Abstract
A comparison of Arabidopsis DNA sequences revealed that the final nucleotides at the 3′ end of approximately half of the Arabidopsis mRNAs, immediately upstream of the poly(A) tail, differ from the corresponding genomic sequences. This suggests that extra nucleotides were added to these mRNAs at their 3′ termini prior to polyadenylation. Among the mRNAs containing additional nucleotides, approximately 65% had a single additional nucleotide, with the nucleotide C added most often. This nontemplated addition before the addition of the poly(A) sequence could be a major contributing factor to the often observed heterogeneity in transcription products. These findings should be helpful in the elucidation of the mechanisms of mRNA 3′-end processing.
Keywords: cleavage/polyadenylation site, nontemplated nucleotides, 3′-end processing
The 3′ ends of most eukaryotic mRNAs contain a poly(A) tail, which is synthesized by poly(A) polymerase at specific pre-mRNA sites after an endonucleolytic cleavage (Colgan and Manley 1997; Zhao et al. 1999). The site of cleavage in the pre-RNA is determined by the position of the regulatory elements and the nucleotide composition of the cleavage region. This region is located 11–24 nt downstream from an AAUAAA element and 10–30 nt upstream of a U/GU-rich element. Several tools have been designed to predict the 3′-processing sites in mRNAs of the yeast Saccharomyces cerevisiae (Graber et al. 2002). However, the precise position and detailed mechanisms of the 3′ processing of mRNAs are often unclear.
In this study, we mapped the 3′ ends of Arabidopsis mRNAs to the corresponding sites in the genome using raw full-length cDNA data (Seki et al. 2002). A total of 4,145 Arabidopsis cDNAs with poly(A) tails were selected (the accession numbers can be obtained at http://www.cls.zju.edu.cn/sub/shsjyf). Alignment of the cDNA sequences to the sequences of the five Arabidopsis thaliana chromosomes (http://signal.salk.edu/dblast.html) revealed that greater than 41% of the 3′-end nucleotides immediately upstream of the poly(A) sequence differed from the genomic sequence. A similar phenomenon was observed in full-length hypothetical cDNAs from chromosome 2 of Arabidopsis (Xiao et al. 2002). In rice, approximately 15% of the cDNAs were found to contain a 3′ nontemplated nucleotide addition prior to the poly(A) sequence, based on a study of 2,100 full-length cDNA sequences (GenBank accession numbers AK064803–AK066902). These observations indicate that this discrepancy occurs in at least some eukaryotic species, although such a high rate of discrepancies was not observed during an analysis of ESTs from mammalian and yeast cells using the same methods.
The above results suggest that extra nucleotides were added at the 3′ termini of the mRNAs in a nontemplated manner prior to polyadenylation. This finding has profound implications for the understanding of the precise position and mechanisms of the 3′ processing of mRNAs. In the maize mitochondrial rps12, cox2, and atp9 mRNAs, one to four nongenomically encoded nucleotides are present at the 3′ termini (Williams et al. 2000). In contrast to these nonpolyadenylated mitochondrial RNAs, most eukaryotic mRNAs contain a poly(A) tail (Colgan and Manley 1997; Zhao et al. 1999), which conceals the 3′-processing steps between the cleavage and the polyadenylation addition. It is likely that significant changes occur between these two steps, including nontemplated addition at the 3′ termini (Fig. 1 ▶). This nontemplated addition prior to polyadenylation could be a major contributing factor to the heterogeneity observed in some eukaryotic transcription products (Fig. 2 ▶).
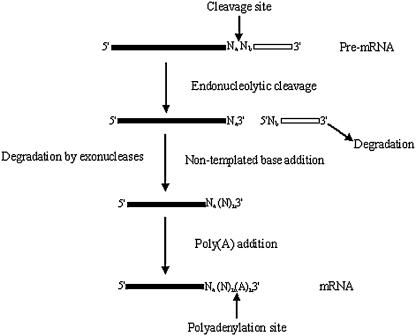
FIGURE 1.
A revised model for the 3′-end processing of mRNA precursors. A primary transcript is cleaved endonucleolytically at the poly(A) site, followed by nontemplated base addition or/and degradation by exonucleases, which is then followed by the addition of adenylate residues to the 3′ end of the upstream fragment to form a poly(A) tail.
FIGURE 2.

Heterogeneity analysis of the 3′ termini of GAPDH mRNAs in Arabidopsis EST libraries. (Δ) The corresponding genomic sequence.
The above ratio for the occurrence of nontemplated addition in Arabidopsis is certainly underestimated, because the addition of nontemplated A nucleotides or nontemplated nucleotides that are identical to the genomic sequence are not included. Therefore, the actual percentage of mRNAs containing nontemplated nucleotide additions prior to polyadenylation is estimated to be higher, approximately 70%. Among the cDNAs observed to have additional nucleotides, approximately 65% contained a single additional nucleotide, one-third contained two additional nucleotides, and a minor fraction contained more than two additional nucleotides. Although the single extra nucleotides could be G, A, T, or C residues, the extra nucleotides on the mRNAs were most often a single C residue (47%). Similarly, mRNAs containing two extra nucleotides were usually two consecutive C residues (27%) (Fig. 3 ▶). Therefore, the nucleotide C is preferentially added to the 3′ ends of mRNAs prior to polyadenylation.
FIGURE 3.
Histogram of distribution of nontemplated nucleotides (shaded bars) and templated nucleotides (no apparent nontemplated residues, open bars) immediately upstream of the poly(A) sequence. (A) the penultimate nucleotide upstream of the poly(A) tail; (B) the binucleotide upstream of the poly(A) tail.
The 50 nt immediately upstream of the poly(A) tail in the cDNAs were also analyzed. As shown in Figure 4 ▶, the A-rich element, frequently AAUAAA, which is approximately centered on position -20, was present in the Arabidopsis cDNAs. T-rich segments often preceded the 3′-end processing site, but C-rich sequences were dominant just upstream of the poly(A) (Fig. 4 ▶). In general, the proportion of C in the 50 nt immediately upstream of the poly(A) tail was less than 20%, but the C content strongly increased near the poly(A) tail (Fig. 4 ▶).
FIGURE 4.
Frequencies of single nucleotides preceding the 3′-end processing sites in Arabidopsis full-length cDNAs containing nontemplated nucleotide additions just prior to the poly(A) sequence. The positions are shown relative to the putative 3′-end cleavage site.
The addition of nontemplated nucleotides to the 3′ termini of mRNAs prior to polyadenylation is a novel observation. However, the mechanism of these additions is unknown. Nontemplated nucleotide addition prior to polyadenylation is probably carried out by a novel low-specificity terminal transferase that is distinct from poly(A) polymerase, along with a tRNA-specific terminal transferase. This novel nucleotidyltransferase probably collaborates with poly(A) polymerase to synthesize a nontemplated sequence at the 3′ termini of mRNAs.
Although the biological significance of this nontemplated addition is unknown at present, much evidence indicates that the function of the nontemplated addition extends beyond simply the addition of nontemplated nucleotides (Miller et al. 1986; Price et al. 1995; Sun et al. 1996; Murphy & Park 1997; Tretheway et al. 2001). The penultimate nucleotide or the last two nucleotides are most often C residues, observed in approximately 50% of all of the genes analyzed. Thus, a CAA or CCA trinucleotide defines the poly(A) site for most genes. Although it is not clear whether the sequences CCA or CAA at the 3′ end constitute a functional box, all tRNAs end in the conserved 3′-CCA, as well as approximately 65% of mature human U2 small nuclear RNAs (snRNAs) and 83% of maize mitochondrial rps12 RNAs (Williams et al. 2000; Cho et al. 2001). Therefore, it is possible that a nontemplated addition prior to polyadenylation is involved in mRNA metabolism, independently or in conjunction with polyadenylation. These findings imply that a system exists that acts midway between the CCA addition in eukaryotic cells and polyadenylation. In any event, the nontemplated nucleotide addition prior to polyadenylation is an interesting and unexpected phenomenon.
Acknowledgments
This work is partially supported by the Nation Natural Science Foundation of China (NO: 30370759).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7610404.
REFERENCES
- Cho, H.D., Tomita, K., Suzuki, T., and Weiner, A.M. 2001. U2 small nuclear RNA is a substrate for the CCA-adding enzyme (tRNA nucleotidyltransferase). J. Biol. Chem. 277: 3447–3455. [DOI] [PubMed] [Google Scholar]
- Colgan, D. and Manley, J. 1997. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 11: 2755–2766. [DOI] [PubMed] [Google Scholar]
- Graber, J.H., McAllister, G.D., and Smith, T.F. 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W.A., Bujarski, J.J., Dreher, T.W., and Hall, T.C. 1986. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J. Mol. Biol. 187: 537–546. [DOI] [PubMed] [Google Scholar]
- Murphy, S.K. and Park, G.D. 1997. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the paramyxovirus simian virus 5. Virology 232: 144–157. [DOI] [PubMed] [Google Scholar]
- Price, S.R., Ito, N., Outbridge, C., Avis, J.M., and Nagai, K. 1995. Crystallization of RNA–protein complexes. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol. 249: 398–408. [DOI] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A., Akiyama, K., Oono, Y., et al. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145. [DOI] [PubMed] [Google Scholar]
- Sun, J.H., Adkins, S., Faurote, G., and Kao, C.C. 1996. Initiation of (2)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: Synthesis of oligonucleotides. Virology 226: 1–12. [DOI] [PubMed] [Google Scholar]
- Tretheway, D.M., Yoshinari, S., and Dreher, T.W. 2001. Autonomous role of 3′-terminal CCCA in directing transcription of RNAs by Qβ replicase. J. Virol. 75: 11373–11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M.A., Johzuka, Y., and Mulligan, R.M. 2000. Addition of non-genomically encoded nucleotides to the 3′-terminus of maize mitochondrial mRNAs: Truncated rps12 mRNAs frequently terminate with CCA. Nucleic Acids Res. 28: 4444–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y.L., Malik, M., Whitelaw, C.A., and Town, C.D. 2002. Cloning and sequencing of cDNAs for hypothetical genes from chromosome 2 of Arabidopsis. Plant Physiol. 130: 2118–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]