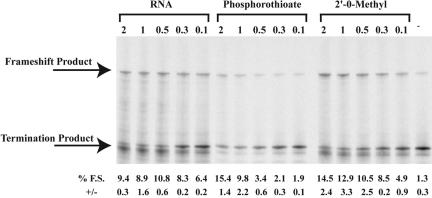
The ability of 2′-O-methyl, phosphorothioate, and RNA antisense oligonucleotides to induce frameshifting at the UUUUUUA shift site was examined to determine if the frameshift stimulatory activity was unique to the morpholino-modified oligonucleotides. Each modified antisense oligonucleotide, with the same sequence as MOAB, was synthesized and added to in vitro transcription/translation reactions of p2lucU6A at 2 μM, 1 μM, 0.5 μM, 0.3 μM, or 0.1 μM. Phosphorothioate and 2′-O-methyl oligonucleotides induced maximal frameshifting levels to ~15% and the RNA antisense oligonucleotide to 10% on the UUUUUUA shift site (Fig. 7 ▶). Although frameshift stimulation was somewhat reduced compared to the morpholino antisense oligonucleotide of the same sequence and size, each antisense oligonucleotide revealed significant frame-shift stimulation.
FIGURE 7.
The effect of antisense oligonucleotide chemistry on frameshift induction. p2lucU6A was transcribed and translated in rabbit reticulocyte lysate in the presence of increasing amounts of complementary RNA, phosphorothioate, or 2′-O-methyl antisense oligonucleotides. SDS PAGE (4%–12% Bistris polyacrylamide gel) of 35S-methionine-labeled protein products from transcription and translation reactions is shown. The concentration of oligonucleotide (μM) is indicated above each gel lane, and average percent frameshifting (% F.S.) and standard deviations (+/−) from the mean are shown below each lane of the gel.
To determine whether the stability of the oligonucleotide:RNA duplex is an important factor in frameshift stimulation, melting temperature analysis of each modified oligonucleotide was determined. A 25-nt sense RNA corresponding to the complementary sequence of MOAB was annealed to each corresponding antisense RNA or modified antisense oligonucleotide and the absorbance over a temperature gradient was followed and melting temperatures calculated as described in Materials and Methods. The melting temperature for a sense RNA hybridized to phosphorothioate, RNA, morpholino, and 2′-O-methyl antisense oligonucleotides was 53°C, 79°C, 77°C, and 84°C, respectively.
DISCUSSION
Several of the heptanucleotide motifs utilized in this study occur naturally in retroviral programmed -1 frameshifting. The efficiency of ribosomal frameshifting in eukaryotes, stimulated by natural RNA structures, varies between about 5% and 30% depending upon the frameshift site (Brierley et al. 1992) and the identity of the downstream RNA structure that stimulates frameshifting (Brierley and Pennell 2001). The data presented here demonstrate that trans-acting morpholino, 2 To determine whether the stability of the oligonucleotide:RNA duplex is an important factor in frameshift stimulation, melting temperature analysis of each modified oligonucleotide was determined. A 25-nt sense RNA corresponding to the complementary sequence of MOAB was annealed to each corresponding antisense RNA or modified antisense oligonucleotide and the absorbance over a temperature gradient was followed and melting temperatures calculated as described in Materials and Methods. The melting temperature for a sense RNA hybridized to phosphorothioate, RNA, morpholino, and 2′-O-methyl -O-methyl, phosphorothioate, and RNA anti-sense oligonucleotides may stimulate −1 frameshifting at levels that are comparable to, or greater than, those observed with natural cis-acting mRNA secondary structures in programmed −1 frameshifting.
For three of the heptanucleotide motifs tested in this study, HIV gag-pol, UUUUUUA; MMTV gag-pro, AAAAAAC; and RSV gag-pol, AAAUUUA frameshift sites, an antisense morpholino oligonucleotide induced substantially greater frameshifting than those reported with the natural RNA stimulator. The MMTV and HIV sites both revealed ~40% morpholino oligonucleotide-induced ribosome frameshifting, whereas, the complete HIV and MMTV frameshift cassettes with their natural stimulators have been reported to induce ribosome frameshifting to ~10% (Grentzmann et al. 1998; Dinman et al. 2002; Dulude et al. 2002) and 20% (Jacks et al. 1987; Chamorro et al. 1992), respectively. Similarly, the MOAB morpholino:RNA hybrid induces nearly 25% frameshifting on the AAAUUUA Rouse Sarcoma Virus gag-pol shift site, where natural levels of frameshifting are only about 5% (Jacks et al. 1988; Marczinke et al. 1998). Two shift sites, the Simian Retrovirus-1 (SRV-1) gag-pro G GGA AAC and IBV UUUAAAC frameshift sites revealed slightly lower morpholino oligonucleotide-induced frame-shifting levels than those induced by the natural stimulators. In this case, morpholino-induced frameshifting occurs at ~15% for both sequences, compared to 25% (ten Dam et al. 1994) and 30% (Brierley et al. 1991) for the natural RNA stimulators of SRV-1 and IBV. Finally, altering the 3′ most nucleotide of the MMTV AAAAAAC shift site to either G or U significantly reduced morpholino oligonucleotide-induced frameshifting. These results are consistent with those obtained using the MMTV shift site and natural stimulatory pseudoknot, whereby altering this position likewise substantially reduced frameshift efficiency (Chamorro et al. 1992). These data demonstrate that the heptanucleotide motif, and in particular the A-site tRNA:mRNA pairing potential, can strongly influence antisense oligonucleotide-induced -1 frameshifting, as has previously been observed for frameshift stimulation by natural RNA structures (Brierley et al. 1992).
Recent structural studies of the ribosome have provided remarkable insight into the mechanism of decoding (Noller et al. 2001; Moore and Steitz 2003; Ogle et al. 2003 and references therein). Despite the many similarities, there are clear structural differences between prokaryotic and eukaryotic ribosomes due primarily to the presence of eukaryotic rRNA expansion elements and additional proteins (Spahn et al. 2001). Nevertheless, the observations that the mRNA begins to enter the ribosome 7–9 nt downstream from the P-site codon (most −1 frameshift stimulators start 5–9 nt downstream from the end of the heptanucleotide motif) and that the ribosome entrance tunnel of the Thermus thermophilus ribosome is unable to accommodate an RNA duplex is of direct relevance to this study, as it provokes the concept that the site of action of frameshift stimulators is likely to be at the location where the eukaryotic counterparts of the ribosomal proteins S3, S4, and S5 are located (Yusupova et al. 2001). The basic side chains of these proteins are oriented such that they may bind to the phosphate backbone of the mRNA as it enters the ribosome. Yusupova and colleagues propose that binding of S3, associated with the head of the small ribosomal subunit, with one strand of an mRNA structure and S4/5 with the other strand, disrupts the structure and pulls the mRNA into the ribosome by about 3 bp at a time due to the movement of the head during translocation (Agrawal et al. 1999). Disruption of this process by pseudoknots or other frameshift stimulators could alter the step size of decoding such that ribosome frameshifting occurs (Yusupova et al. 2001).
It is surprising that the optimal distance for the morpholino:mRNA hybrids demonstrated in this article is 0–5 nt, and that morpholinos hybridized further downstream from the shift site exhibit reduced frameshifting potential. When the frameshift site is located in the P- and A-sites of the ribosome, the portion of the oligonucleotide located closer than 7 nt must be partially unwound or contained within the ribosome. As the morpholino oligonucleotides used in this study lack the negatively charged phosphate backbone of RNA, the issue arises as to how the morpholino:RNA hybrid is dissociated during translation. By the above model (Yusupova et al. 2001), the acidic side chains of the eukaryotic counterparts to S3, S4, or S5 would be unable to bind to the morpholino. Either this unwinding model must be altered to account for unwinding of duplexes where one strand is uncharged or another mechanism is involved. Once the shift site reaches the P- and A-sites of the ribosome, the remaining duplexed morpholino oligonucleotide outside the ribosome may exert the frameshifting effect. However, it cannot at the current time be ruled out that the ribosome entry tunnel is dynamic—capable of opening during translation to accommodate the duplex—or that the eukaryotic ribosome has a larger entrance for mRNA than the T. thermophilus ribosome. Under these conditions, the antisense:RNA duplexes might exert their effects within the ribosome.
Several models of pseudoknot stimulation of programmed frameshifting have been proposed (for reviews, see Giedroc et al. 2000; Brierley and Pennell 2001; Plant et al. 2003). Nearly all of these invoke a pausing mechanism whereby the downstream stimulator blocks the ribosome over the shift site such that time is allowed for the tRNAs to reposition in the new reading frame. The observations presented in this article demonstrate that a perfect duplex formed by complementary oligonucleotides is sufficient to induce high-level eukaryotic −1 frame-shifting, and that elaborate tertiary structures are not required. The simplest explanation is that the act of unwinding these simple duplex structures provides a barrier to ribosome movement on the mRNA, which drives slippage of the A- and P-site tRNAs relative to the zero frame codons. For this model to accommodate the observation that not all secondary structures that pause the ribosome can cause frameshifting (Tu et al. 1992; Somogyi et al. 1993; Kontos et al. 2001), it must be assumed that the pausing event being measured in these experiments is either not the direct mediator of frameshifting or is not sufficient to induce frameshifting. One compelling possibility is that the critical events for frameshift stimulation occur during one round of ribosome decoding and translocation, when the shift site is located in the A- and P-sites of the ribosome. Subsequently, nonproductive pausing that occurs as the ribosome unwinds the remaining structure is irrelevant to frameshift stimulation, and may account for most (or all) of the pausing detected in these experiments. The possibility of two pausing events, one productive and one unproductive for frameshifting, has been presented previously (Kontos et al. 2001). It may be that natural frameshift stimulators require specific structures only as a means to achieve the appropriate barrier to unwinding at the crucial time, when the ribosome is decoding the shift site.
This model is consistent with our results that the sequence composition of the 3′ most end of the morpholino:RNA hybrid can dramatically effect frameshift levels. We find that an intermediate G:C sequence composition of the hybrid nearest the shift site results in the highest levels of frameshifting. Likewise, oligonucleotides of lesser stability (phosphorothioate) and greater stability (2′-O-methyl) are less efficient frameshift stimulators than morpholino oligonucleotides of the same sequence composition. However, an RNA antisense oligonucleotide of nearly equal stability, as determined by melting temperature analysis, compared to a morpholino oligonucleotide induced far less frameshifting. Although the melting temperatures are nearly the same, the uncharged nature of the morpholino oligonucleotide may create a unique barrier to unwinding, as described above, which could account for its ability to induce higher levels of frameshifting.
Antisense oligonucleotides have been used previously in a limited study to modify frameshift levels at the HIV frame-shift site (Vickers and Ecker 1992). In this case, a stem loop stimulator was present and it was found that 2′-O-methyl oligonucleotides that anneal adjacent to and downstream from the natural stem loop can increase frameshifting levels by up to sixfold. The rational of these experiments was to modify programmed frameshift levels such that the ratio of Gag to Pol would be altered as a therapeutic approach to inhibiting HIV replication. In contrast to these results, which demonstrate the ability of antisense oligonucleotides to modify the activity of an existing stimulator of frame-shifting, our results demonstrate that antisense oligonucleotides can induce substantial frameshifting even in the absence of natural 3′ frameshift stimulators.
Finally, the ability to direct site-specific translational frameshifting suggests a novel therapeutic application for antisense oligonucleotides. Directed translational frame-shifting near a disease-causing frameshift mutation would revert some ribosomes back to the wild-type open reading frame, potentially restoring the production of full-length protein from mutant alleles.
MATERIALS AND METHODS
Frameshift reporter constructs and morpholino oligonucleotides
Complementary oligonucleotides, to construct the sequences listed below, were synthesized at the University of Utah DNA/Peptide Core Facility such that, when annealed, they would have appropriate ends to ligate into the SalI/BamHI sites of the dual luciferase vector, p2luc (Grentzmann et al. 1998). Dual luciferase constructs were prepared and sequence verified as described previously (Howard et al. 2000).
p2Luc inserts are listed below (0 reading frame is shown):
U6A-0: TCGACGAATTTTTTATGG U6A: TCGACGAATTTTTTAGGGTGG U6A-6: TCGACGAATTTTTTAGGGCAGTGG U6A-9: TCGACGAATTTTTTAGGGCAGAGCTGG U6A A:T: TCGAATTTTTTAGGGATATAA U6A G:C: TCGAATTTTTTAGGGGCGGGC A6C: TCGTCAAAAAACTTGTGG A6G: TCGTCAAAAAAGTTGTGG A6U: TCGTCAAAAAATTTGTGG UUUAAAC: TCGCCTTTAAACCAGTGG GGGAAAC: TCGCAGGGAAACGGATGG AAAUUUA: TCGACAAATTTATAGTGG
Morpholino oligonucleotides were synthesized by Gene Tools,LLC. Oligonucleotides were resuspended in H2O and added directly to in vitro translations. Mismatch nucleotides are indicated in bold for dmm3, -4, and -5. 2′-O-Methyl, phosphorothioate, and RNA oligonucleotides were synthesized by Integrated DNA Technologies.
The following is a list of antisense oligonucleotides:
Control MOA: ATCCTTCAACTTCCCTGAGCTCGAA MOA-1: CAGGGAAGTTGAAGGATCCCACCCT MOAA: CTCAGGGAAGTTGAAGGATCCCACC MOAB: AGCTCAGGGAAGTTGAAGGATCCCA MOAC: CGAGCTCAGGGAAGTTGAAGGATCC MOAD: TTCGAGCTCAGGGAAGTTGAAGGAT MOAE: TCTTCGAGCTCAGGGAAGTTGAAGG MOA dmm3: ACCTCAGCGAAGTTGAAGCATCCCA MOA dmm4: ACCTCAGCGAAGTTGAAGCATCGCA MOA dmm5: ACCTCAGCGAACTTGAAGCATCGCA MOA A:T: TCAGGGAAGTTGAAGGATCTTATAT MOA G:C: TCAGGGAAGTTGAAGGATCGCCCGC
In vitro transcription and translation
The dual luciferase constructs described above were either added directly to TNT coupled reticulocyte lysate reactions as described (Promega) or linearized with Pml-1 restriction enzyme prior to capped in vitro transcription reactions utilizing the mMessage mMachine Kit obtained from Ambion, Inc. In the latter case, 0.2 μg of capped mRNA was added to 6.6 μL of rabbit reticulocyte lysate, 70 mM KCL, 0.02 mM each amino acid except methionine, 4 μCi of 35S methionine (1000 Ci/mmole) in a total of 10 μL. Proteins were separated by SDS polyacrylamide gel electrophoresis and the gels were fixed with 7.5% acetic acid and methanol for 20 min. After drying under vacuum, the gels were visualized using a Storm 860 phosphorimager (Molecular Dynamics) and radioactive bands quantified using ImageQuant software. Percent frame-shifting was calculated as the percent of full-length (frameshift) product relative to the termination product and the full-length product combined. The value of each product was corrected for the number of methionine codons present in the coding sequence. The reported values are the average of three independent points from a representative experiment.
Melting temperatures analysis
A 25-nt sense RNA corresponding to the complementary sequence of MOAB was synthesized by Integrated DNA Technologies. Complementary RNA, phosphorothioate, 2′-O-methyl, and morpholino antisense oligonucleotides were hybridized with the sense RNA in analysis buffer (10 mM Na cacodylate at pH 6.8, 100 μM EDTA, 50 mM NaCl, 5 mM MgCl2) by heating to 95°C for 2 min and cooled to room temperature for 15 min.
Melting temperatures analysis was performed by following the absorbance of the oligonucleotides in a Beckman DU4700 spectrophotometer over a temperature gradient from 25°C to 95°C (1°C/min). Absorbance values were then normalized following subtraction of the buffer controls and melting temperatures estimated by taking the first derivative of the melting profile.
NOTE ADDED IN PROOF
While this manuscript was in final preparation, we learned that an independent study has demonstrated efficient frameshift stimulation at a shift prone site by an RNA oligonucleotide in in vitro translations (R.C.L. Olsthoorn, M. Laurs, F. Sohet, H.A. Heus, C.W.A. Pleij, pers. comm.). The ability of natural trans-acting nucleic acids to stimulate frameshifting has implications for small noncoding RNA regulation of gene expression.
Acknowledgments
This project was funded by an MDA Development grant to M.T.H. and NIH R01 NS43264 (to K. Flanigan and J.F.A.). J.F.A. was supported by NIH grant GM48152 and in the final stages by Science Foundation Ireland.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7810204.
REFERENCES
- Agrawal, R.K., Heagle, A.B., Penczek, P., Grassucci, R.A., and Frank, J. 1999. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 6: 643–647. [DOI] [PubMed] [Google Scholar]
- Baranov, P.V., Gesteland, R.F., and Atkins, J.F. 2002. Recoding: Translational bifurcations in gene expression. Gene 286: 187–201. [DOI] [PubMed] [Google Scholar]
- Bidou, L., Stahl, G., Grima, B., Liu, H., Cassan, M., and Rousset, J.P. 1997. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA 3: 1153–1158. [PMC free article] [PubMed] [Google Scholar]
- Brierley, I. and Pennell, S. 2001. Structure and function of the stimulatory RNAs involved in programmed eukaryotic -1 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 66: 233–248. [DOI] [PubMed] [Google Scholar]
- Brierley, I., Digard, P., and Inglis, S.C. 1989. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell 57: 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley, I., Rolley, N.J., Jenner, A.J., and Inglis, S.C. 1991. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 220: 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley, I., Jenner, A.J., and Inglis, S.C. 1992. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 227: 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro, M., Parkin, N., and Varmus, H.E. 1992. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl. Acad. Sci. 89: 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Chamorro, M., Lee, S.I., Shen, L.X., Hines, J.V., Tinoco Jr., I., and Varmus, H.E. 1995. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: Nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 14: 842–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Kang, H., Shen, L.X., Chamorro, M., Varmus, H.E., and Tinoco Jr., I. 1996. A characteristic bent conformation of RNA pseudoknots promotes −1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 260: 479–483. [DOI] [PubMed] [Google Scholar]
- Dinman, J.D., Richter, S., Plant, E.P., Taylor, R.C., Hammell, A.B., and Rana, T.M. 2002. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl. Acad. Sci. 99: 5331–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulude, D., Baril, M., and Brakier-Gingras, L. 2002. Characterization of the frameshift stimulatory signal controlling a programmed -1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 30: 5094–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli, M., Minasov, G., Su, L., and Rich, A. 2002. Metal ions and flexibility in a viral RNA pseudoknot at atomic resolution. Proc. Natl. Acad. Sci. 99: 4302–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh, P.J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60: 103–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower, A.M. and McHenry, C.S. 1990. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl. Acad. Sci. 87: 3713–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant, J.A. and Lindsley, D. 1998. Ribosomes can slide over and beyond “hungry” codons, resuming protein chain elongation many nucleotides downstream. Proc. Natl. Acad. Sci. 95: 13771–13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland, R.F. and Atkins, J.F. 1996. Recoding: Dynamic reprogramming of translation. Annu. Rev. Biochem. 65: 741–768. [DOI] [PubMed] [Google Scholar]
- Gesteland, R.F., Weiss, R.B., and Atkins, J.F. 1992. Recoding: Reprogrammed genetic decoding. Science 257: 1640–1641. [DOI] [PubMed] [Google Scholar]
- Giedroc, D.P., Theimer, C.A., and Nixon, P.L. 2000. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 298: 167–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramstat, A., Prufer, D., and Rohde, W. 1994. The nucleic acid-binding zinc finger protein of potato virus M is translated by internal initiation as well as by ribosomal frameshifting involving a shifty stop codon and a novel mechanism of P-site slippage. Nucleic Acids Res. 22: 3911–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann, G., Ingram, J.A., Kelly, P.J., Gesteland, R.F., and Atkins, J.F. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486. [PMC free article] [PubMed] [Google Scholar]
- Howard, M.T., Shirts, B.H., Petros, L.M., Flanigan, K.M., Gesteland, R.F., and Atkins, J.F. 2000. Sequence specificity of aminoglycoside-induced stop codon readthrough: Potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 48: 164–169. [PubMed] [Google Scholar]
- Jacks, T., Townsley, K., Varmus, H.E., and Majors, J. 1987. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc. Natl. Acad. Sci. 84: 4298–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks, T., Madhani, H.D., Masiarz, F.R., and Varmus, H.E. 1988. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell 55: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H. and Tinoco Jr., I. 1997. A mutant RNA pseudoknot that promotes ribosomal frameshifting in mouse mammary tumor virus. Nucleic Acids Res. 25: 1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H., Hines, J.V., and Tinoco Jr., I. 1996. Conformation of a non-frameshifting RNA pseudoknot from mouse mammary tumor virus. J. Mol. Biol. 259: 135–147. [DOI] [PubMed] [Google Scholar]
- Kim, Y.G., Su, L., Maas, S., O’Neill, A., and Rich, A. 1999. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc. Natl. Acad. Sci. 96: 14234–14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmus, H., Honigman, A., Panet, A., and Hauser, H. 1994. The sequences of and distance between two cis-acting signals determine the efficiency of ribosomal frameshifting in human immunodeficiency virus type 1 and human T-cell leukemia virus type II in vivo. J. Virol. 68: 6087–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos, H., Napthine, S., and Brierley, I. 2001. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 21: 8657–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, B., Gesteland, R.F., Atkins, J.F. 1997. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50%. J. Mol. Biol. 271: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt, J., Napthine, S., Kontos, H., and Brierley, I. 1999. Evidence for an RNA pseudoknot loop-helix interaction essential for efficient -1 ribosomal frameshifting. J. Mol. Biol. 288: 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopinski, J.D., Dinman, J.D., and Bruenn, J.A. 2000. Kinetics of ribosomal pausing during programmed -1 translational frameshifting. Mol. Cell. Biol. 20: 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczinke, B., Fisher, R., Vidakovic, M., Bloys, A.J., and Brierley, I. 1998. Secondary structure and mutational analysis of the ribosomal frameshift signal of rous sarcoma virus. J. Mol. Biol. 284: 205–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels, P.J., Versleijen, A.A., Verlaan, P.W., Pleij, C.W., Hilbers, C.W., and Heus, H.A. 2001. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 310: 1109–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, P.B. and Steitz, T.A. 2003. The structural basis of large ribosomal subunit function. Annu. Rev. Biochem. 72: 813–850. [DOI] [PubMed] [Google Scholar]
- Namy, O., Rousset, J.P., Napthine, S., and Brierley, I. 2004. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell 13: 157–168. [DOI] [PubMed] [Google Scholar]
- Napthine, S., Liphardt, J., Bloys, A., Routledge, S., and Brierley, I. 1999. The role of RNA pseudoknot stem 1 length in the promotion of efficient -1 ribosomal frameshifting. J. Mol. Biol. 288: 305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller, H.F., Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Lancaster, L., Dallas, A., Fredrick, K., Earnest, T.N., and Cate, J.H. 2001. Structure of the ribosome at 5.5 Å resolution and its interactions with functional ligands. Cold Spring Harbor Symp. Quant. Biol. 66: 57–66. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Carter, A.P., and Ramakrishnan, V. 2003. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28: 259–266. [DOI] [PubMed] [Google Scholar]
- Parkin, N.T., Chamorro, M., and Varmus, H.E. 1992. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: Demonstration by expression in vivo. J. Virol. 66: 5147–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant, E.P., Jacobs, K.L., Harger, J.W., Meskauskas, A., Jacobs, J.L., Baxter, J.L., Petrov, A.N., and Dinman, J.D. 2003. The 9-Å solution: How mRNA pseudoknots promote efficient programmed -1 ribosomal frameshifting. RNA 9: 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij, C.W., Rietveld, K., and Bosch, L. 1985. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 13: 1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L.X. and Tinoco Jr., I. 1995. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 247: 963–978. [DOI] [PubMed] [Google Scholar]
- Somogyi, P., Jenner, A.J., Brierley, I., and Inglis, S.C. 1993. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 13: 6931–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, C.M., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., and Frank, J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell 107: 373–386. [DOI] [PubMed] [Google Scholar]
- Su, L., Chen, L., Egli, M., Berger, J.M., and Rich, A. 1999. Minor groove RNA triplex in the crystal structure of a ribosomal frame-shifting viral pseudoknot. Nat. Struct. Biol. 6: 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam, E., Brierley, I., Inglis, S., and Pleij, C. 1994. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucleic Acids Res. 22: 2304–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi, Z. and Kornberg, A. 1990. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. 87: 2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, C., Tzeng, T.H., and Bruenn, J.A. 1992. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl. Acad. Sci. 89: 8636–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, T.A. and Ecker, D.J. 1992. Enhancement of ribosomal frame-shifting by oligonucleotides targeted to the HIV gag-pol region. Nucleic Acids Res. 20: 3945–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Wills, N.M., Du, Z., Rangan, A., Atkins, J.F., Gesteland, R.F., and Hoffman, D.W. 2002. Comparative studies of frameshifting and nonframeshifting RNA pseudoknots: A mutational and NMR investigation of pseudoknots derived from the bacteriophage T2 gene 32 mRNA and the retroviral gag-pro frameshift site. RNA 8: 981–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, R.B., Dunn, D.M., Atkins, J.F., and Gesteland, R.F. 1987. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 52: 687–693. [DOI] [PubMed] [Google Scholar]
- ———. 1990. Ribosomal frameshifting from −2 to +50 nucleotides. Prog. Nucleic Acid Res. Mol. Biol. 39: 159–183. [DOI] [PubMed] [Google Scholar]
- Yusupova, G.Z., Yusupov, M.M., Cate, J.H., and Noller, H.F. 2001. The path of messenger RNA through the ribosome. Cell 106: 233–241. [DOI] [PubMed] [Google Scholar]