Abstract
Small RNAs play an important role in regulation of gene expression in eukaryotic and eubacterial cells by modulating gene expression both at the level of transcription and translation. Here, we show that short complementary RNAs can also affect gene expression by stimulating ribosomal frameshifting in vitro. This finding has important implications for understanding the process of ribosomal frameshifting and for the potential application of small RNAs in the treatment of diseases that are due to frameshift mutations.
Keywords: frameshifting, slippery sequence, small RNA, antisense, RNAi
Frameshifting requires that a ribosome be paused at a so-called slippery sequence by the presence of a 3′-positioned structural element, a pseudoknot, or a stem–loop structure (for reviews, see Giedroc et al. 2000; Brierley and Pennell, 2001; Harger et al. 2002). The ribosome then has the opportunity to slip either forward or backward, leading to, for example, a +1 or −1 shift of the reading frame. This process is commonly used by RNA viruses like HIV and SARS coronavirus to express multiple genes at a fixed ratio from a single mRNA. Besides the case of the antizyme, frameshifting is rarely found in cellular mRNAs of eukaryotic cells (Ivanov et al. 2004).
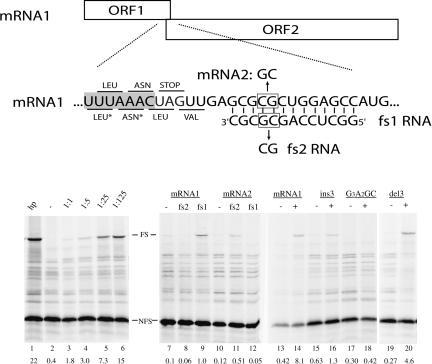
We have found that frameshifting can also occur when the structural element is divided over two RNAs. On the mRNA shown in Figure 1 ▶, the first open reading frame (ORF) is followed by a second ORF that is in the −1 frame with respect to ORF1. Ribosomes translating mRNA1 stop synthesizing the 19-kD protein when they reach the UAG stopcodon at the end of ORF1. In an in vitro translation system, ~0.4% of ribosomes slipped into the −1 frame, due to the inherent slipperiness of the UUUAAAC sequence (Brierley et al. 1992) and produced the longer fusion protein (Fig. 1 ▶, lanes 2,13). Addition of increasing amounts of a 13-nt RNA (fs1) that is complementary to a region downstream of the stopcodon led to a steady increase in the level of the fusion protein (lanes 3–6). At 125-fold molar excess of fs1, ~15% of the translating ribosomes shifted into the −1 frame. By comparison, a 12-bp hairpin of similar base composition showed 22% frameshifting (lane 1). Similar results were obtained with other known slippery sequences (data not shown), whereas a nonslippery sequence (GGGAAGC) was unaffected by the addition of a 40-fold molar excess of fs1 (lanes 17,18). The degree of frameshifting was dependent on base complementarity; frameshifting dropped more than 10-fold when oligonucleotide fs2, having two mismatches, was used (Fig. 1 ▶, cf. lanes 8 and 9). Introduction of compensatory changes in the mRNA restored frameshift enhancement by fs2, but not by fs1 (lanes 11,12). The lower frameshifting activity obtained with the fs2/mRNA2 couple may be due to the formation of intramolecular structures in these RNAs and/or lower thermodynamic stability (ΔΔG37) = + 1.5 kcal/mol) according to Turner rules.
FIGURE 1.
Schematic representation of the frameshift reporter construct. ORF1 (19 kD) is in the 0-frame, ORF2 (46 kD) is in the −1-frame with respect to ORF1. The −1 frameshifting is monitored by the appearance of the 65-kD fusion product. The UUUAAAC slippery sequence is indicated by shading. The 0-reading frame codons are indicated above the sequence, the −1 frame codons below the sequence. (*) Note that during frameshifting, tRNAs for Leu and Asn simultaneously slip into the −1 frame. In mRNA2, two mutations were introduced to restore base-pairing with fs2 RNA. mRNAs were synthesized by SP6 polymerase from a DNA template. SDS–polyacrylamide gel showing [35S]methionine proteins obtained after in vitro translation using rabbit reticulocyte lysate (Promega) as described previously (ten Dam et al. 1994). (Lane 1) A 12-bp hairpin reference; (lanes 2–6: 0, 0.25, 1.25, 6.25, 31.25) pmol of fs1 RNA (IBA GmbH), respectively, were added to 0.25 pmol of mRNA1 and kept at room temperature for 10 min. Reticulocyte lysate was added and incubation was continued for 1 h at 28°C; (lanes 7–9) mRNA1 without, with 40-fold excess fs2 or fs1 RNA, respectively; (lanes 10–12) mRNA2, without, with 40-fold excess fs2 or fs1 RNA, respectively. Note that overall frameshift values for lanes 7–12 are low due to a 30-min incubation time. (Lanes 13,14) mRNA1 without and with 40-fold excess of fs1, respectively; (lanes 15,16) as in lanes 13 and 14, but with insertion of 3 nt downstream of stopcodon in mRNA1; (lanes 17,18) as in lanes 13 and 14, but with slippery sequence mutant GGGAAGC; (lanes 19,20) as in lanes 13 and 14, but with deletion of 3 nt downstream of stopcodon in mRNA1. Migration of 0-frame and frameshifted product are indicated by NFS and FS, respectively. Band intensities were measured by phosphorimaging (Bio-Rad). Frameshifting percentages were calculated as the fraction of FS product divided by the sum of FS and NFS products after correction for methionine content of both products. Indicated values are the average percentage of at least two independent assays.
As found previously for conventional frameshift enhancing elements, both position and thermodynamic stability of the RNA structure were important for frameshifting (ten Dam et al. 1994). Moving the duplex 3 nt farther away (ins3) from the slip site decreased frameshifting (Fig. 1 ▶, cf. lanes 14 and 16), while moving it 3 nt more upstream only partially reduced frameshifting (lane 20), suggesting that in the latter case, still 9 or 10 bp can form when the ribosome is at the slippery sequence. A similar decrease in frameshifting was observed when the complementarity was reduced to 9 bp, whereas a 6-bp duplex resulted in just 1% of frame-shifting activity (data not shown). A DNA oligonucleotide was less capable of enhancing frameshifting, in agreement with the lower stability of RNA–DNA duplexes (data not shown).
Our data show that the duplex mainly serves as a physical barrier for an elongating ribosome, and that there are no base-specific interactions with the ribosome in this system. The utilization of elaborated structures, for example, pseudoknots, as frameshift stimulators by RNA viruses may have other reasons, such as to provide locally a higher stability to obtain higher frameshifting, or to escape the RNAi pathway which may be triggered by a long, continuous duplex.
The separation of frameshift-enhancing elements over two molecules may have certain advantages over a cis-acting signal, as it provides a higher level of regulation, including coactivation of a set of mRNAs. The apparent lack of genes found so far to be regulated by frameshifting in eukaryotic cells might be due to splitting of frameshift signals over two molecules, making it inevitably more difficult to identify them.
Finally, the apparent ease at which small oligonucleotides induce frameshifting has implications for therapeutic use. Many diseases are caused by base deletions or insertions which result in the production of truncated or aberrant proteins due to ribosomes ending up in the wrong reading frame (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM). Binding of a small synthetic oligonucleotide can bring ribosomes back on the right track, provided that a suitable slippery sequence is located nearby. Fortunately, base deletions and insertions often take place at homopolymeric runs of nucleotides which are slippery for ribosomes too.
NOTE ADDED IN PROOF
During the final preparation of this manuscript it came to our attention that similar results were obtained by Howard et al. (2004).
Acknowledgments
We thank Drs. de Smit and Backendorf for useful comments. This research was supported by The Netherlands Organization for Scientific Research (NWO), Chemical Sciences division (CW).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7139704.
REFERENCES
- Brierley, I. and Pennell, S. 2001. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harbor Symposia on Quantitative Biology, pp. 233–248. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Brierley, I., Jenner, A.J., and Inglis, S.C. 1992. Mutational analysis of the “slippery sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 227: 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc, D.P., Theimer, C.A., and Nixon, P.L. 2000. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 298: 167–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger, J.W., Meskauskas, A., and Dinman, J.D. 2002. An “integrated model” of programmed ribosomal frameshifting.Trends Biochem. Sci. 27: 448–454. [DOI] [PubMed] [Google Scholar]
- Howard, M.T., Gesteland, R.F., and Atkins, J.F. 2004. Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides. RNA 10: 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, I.P., Anderson, C.B., Gesteland, R.F., and Atkins, J.F. 2004. Identification of a new antizyme mRNA +1 frameshifting stimulatory pseudoknot in a subset of diverse invertebrates and its apparent absence in intermediate species. J. Mol. Biol. 339: 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam, E.B., Brierley, I., Inglis, S.C., and Pleij, C.W.A. 1994. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucleic Acids Res. 22: 2304–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]