Abstract
Immuno- and other affinity-purification approaches are commonly used to characterize the composition of ribonucleoprotein complexes. While associations detected by these procedures are often interpreted as reflecting in vivo interactions, it is also possible that they arise from reassociation of molecules after cell lysis. Here we used an experimental approach that allowed us to distinguish between these possibilities. Surprisingly, we show that the association of the RNA-binding protein HuR with its target mRNA, c-fos, as detected by co-immunoprecipitation, results largely from reassociation of molecules subsequent to cell lysis. The existence of such postlysis reassortments thus demonstrates that co-immunoprecipitation does not always recapitulate the in vivo state of ribonucleoprotein complexes.
Keywords: immunoprecipitation, RNA-binding, HuR, re-association, ribonucleoprotein
An increasingly used approach to characterize ribonucleoprotein complexes associated with specific RNA-binding proteins relies on immuno- or affinity-purification procedures followed by identification of the copurifying RNA or protein molecules. The interpretation of such experiments is often based on the assumption that during the experimental manipulations the integrity of in vivo assembled RNA–protein complexes is preserved and that the observed associations therefore reflect in vivo interactions. However, the validity of this assumption has not been tested experimentally. Here we asked whether an RNA-binding protein expressed in one cell population can associate after cell lysis with a target RNA expressed in a different cell population. Counter to common expectation, significant levels of binding were observed, indicating that co-immunoprecipitation may not always recapitulate the in vivo state of ribonucleoprotein complexes.
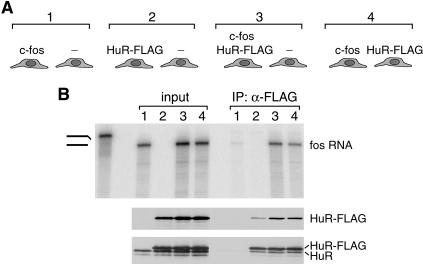
For analysis, we focused on the interaction between the well-characterized RNA-binding protein HuR and its target mRNA c-fos (Brennan and Steitz 2001). The experimental design is shown schematically in Figure 1A ▶. Specifically, plasmids expressing HuR or c-fos were transfected together or separately into HEK293 cells. The transfected HuR carried a FLAG tag at its C-terminus to distinguish the exogenously expressed protein from that endogenous to the transfected cells. Endogenous c-fos mRNA is not detectable in HEK293 cells under the conditions used and therefore does not interfere with the analysis (Fig. 1B ▶, lane 2). The indicated cell populations (Fig. 1A ▶) were mixed, lysed, and the extracts subjected to immunoprecipitation with anti-FLAG antibodies under conditions similar to those generally used in these types of experiments (Steitz 1989; Pinol-Roma et al. 1990; Tenenbaum et al. 2002).
FIGURE 1.
A. Schematic diagram of the populations of HEK293 cells examined in the mixing experiments. Cells were transfected (using TransIT 293 transfection reagent, Mirus) with the indicated plasmids 24 h before harvesting: c-fos: plasmid containing the c-fos gene under control of the CMV promoter (kindly provided by Dr. Nicholas K. Conrad); HuR-FLAG: plasmid expressing FLAG-tagged HuR under control of the CMV promoter (Fan and Steitz 1998); -: pcDNA3 empty vector (Invitrogen). B. The cells described in A were scraped in PBS and the populations indicated in brackets in A were mixed just before lysis. Cells were pelleted by centrifugation and cell pellets were resuspended in a buffer containing 10 mM Tris-Cl (pH 7.5), 100 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100 and disrupted by brief sonication (2 × 5 sec at setting 2.5) on ice. Extracts were centrifuged at 10,000 g for 10 min and subjected to IP with the monoclonal M2 anti-FLAG antibody (Sigma) for 1.5 h at 4°C. The immunoprecipitated material was released from the beads in a buffer containing 1% SDS and 10 mM DTT. A fraction of the input and immunoprecipitated material was analyzed by RNase protection essentially as described (Tymms 1995) with a probe hybridizing to the last exon of the human c-fos RNA (upper panel). First two lanes: undigested probe (0.025% loaded) and digested probe (100% loaded), respectively. The remaining material was analyzed by western blot with rabbit polyclonal anti-FLAG antibody (Sigma) (middle panel); the membrane was stripped and reprobed with the 3A2 monoclonal antibody against HuR (Gallouzi et al. 2000) (lower panel).
As expected, significant levels of c-fos mRNA co-immunoprecipitate with HuR-FLAG when the two components were transfected into the same cells (Fig. 1B ▶, lane 3). Remarkably, significant levels of co-immunoprecipitation are also observed when the RNA-binding protein and the target RNA were expressed in different cells (Fig. 1B ▶, lane 4). It should be noted that the apparent reduction in the amount of coprecipitating c-fos RNA when expressed in a different cell population than HuR-FLAG (compare IP, lanes 3,4) is likely due to the reduced relative expression of c-fos in these cells (compare input, lanes 3,4). This probably results from the stimulatory effect of HuR on expression of c-fos (Fan and Steitz 1998; Peng et al. 1998). This result unambiguously indicates that HuR expressed in one cell population efficiently binds to c-fos mRNA expressed in a different cell population after cell breakage.
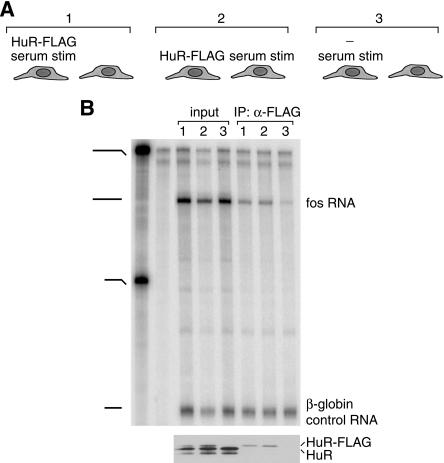
In the experiment described above, the analyzed associations occur between a protein and a target RNA that are both expressed at levels higher than those of their endogenous counterparts. We therefore asked whether the extensive degree of reassociations was due to overexpression of the two binding partners. In NIH/3T3 cells, HuR-FLAG can be expressed at levels significantly lower than the endogenous protein (Fig. 2B ▶, lower panel) and endogenous c-fos mRNA transcription can be induced by serum stimulation. Figure 2B ▶ shows that similar amounts of endogenous c-fos mRNA associate with HuR-FLAG regardless of whether the protein and RNA were expressed in the same or in different populations of NIH/3T3 cells, thus ruling out a role for overexpression as a causative factor of the observed reassociations.
FIGURE 2.
A. Schematic diagram depicting treatments of specific populations of NIH/3T3 cells. HuR-FLAG: transfection (using Effectene transfection reagent, Qiagen) with a plasmid expressing FLAG-tagged HuR; -: transfection with pcDNA3 empty vector. Efficiency of transfection was ~30%–40%. Twenty-four hours after transfection all cells were serum-starved for 24 h in media containing 0.3% bovine serum. Where indicated by “serum stim”, cells were stimulated with 20% bovine serum for 45 min. B. Cells treated as described in A were scraped in PBS and the populations indicated in brackets in A were mixed just before lysis. Lysis, immunoprecipitation, and analysis of the samples were carried out as described in Figure 1 ▶, except that after immunoprecipitation, the same amount of in vitro transcribed β-globin RNA was added to each sample to control for differences in recovery of the RNA during the subsequent steps. Upper panel: RNase protection assay performed with a probe hybridizing to the last exon of the mouse c-fos RNA and a probe complementary to the exogenously added β-globin RNA. First two lanes: undigested probes (0.025% loaded) and digested probes (100% loaded) respectively. Lower panel: Western blot using the 3A2 monoclonal antibody against HuR. Note that under the conditions used, c-fos mRNA from non-stimulated cells is not detectable (data not shown).
These data therefore demonstrate that HuR can reassociate with c-fos mRNAs after cell lysis and that the observed association between HuR and c-fos mRNA results largely from interaction of molecules in the cell extract. Whether and the extent to which other RNA–protein complexes undergo similar reassortments will likely vary and depend on the affinity of each particular protein for its RNA target site(s), the existence of factors that influence this binding (e.g., through cooperative interactions), as well as on the specific experimental conditions used. For instance, a similar experimental approach indicated that ribonucleoprotein complexes associated with the RNA-binding protein nucleolin do not undergo significant rearrangements after cell lysis (F. Triolo and S. Pinol-Roma, pers. comm.).
Immuno- or affinity-purification approaches can provide valuable information regarding the specificity and potential for formation of a particular interaction. However, the data presented here demonstrate that, at least in certain cases, such interactions might not reflect the in vivo state of complexes but might arise from associations generated after cell lysis. While such interactions may be indicative of an in vivo association, they could also be artificially generated in vitro. For example, since molecules in an extract can freely diffuse, interactions that were prevented in vivo due to differential compartmentalization might occur. Furthermore, the RNA-binding activity of proteins with overlapping binding specificities could be differentially affected in an extract (because of differential sensitivity of each interaction to the specific lysis conditions, requirements for cofactors, posttranslational modifications, etc). This could lead to the exposure of binding sites that were not accessible in vivo or, conversely, to the masking of true interaction sites. Taking these possibilities into account, we therefore recommend that in the absence of additional evidence, caution should be exercised in interpreting associations detected in immunopurification experiments as reflecting true in vivo interactions.
Acknowledgments
We thank S. Pinol-Roma and F. Triolo for helpful discussions and their influence in the design of the experimental approach, K. Tykowski and V. Fok for helpful suggestions on the manuscript, N.K. Conrad for generously providing reagents, and Mei-Di Shu for valuable technical advice. This work was supported by grant GM 26154 from the NIH. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7151404.
REFERENCES
- Brennan, C.M. and Steitz, J.A. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58: 266–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X.C. and Steitz, J.A. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17: 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi, I.E., Brennan, C.M., Stenberg, M.G., Swanson, M.S., Eversole, A., Maizels, N., and Steitz, J.A. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. 97: 3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S.S., Chen, C.Y., Xu, N., and Shyu, A.B. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17: 3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma, S., Choi, Y.D., and Dreyfuss, G. 1990. Immunological methods for purification and characterization of heterogeneous nuclear ribonucleoprotein particles. Meth. Enzymol. 181: 317–325. [DOI] [PubMed] [Google Scholar]
- Steitz, J.A. 1989. Immunoprecipitation of ribonucleoproteins using autoantibodies. Meth. Enzymol. 180: 468–481. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, S.A., Lager, P.J., Carson, C.C., and Keene, J.D. 2002. Ribonomics: Identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods 26: 191–198. [DOI] [PubMed] [Google Scholar]
- Tymms, M.J. 1995. Quantitative measurement of mRNA using the RNase protection assay. Methods Mol. Biol. 37: 31–46. [DOI] [PubMed] [Google Scholar]