Abstract
RNA interference involves endonucleolytic cleavage of mRNAs at a site determined by complementary siRNAs. Initial cleavage leads to rapid degradation of the message, resulting in a corresponding reduction in the level of the encoded protein. Despite intensive study, the identity of the endonucleolytic activity (designated slicer) has remained obscure. Now, a combination of structural and biochemical analyses provide compelling evidence that human Argonaute2 (Ago2), a protein already known to be a key player in the RNAi pathway, is in fact the missing endonuclease.
Keywords: RNA interference, RISC, Argonaute proteins, Piwi domain, endonuclease, slicer
RNA interference (RNAi) is an evolutionarily conserved cellular response to the presence of double-stranded RNA (dsRNA). As such, RNAi is thought to represent an ancient defense mechanism that protects organisms from viral infections and mobile genetic elements (Plasterk 2002). Although first described in Caenorhabditis elegans (Fire et al. 1998), further studies revealed that RNAi is related to a previously described phenomenon called post-transcriptional gene silencing (PTGS) in plants. Since then it has become clear that the RNAi pathway belongs to a family of related processes that lead to sequence-specific gene silencing triggered by dsRNA. These pathways act not only at the post-transcriptional level by eliciting mRNA decay or inhibiting translation, but also at the transcriptional level by altering the conformation of the chromatin (for review, see Denli and Hannon 2003; Dykxhoorn et al. 2003; Bartel 2004; Novina and Sharp 2004).
The process of RNAi is triggered by the presence of long dsRNA molecules in the cells. During the initiation step of RNAi, these dsRNA molecules are cleaved into 21–23 nucleotide (nt) small-interfering RNA duplexes (siRNAs) by Dicer, a conserved family of enzymes containing two RNase III-like domains (Bernstein et al. 2001; Elbashir et al. 2001). The siRNAs are characterized by a 19–21-bp duplex region and 2-nt 3′ overhangs on each strand. During the effector step of RNAi, the siRNAs become incorporated into a multimeric protein complex called RNA-induced silencing complex (RISC), where they serve as guides to select fully complementary mRNA substrates for degradation. Degradation is initiated by endonucleolytic cleavage of the mRNA within the region complementary to the siRNA. More precisely, the mRNA is cleaved at a position 10 nt from the 5′ end of the guiding siRNA (Elbashir et al. 2001; Nykanen et al. 2001; Martinez et al. 2002). The endonuclease responsible for this cleavage was termed slicer, but its identity has remained to be determined.
In this regard, a protein called Tudor-SN was previously identified as a component of RISC in Drosophila, C. elegans, and mammals (Caudy et al. 2003). Because Tudor-SN contains several domains with homology to Staphylococcus nuclease, it was proposed as a candidate for the endonucleolytic activity of RISC. Staphylococcus nuclease homologs are known to require Ca2+ ions and to generate 3′-phosphorylated termini. Mechanistic studies of RISC-mediated cleavage, however, revealed that the reaction requires Mg2+ ions and that the released 5′ product has a hydroxyl group at its 3′ terminus, effectively ruling out Tudor-SN as slicer (Martinez and Tuschl 2004; Schwarz et al. 2004).
Genetic and biochemical studies have shown that the Argonaute proteins are essential for RNAi and that they constitute core components of RISC (Tabara et al. 1999; Hammond et al. 2001; Martinez et al. 2002). They form a conserved family of ~100 kDa, highly basic proteins characterized by an N-terminal PAZ domain and a C-terminal Piwi domain (Cerutti et al. 2000; Carmell et al. 2002).
The first structural insights into the Argonaute proteins came from NMR and X-ray analyses of the Drosophila melanogaster Argonaute2 (DmAgo2) PAZ domain (Lingel et al. 2003; Song et al. 2003), and from the solution structure of the Drosophila Ago1 PAZ domain (Yan et al. 2003). These studies revealed that the PAZ domain contains a nucleic acid-binding fold. Subsequently, the structures of the DmAgo2 and the human Ago1 PAZ domains in complex with RNA or DNA showed that the domain binds specifically to single-stranded 3′ ends of nucleic acids (Lingel et al. 2004; Ma et al. 2004), suggesting that within RISC, the PAZ domain of Argonaute proteins could be involved in the specific recognition of the 2-nt 3′ overhangs of siRNAs. The other signature domain of Argonaute proteins, the Piwi domain, remained largely unexplored at either the structural or functional level.
An archebacterial protein is an Argonaute protein
To gain insight into the possible function of the Piwi domain, Song et al. (2004) performed structural analysis of a multidomain protein from the archebacterium Pyrococcus furiosus. By sequence homology, it was predicted that this protein contained a Piwi domain. Remarkably, the crystal structure of the protein revealed that it also contains an unanticipated PAZ domain. Thus, the protein (now known as PfAgo) represents a bona fide Argonaute protein, indicating that this protein family is not restricted to eukaryotes.
PfAgo consists of four distinct domains held together by an interdomain connector: an N-terminal domain, a PAZ-like domain, a middle domain structurally related to the Lac repressor, and a C-terminal Piwi domain. The N-terminal, middle, and Piwi domains form a crescent-shaped structure, with the Piwi domain sitting in the center of the crescent. The N-terminal domain forms a stalk that holds the PAZ domain on top of the concave face of the crescent, placing it in opposition to the Piwi domain. Overall, the structure is quite compact, considering the length of the polypeptide chain (i.e., 770 residues).
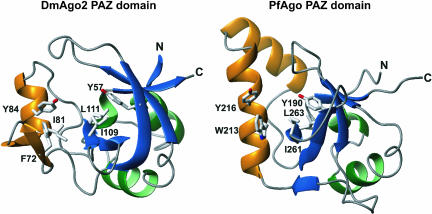
Despite very low sequence identity (<10%), the PAZ-like domain of PfAgo protein superimposes well with the known structures of eukaryotic PAZ domains. However, the structural similarity is not complete, and there are structural elements unique to the PfAgo PAZ domain. Eukaryotic PAZ domains are composed of a central β-barrel, which is flanked on one side by two N-terminal helices and on the other side by a conserved module comprising a β-hairpin and a α-helix (Fig. 1 ▶, the module is shown in orange) (Lingel et al. 2003, 2004; Song et al. 2003; Ma et al. 2004; Yan et al. 2004). In the PfAgo, however, the β-hairpin/α-helix module is replaced by two α-helices (Fig. 1 ▶). In addition, some loop conformations are different. Despite these variations, amino acids previously reported to be important for binding to the 3′ end of nucleic acids, including those in the β-hairpin/α-helix module, are conserved. Their side chains are located in similar spatial positions in the Pyrococcus protein, although some of these residues are at different positions in the protein sequence and protrude from different secondary structural elements (Fig. 1 ▶).
FIGURE 1.
Comparison of the Drosophila melanogaster Argonaute2 PAZ domain (DmAgo2, left) and the PAZ domain of the Argonaute protein from Pyrococcus furiosus (PfAgo, right). Structures are shown as ribbon diagrams, with helices colored green and β-strands shown in blue. The β-hairpin/α-helix module is shown in orange. This module contains two β-helices in the PAZ domain of PfAgo. Side chains of residues shown to contact nucleic acids in structures of the DmAgo2 PAZ domain in complex with nucleic acids are shown in gray and are labeled (Lingel et al. 2004). Residues in equivalent positions in the PfAgo protein are indicated.
Key features of nucleic acid recognition by the PAZ domain involves base interactions with conserved aromatic residues. Residues Y190 and Y216 of PfAgo are equivalent to Y57 and Y84 of DmAgo2 PAZ domain. Residue F72 of DmAgo2 PAZ domain is critical for nucleic acid recognition, as mutation of this residue to alanine abolishes nucleic acid binding (Lingel et al. 2003, 2004). This position is occupied by another aromatic residue, W213, in PfAgo. On the basis of these similarities, Song et al. (2004) suggest that the PAZ domain of PfAgo could also bind to single-stranded 3′ ends of RNAs. However, the function of this Argonaute-like protein in archea remains to be established.
The Piwi domain adopts an RNase H fold
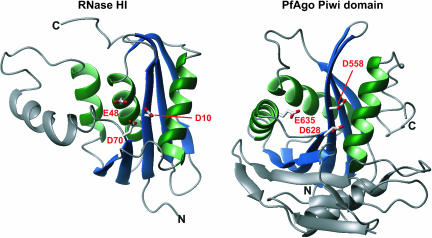
The structure of the full-length PfAgo also provides the first structural view of a Piwi domain (Song et al. 2004). The Piwi domain of PfAgo shows a striking structural similarity and conserved secondary structure topology with the family of RNase H enzymes (Fig. 2 ▶). This class of enzymes is known to cleave the RNA strand of an RNA/DNA hybrid (Yang and Steitz 1995). The RNase H fold consists of a five-stranded mixed β-sheet surrounded by α-helices on both sides (Fig. 2 ▶). The active site is comprised of a so-called DDE-motif, three acidic amino acids with the side-chain carboxylates positioned to catalyze the cleavage reaction. The reaction is known to require the presence of divalent cations, such as Mg2+ or Mn2+. The reaction mechanism differs from most ribonucleases, but resembles deoxyribo-nucleases, leaving 3′-OH and 5′ phosphate termini. Notably, these are the same products resulting from slicer activity (Martinez and Tuschl 2004; Schwarz et al. 2004). In the Piwi domain of the PfAgo protein, two of the three potentially catalytic carboxylates (D558 and D628) are in equivalent secondary structure positions as those found in the RNase H fold. Specifically, these aspartates are equivalent to D10 and D70 in Escherichia coli RNase H1 (Fig. 2 ▶; Katayanagi et al. 1993). The location of the third carboxylate (E48 in RNase H1) is not conserved, but another carboxylate (E635) is found in close spatial distance to the two aforementioned side chains, completing a potential active site. Thus, D558, D628, and E635 form the DDE motif in PfAgo. Strikingly, this putative catalytic center is placed in a cleft in the concave side of the crescent, facing the PAZ domain.
FIGURE 2.
The Piwi domain of the PfAgo protein comprises an RNase H fold. The structure of RNase HI from E. coli is shown on the left (RCSB protein data bank accession no. 1RDD) (Katayanagi et al. 1993), the Piwi domain of the PfAgo protein on the right. Secondary structure elements that constitute the RNase H fold are colored as in Figure 1 ▶; additional structural elements are shown in gray. Catalytic side chains of RNase HI and the corresponding side chains in the PfAgo protein are indicated and labeled. Figures 1 ▶ and 2 ▶ were prepared with the programm MOLMOL (Koradi et al. 1996).
Human Ago2-containing RISCs mediate siRNA-guided cleavage of mRNA
The structure of PfAgo clearly suggested the possibility that Argonaute proteins were the likely nucleases in RISC. To explore this notion, Liu et al. (2004) investigated the roles of different human Argonaute proteins in RISC activity. Myc-tagged versions of Ago1 (eIF2C1), Ago2 (eIF2C2), or Ago3 (nomenclature according to Carmell et al. 2002) were transiently expressed in human 293T cells. Cells were cotransfected with a siRNA. Following immunoprecipitation, the different complexes were tested for siRNA-guided cleavage of a complementary synthetic mRNA. Only Ago2-associated RISC was able to catalize cleavage, even though all Argonaute proteins bound the cotransfected siRNA in similar amounts.
To investigate further the role of Ago2 in the cleavage reaction, the authors then attempted to reconstitute RISC-mediated cleavage activity in vitro. To this end, myc-tagged Ago2 was immunopurified from transiently transfected 293T cells; in this case, from cells that were not cotransfected with siRNAs. The cleavage assays were then performed after addition of siRNAs in vitro. Addition of a double-stranded siRNA did not result in cleavage of the target mRNA, but addition of the anti-sense strand of the siRNA generated RISC activity. To test whether the cleavage activity was an intrinsic feature of Ago2, or whether it was contributed by a copurified Ago2-associated factor, the authors introduced mutations into the Ago2 protein. As described above, two conserved aspartates were identified as potential catalytical residues in the active site of the RNase H-like Piwi domain of PfAgo (i.e., D558 and D628). When each of the aspartates present at equivalent positions in human Ago2 were changed into alanines, the purified mutant proteins failed to assemble into cleavage-competent RISCs, either after cotransfection of cells with siRNAs or after reconstituting RISC in vitro. Importantly, the mutant proteins were expressed at wild-type levels, and associated with siRNAs to a similar extent, a result that showed that the failure to cleave the mRNA was not due to impaired siRNA binding. Recent results from Tuschl and coworkers add additional support for a role of Ago2 in cleavage (Meister et al. 2004). They showed that purified Ago2 complexes, but not Ago1, Ago3, or Ago4 complexes, had RISC activity (Meister et al. 2004).
Together, the structural homology between the Piwi domain and RNase H, the observation that mutation of the predicted catalytic carboxylates in Ago2 abolishes siRNA-guided cleavage activity, and the similar features of the cleavage reaction (Mg2+ dependence, product chemistry) provide compelling evidence that human Ago2 is the source of the slicer activity in the RNAi pathway.
Argonaute proteins as slicer
On the basis of insights gained from the structure of PfAgo, Song et al. (2004) propose a molecular model for the RISC cleavage reaction. To model the RNA on the protein, they aligned the PAZ domain of PfAgo with the structure of the human Ago1 PAZ domain in complex with an siRNA-like duplex (Ma et al. 2004). The strand with its 3′-end bound by the PAZ domain was considered to represent the siRNA guide (i.e., the strand complementary to the mRNA). The mRNA was modeled by extending the second strand of the duplex. By doing so, the mRNA strand is positioned directly above the putative active site of the Piwi domain. Moreover, hydrolysis would occur at a position that was shown in previous experiments to be the scissile phosphate of the mRNA (Elbashir et al. 2001). This model fits nicely with the slicer activity of Argonaute proteins.
An unsolved question is why, despite conservation of the two putative catalytic aspartates among known Piwi domains, not all Argonaute proteins exhibit cleavage activity. As shown by Liu et al. (2004) and Meister et al. (2004), human Ago1- and Ago3-associated RISCs fail to mediate the cleavage reaction. Lack of activity might correlate with amino-acid differences within the Piwi domain, or perhaps other regions of the Argonaute protein. Alternatively, ancillary RISC components could contribute to substrate recognition and to the formation of a catalytically active conformation. Such structural variations could determine whether an active enzyme is generated or not. Remarkably, there is precedent for such variations in other RNase H homologs (e.g., the Tn5 transposase inhibitor [Davies et al. 2000] and HIV-1 reverse transcriptase [Sarafianos et al. 2001]). Moreover, variations in substrate specificity between RNase H type 1 and type 2 proteins have been linked to the presence of an additional C-terminal domain in type 2 enzymes (Lai et al. 2000; Chapados et al. 2001).
Interestingly, human Ago1- through Ago3-associated complexes have the ability to bind siRNAs, and similarly, human Ago1- through Ago4-associated complexes contain miRNAs (Meister et al. 2004). The fact that Argonaute proteins could have evolved to serve different functions in different complexes fits well with the known fact that silencing phenomena are diverse and are not only based on mRNA degradation, but also on translational inhibition by micro-RNAs and heterochromatin formation. Future structural and biochemical studies will undoubtedly provide further understanding for the molecular basis of functional heterogeneity among Argonaute proteins.
Acknowledgments
We thank Elena Conti and Michael Sattler for critical reading of the manuscript and Leemor Joshua-Tor for providing the coordinates of the PfAgo protein prior to release.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7175704.
REFERENCES
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. 2002. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 16: 2733–2742. [DOI] [PubMed] [Google Scholar]
- Caudy, A.A., Ketting, R.F., Hammond, S.M., Denli, A.M., Bathoorn, A.M., Tops, B.B., Silva, J.M., Myers, M.M., Hannon, G.J., and Plasterk, R.H. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414. [DOI] [PubMed] [Google Scholar]
- Cerutti, L., Mian, N., and Bateman, A. 2000. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25: 481–482. [DOI] [PubMed] [Google Scholar]
- Chapados, B.R., Chai, Q., Hosfield, D.J., Qiu, J., Shen, B., and Tainer, J.A. 2001. Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J. Mol. Biol. 307: 541–556. [DOI] [PubMed] [Google Scholar]
- Davies, D.R., Goryshin, I.Y., Reznikoff, W.S., and Rayment, I. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289: 77–85. [DOI] [PubMed] [Google Scholar]
- Denli, A.M. and Hannon, G.J. 2003. RNAi: An ever-growing puzzle. Trends Biochem. Sci. 28: 196–201. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn, D.M., Novina, C.D., and Sharp, P.A. 2003. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell. Biol. 4: 457–467. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Katayanagi, K., Okumura, M., and Morikawa, K. 1993. Crystal structure of Escherichia coli RNase HI in complex with Mg2+ at 2.8 A resolution: Proof for a single Mg(2+)-binding site. Proteins 17: 337–346. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- Lai, L., Yokota, H., Hung, L.W., Kim, R., and Kim, S.H. 2000. Crystal structure of archaeal RNase HII: A homologue of human major RNase H. Structure Fold Des. 8: 897–904. [DOI] [PubMed] [Google Scholar]
- Lingel, A, Simon, B., Izaurralde, E., and Sattler, M. 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 11: 576–577. [DOI] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. 2004. Argonaute2 Is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Ma, J.B., Ye, K., and Patel, D.J. 2004. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J. and Tuschl, T. 2004. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes & Dev. 18: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., Tuschl, and T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15: 185–197. [DOI] [PubMed] [Google Scholar]
- Novina, C.D. and Sharp, P.A. 2004. The RNAi revolution. Nature 430: 161–164. [DOI] [PubMed] [Google Scholar]
- Nykanen, A., Haley, B., and Zamore, P.D. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321. [DOI] [PubMed] [Google Scholar]
- Plasterk, R.H. 2002. RNA silencing: The genome’s immune system. Science 296: 1263–1265. [DOI] [PubMed] [Google Scholar]
- Sarafianos, S.G., Das, K., Tantillo, C., Clark Jr., A.D., Ding, J., Whit-comb, J.M., Boyer, P.L., Hughes, S.H., and Arnold, E. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20: 1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D.S., Tomari, Y., and Zamore, P.D. 2004. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr. Biol. 14: 787–791. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Liu, J., Tolia, N.H., Schneiderman, J., Smith, S.K., Martienssen, R.A., Hannon, G.J., and Joshua-Tor L. 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10: 1026–1032. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Smith, S.K., Hannon, G.J., and Joshua-Tor, L. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132. [DOI] [PubMed] [Google Scholar]
- Yan, K.S., Yan, S., Farooq, A., Han, A., Zeng, L., and Zhou, M.M. 2003. Structure and conserved RNA binding of the PAZ domain. Nature 426: 468–474. [DOI] [PubMed] [Google Scholar]
- Yang, W. and Steitz, T.A. 1995. Recombining the structures of HIV integrase, RuvC and RNase H. Structure 3: 131–134. [DOI] [PubMed] [Google Scholar]