Abstract
All eukaryotic mRNAs possess a 5′-cap (m7GpppN) that is recognized by a family of cap-binding proteins. These participate in various processes, such as RNA transport and stabilization, as well as in assembly of the translation initiation complex. The 5′-cap of trypanosomatids is complex; in addition to 7-methyl guanosine, it includes unique modifications on the first four transcribed nucleotides, and is thus denoted cap-4. Here we analyze a cap-binding protein of Leishmania, in an attempt to understand the structural features that promote its binding to this unusual cap. LeishIF4E-1, a homolog of eIF4E, contains the conserved cap-binding pocket, similar to its mouse counterpart. The mouse eIF4E has a higher Kas for all cap analogs tested, as compared with LeishIF4E-1. However, whereas the mouse eIF4E shows a fivefold higher affinity for m7GTP than for a chemically synthesized cap-4 structure, LeishIF4E-1 shows similar affinities for both ligands. A sequence alignment shows that LeishIF4E-1 lacks the region that parallels the C terminus in the murine eIF4E. Truncation of this region in the mouse protein reduces the difference that is observed between its binding to m7GTP and cap-4, prior to this deletion. We hypothesize that variations in the structure of LeishIF4E-1, possibly also the absence of a region that is homologous to the C terminus of the mouse protein, promote its ability to interact with the cap-4 structure. LeishIF4E-1 is distributed in the cytoplasm, but its function is not clear yet, because it cannot substitute the mammalian eIF4E in a rabbit reticulocyte in vitro translation system.
Keywords: cap-4, cap-binding protein, Leishmania, eIF4E, fluorescence
INTRODUCTION
In eukaryotes, the 5′-cap structure on mRNAs consists of a 7-methyl guanosine that is linked via a triphosphate bridge to the 5′-end of the first transcribed nucleotide, resulting in m7G(5′)ppp(5′)N, where N is any nucleotide (Shatkin 1976). The mRNA cap plays an important role in gene expression. It protects the mRNAs from degradation (Tucker and Parker 2000), enables transport of RNAs from the nucleus to the cytoplasm (Izaurralde et al. 1995), and participates in assembly of the translation initiation complex (Gingras et al. 1999b). The eukaryote initiation factor 4E (eIF4E) binds the 5′-cap, and is part of the trimeric eIF4F complex that also contains eIF4A, an RNA helicase that unwinds the 5′-UTR, and eIF4G, a scaffold protein that interacts with the eIF4F subunits (Gingras et al. 1999b). In mammalian cells, binding of eIF4E and eIF4G can be competed out by a family of 4E-binding proteins (4E-BP) that are involved in repression of translation (Haghighat et al. 1995). A hypermethylated form of the 5′-cap structure, m32,2,7GpppN, denoted trimethyl guanosine (TMG), is found on some U-rich small nuclear RNAs (snRNAs), and functions as a nuclear targeting signal for the corresponding nuclear ribonucleoprotein particles, U snRNPs (Lamond 1990; Gorlich and Mattaj 1996).
Protozoans from the order Kinetoplastida are ancient eukaryotes that diverged rather early in evolution. Transcription of protein-encoded genes in Leishmania and other try-panosomatids is polycistronic, and the resulting pre-mRNAs are processed into mature transcripts by trans-splicing (Kooter et al. 1984; Sutton and Boothroyd 1986) that is coupled with polyadenylation (LeBowitz et al. 1993; Matthews et al. 1994). Trans-splicing involves the joining of a conserved exon (39 nt) derived from the spliced leader (SL) RNA, to the 5′-end of all transcripts. The SL RNA itself is capped, thus processing of the polycistronic pre-mRNA molecules generates capped monocistronic mature mRNAs. In trypanosomatids, the 5′-cap of mRNAs has a unique structure. In addition to 7-methyl guanosine, it includes base methylations on the first (A) and fourth (U) nucleotides, and 2′-O-methylations on the ribose moieties of the first four transcribed nucleotides, resulting in m7Gpppm3 6,6,2′ Apm2′ Apm2′ Cpm23,2′, U, denoted as cap-4 (Bangs et al. 1992). Similar modifications in the 5′-cap were also observed in a cell-free system of Trypanosoma brucei cells (Ullu and Tschudi 1995).
The cap-binding translation factor eIF4E has been studied in a variety of organisms, revealing the existence of multiple homologs. Mammalian cells contain three different isoforms of eIF4E. All three proteins bind m7GTP, but they vary in their ability to interact with eIF4G and with the members of the 4E-BP family of repressors. The exact function of all the different eIF4E isoforms is not fully resolved. They vary in terms of tissue distribution, and only one of them, eIF4E-1, can rescue the growth of a yeast mutant that lacks a functional eIF4E gene (Joshi et al. 2004). Two eIF4E-related genes were found in zebrafish, but only one of them is capable of binding m7GTP. The function of the second protein is yet unknown (Robalino et al. 2004). Differences in the ability to complement the yeast eIF4E were also reported for the two isoforms from Arabidopsis thaliana (Rodriguez et al. 1998). The three-dimensional (3D) structures of the mouse, human, and yeast eIF4E translation factor (Marcotrigiano et al. 1997; Matsuo et al. 1997; Tomoo et al. 2003) have been solved, showing considerable phylogenetic conservation, especially of the cap-binding pocket. In view of the unique structure of the trypanosomatid cap-4, we were interested to examine the cap-binding protein homologs in these organisms. We describe a molecular and biochemical analysis of LeishIF4E-1, an eIF4E homolog from Leishmania, and its ability to bind different cap structures, including cap-4 that was synthetically prepared for this purpose (Lewdorowicz et al. 2004).
RESULTS
Cloning of an eIF4E homolog from Leishmania
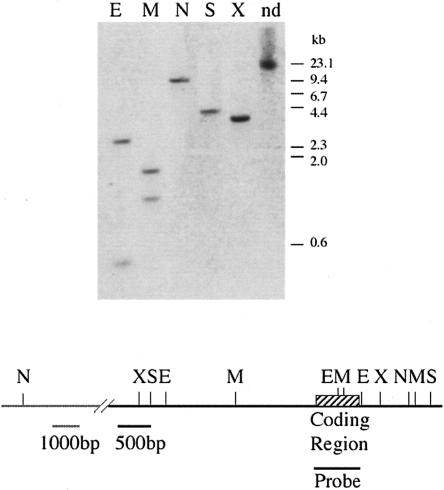
A computerized survey of the Leishmania genome database (LeishDB; http://www.sanger.ac.uk/Projects/L_major/) revealed the presence of an eIF4E homolog (CAB77676), based on its sequence conservation with the mouse eIF4E. The protein was denoted LeishIF4E-1. As protein-coding genes in Leishmania do not have introns within their coding regions, the open reading frame (ORF; 214 amino acids) of LeishIF4E-1 was amplified from genomic DNA. The amino acid sequence of this protein shows 43% similarity (with an E-value of 5e-13) to its mouse counterpart. However, its alignment against the mouse and yeast homologs using Clustal X showed that residues located within the binding pocket are conserved in the Leishmania protein (Fig. 1 ▶). Southern analysis indicated that the cloned gene was present as a single genomic copy (Fig. 2 ▶).
FIGURE 1.
Amino acid sequence alignment of LeishIF4E-1 with the mouse and yeast eIF4E. Sequences of eIF4E from Leishmania (CAB77676), mouse (P20415), and yeast (P07260) were aligned by CLUSTAL X. Residues that are conserved among all three organisms, or between Leishmania and one additional organism, are marked with stars and dots, respectively, below the lines. The conserved residues that are expected to play a role in ligand binding are marked above the lines and are also shadowed.
FIGURE 2.
LeishIF4E-1 is encoded by a single-copy gene. Genomic DNA was digested with different restriction enzymes and separated over a 0.8% agarose gel. The blot was hybridized with a probe derived from the complete coding region of LeishIF4E-1 (645 bp). A map describing the genomic locus is shown below the blot. (E) EcoRI; (M) MspI; (N) NruI; (S) SacII; (X) XhoI; (nd) nondigested.
Computer-based prediction of LeishIF4E-1 structure
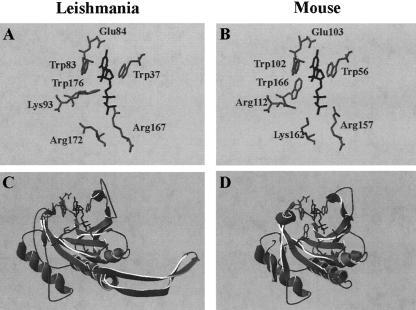
The 3D structure of the mouse and human eIF4E proteins has been determined by X-ray crystallography (Marcotrigiano et al. 1997; Tomoo et al. 2003), and that of the yeast protein by NMR (Matsuo et al. 1997). The murine protein has a shape of a cupped hand, with eight-stranded antipa-rallel β-sheets, backed by three α-helices on its convex side (Marcotrigiano et al. 1997). The interaction between eIF4E and m7GDP occurs in a pocket that contains conserved Trp residues at positions 56, 102, and 166 and a glutamic acid at position 103; these make contacts with the 7-methyl-guanine. The interaction with the negatively charged phosphate chain is stabilized by several basic residues. Homology modeling of LeishIF4E-1 predicts that its 3D structure is compatible with that of the murine protein, except for two short sequence gaps caused by unique nonhomologous sequences in the Leishmania protein, whose structure cannot be predicted, and the lack of a region that is compatible with the C terminus of the mouse and yeast eIF4E (Fig. 3C,D ▶). The cap-binding pocket is highly conserved, as indicated by the conservation between Trp residues 37, 83, and 176 in LeishIF4E-1 and Trp residues 56, 102, and 166 in the murine protein. Similarly, the Leishmania Glu 84 is conserved with murine Glu 103, and the basic amino acids Lys 93, Arg 167, and Lys 172 in LeishIF4E-1 are conserved with the murine Arg 112, Arg 157, and Lys 162 (Fig. 3A,B ▶).
FIGURE 3.
Computer-predicted structure of LeishIF4E-1. The tertiary structure of LeishIF4E-1 (A,C) was predicted by homology modeling using PDBView (B,D), based on the published crystal structure of the mouse eIF4E (Marcotrigiano et al. 1997), pdb 1EJ1a—chain a (this published 3D structure contains several minor discontinuities in the polypeptide chain). The side residues of amino acids that comprise the ligand-binding site are shown in the top panels. The ligand (m7GDP) is marked in black.
LeishIF4E-1 can be affinity-purified over m7GTP-Sepharose
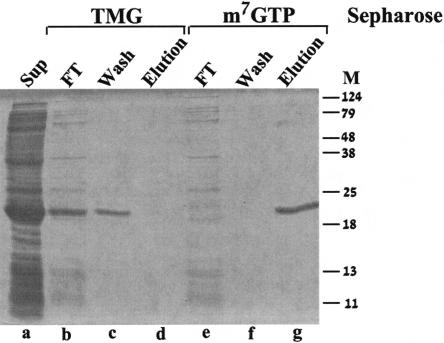
To provide biochemical insight on the function of the Leish-mania IF4E-1, we tested whether recombinant LeishIF4E-1 can bind m7GTP, which is also part of the Leishmania cap-4 structure (Ullu and Tschudi 1995). The soluble crude fraction of bacteria expressing the recombinant LeishIF4E-1 was loaded on an m7GTP-Sepharose column and eluted with high salt (Fig. 4 ▶, lanes e–g). Elution with free m7GTP, but not with GTP, was shown elsewhere (Lewdorowicz et al. 2004). The eluate contained highly purified LeishIF4E-1, indicating that the recombinant protein efficiently bound to the affinity matrix, enabling a single-step purification process.
FIGURE 4.
LeishIF4E-1 binds only to m7GTP and not to TMG. The soluble fraction of E. coli BL21 cells induced by IPTG (lane a) was applied to a TMG-Sepharose column. After collection of the flow-through fraction (lane b), the column was washed with equilibration buffer (lane c) and eluted with the same buffer containing 200 mM NaCl (lane d). The same extract (lane a) was loaded over m7GTP-Sepharose, the flow-through fraction was collected (lane e), and the column was washed with equilibration buffer (lane f) and eluted with the same buffer containing 200 mM NaCl (lane g).
To examine whether LeishIF4E-1 could also bind TMG, a structure found on some U snRNAs that are involved in splicing (Mattaj 1986; Gunzl et al. 2000), the crude bacterial extract containing the induced LeishIF4E-1 was applied onto a TMG-Sepharose column. As shown in Figure 4 ▶, lanes b–d, LeishIF4E-1 was found exclusively in the flow-through fraction, indicating that it did not bind TMG.
LeishIF4E-1 binds cap-4 and m7GTP with similar affinities, whereas the mouse IF4E differentiates between the two compounds
Although the 3D structure of the protein gives important information on the mode of cap binding, its affinity to different analogs can be measured only in solution. Sequence alignment of eIF4E polypeptides from different species showed that the number and positions of tryptophan residues are highly conserved (Marcotrigiano et al. 1997). The mouse eIF4E possesses eight tryptophans—three of them are exposed to the solvent (Trp56, Trp73, Trp102) and the other five residues are buried inside the protein. In LeishIF4E-1, seven of the eight Trp residues are conserved, with replacement of the murine Trp 73 by a Phe residue. However, the emission spectra of both proteins are very similar (data not shown).
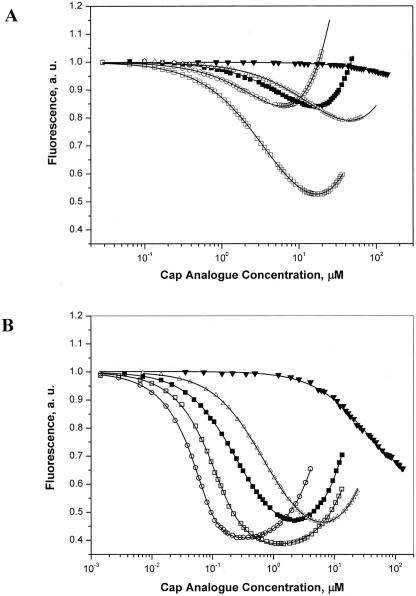
To establish the binding affinity between LeishIF4E-1 and cap-4, as well as with its mono- and dinucleotide cap analogs, we applied a fast and accurate time-synchronized fluorescence titration method, TST, that measures the quenching of intrinsic Trp fluorescence (Niedzwiecka et al. 2002). This method takes into account the emission of the free ligand and the concentration of the active protein, as free parameters of the numerical fitting (see Materials and Methods). Fluorescence measurements with the mouse eIF4E were performed with a truncated recombinant polypeptide (extending between positions 28 and 217) that was previously used for crystallographic studies (Marcotrigiano et al. 1997). To provide a better basis for comparison with the truncated mouse protein, the binding analysis of LeishIF4E-1 was performed with the recombinant protein that initiated at the second methionine residue, located at position 12. The Kass that were determined for complexes of the truncated mouse proteins eIF4E(28–217), eIF4E(28–204), and of LeishIF4E-1(12–214), with different cap structures including m7GTP, the trypanosomatid cap-4, and selected analogs, are given in Table 1 ▶. The mouse eIF4E has a higher for all Kas cap analogs tested, as compared with LeishIF4E-1. However, the binding affinity of the mouse eIF4E(28–217) for the trypanosomatid cap-4 (Kas = 26.1 ± 0.7 μM−1) was fivefold weaker than that measured for m7GTP (Kas = 121.4 ± 3.0 μM−1), whereas the mouse protein LeishIF4E-1(12–214) bound the cap-4 and m7GTP with almost equal affinities, with Kas = 0.230 ± 0.002 μM−1 for cap-4, and for m7GTP. Similar Kas = 0.167 ± 0.003 μM−1 values were obtained for the nontruncated LeishIF4E-1 (data not shown). The binding affinity of LeishIF4E-1 for a capped nonmethylated tetranucleotide (m7GpppApApCpU, cap-0) as compared with m7GTP alone is higher by about threefold, unlike the mouse protein (Table 1 ▶; Fig. 5 ▶). This suggests that although a general similarity in the ligand-binding pocket of both proteins was observed, LeishIF4E-1 possesses additional structural features that promote its ability to interact with extended cap analogs (cap-0, cap-4), as compared with m7GTP.
TABLE 1.
The equilibrium association constants (Kas) for complexes of eIF4E proteins with cap-4 and other cap analogs
| Leishmania eIF4E(12–214) | Mouse eIF4E(28–217) | Mouse eIF4E(28–204)a | |
| Cap analogs | Kas (μM−1)b | ||
| m7GTP | 0.167 ± 0.003 | 121.4 ± 3.0 | 25.3 ± 0.7 |
| m7GpppA | 0.065 ± 0.002 | 4.4 ± 0.1 | 2.85 ± 0.02 |
| m7 Gpppm36,6,2′ A | 0.054 ± 0.004 | 1.45 ± 0.03 | 1.13 ± 0.10 |
| m7GpppApApCpU | 0.467 ± 0.005 | 73.3 ± 1.8 | 20.7 ± 0.6 |
| cap-4 | 0.230 ± 0.002 | 26.1 ± 0.7 | 9.2 ± 0.2 |
| GTP | <0.003 | 0.032 ± 0.004 | <0.007 |
aKCl instead of NaCl was used in fluorescence measurements.
bKas values were measured by quenching of intrinsic Trp fluorescence during titration with the indicated cap analog, at 20°C.
FIGURE 5.
Fluorescence titration measurements on binding of LeishIF4E-1 and mouse eIF4E(28–217) to cap analogs. LeishIF4E-1 protein at a concentration of 0.3 μM (A) and mouse eIF4E (28–217) at a concentration of 0.1 μM (B), were titrated by the cap analogs: cap-4 (open squares), m7GpppApApCpU (black circles), m7GTP (open circles), and GTP (black triangles), at 20°C in 50 mM HEPES/ NaOH, 100 mM NaCl, 1 mM EDTA, and 1 mM DTT. Protein fluorescence, presented as relative values, was excited at 295 nm and observed at 345 nm. The shift of the titration curves toward the higher ligand concentrations indicates a weaker binding between the specific cap analogs and the corresponding protein. The observed increasing fluorescence intensity at higher concentration of ligand originates from the emission of the free cap analogs in solution.
The binding affinities of the dinucleotide cap analogs m7GpppA and m7Gpppm36,6,2′ A to LeishIF4E-1 are very similar (Kas = 0.065 ± 0.002 μM−1, and Kas = 0.054 ± 0.004 μM−1, respectively), and both ligands bind ~2.5–3-fold weaker, in comparison with m7GTP (Table 1 ▶). Similarly, the dinucleotide analog of cap-4, m7Gpppm36,6,2′A, binds very poorly to the mouse eIF4E(28–217) (Table 1 ▶), compared with m7GTP. The destabilization effect conferred by the second base was also observed for the mouse protein with other dinucleotide analogs (Niedzwiecka et al. 2002). The presence of the second base in the dinucleotide cap analogs is responsible for the partial reduction in the negative charge of the phosphate chain, compared with the mono-nucleotide analogs. In the mononucleotide, there is an additional OH group on the last phosphate that is absent from the dinucleotide. This group is partially ionized and could therefore strengthen its interaction with the positively charged amino acids at the entrance of the cap-binding pocket, compared with the dinucleotides. The analog containing a double-methylated second adenosine base binds the mouse protein even less strongly (m7 Gpppm36,6,2′A, Kas = 1.45 ± 0.03 μM−1) than the nonmethylated dinucleotide (m7GpppA, Kas = 4.4 ± 0.1 μM−1). This weaker inter- action with the dinucleotide analogs can be explained by destabilization of the interaction between the adenosine and the C-terminal loop of the mammalian eIF4E (Tomoo et al. 2003). In the crystal complex of the human eIF4E with m7GpppA, the adenine is bound through a hydrogen bond to Thr 205, and via a water-mediated hydrogen bond to Thr 211. In addition, binding of the second base of the cap with the protein is stabilized by van der Waals contacts with Ser 207 and its neighboring residues (Tomoo et al. 2003). Methylations of the adenine base might destabilize the hydrogen bond between the NH6 of the adenine and Thr 205.
It is interesting to note that truncation of the murine C terminus, eIF4E(28–204), reduced the Kas of m7GTP binding by fivefold (Kas = 25.3 ± 0.7 μM−1). This suggests that the C terminus of the protein contributes to its high affinity for the cap, particularly in the formation of molecular contacts with the γ phosphate. Truncation of the C terminus generates a protein that is less sensitive to the presence of the second nucleotide in the cap analog. This is reflected by the relatively small reduction in the Kas of eIF4E(28–204) with the dinucleotides and with cap-4, as compared with m7GTP. The presence of methyl groups on the second base in the cap analog further reduced its affinity to eIF4E(28–217); however, the effect of this methylation on binding to the protein that lacks the C terminus was less profound. It appears from the sequence alignment (Fig. 1 ▶) that the primary structure of LeishIF4E-1 is devoid of the C-terminal loop, and therefore this protein binds with similar affinities to both m7GpppA and m7 Gpppm36,6,2′A. The binding affinity measured for the control analog that consisted of a capped, but nonmethylated tetranucleotide, cap-0, also supports the possibility that the C-terminal domain in the murine protein has an inhibitory effect on binding of methylated cap analogs. While the absence of the methyl groups in cap-0 increases the for complexes of the mouse Kas eIF4E(28–217) with this analog by threefold, as compared with cap-4, it has a smaller effect on the binding of LeishIF4E-1 and the truncated murine eIF4E(28–204), with an increase of only twofold in the measured Kas for both proteins. These observations suggest that in addition to the conserved binding pocket, LeishIF4E-1 may possess other features that promote its interaction with cap-4, such as the absence of the C terminus.
Subcellular distribution of LeishIF4E-1
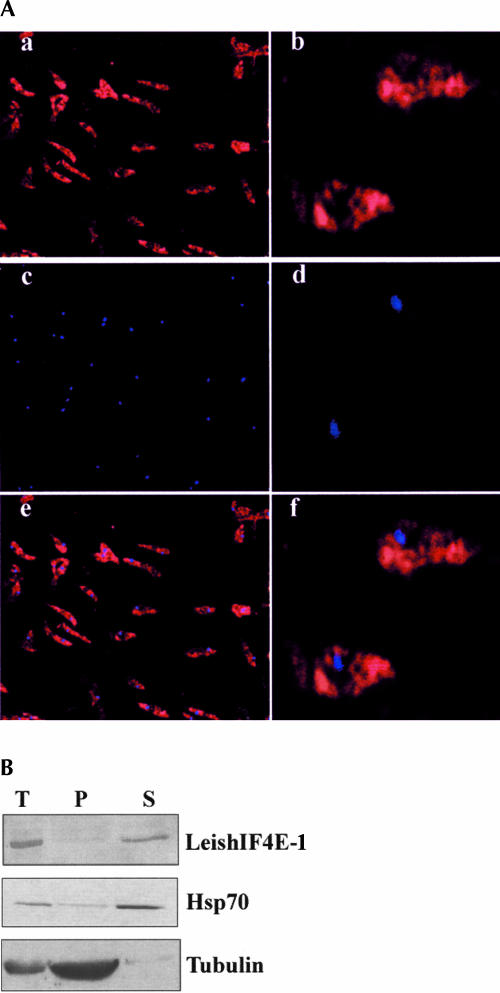
In higher eukaryotes, eIF4E is distributed mainly in the cytoplasm, although small amounts are present in the nucleus (Lejbkowicz et al. 1992). We therefore raised poly-clonal antibodies against LeishIF4E-1 and used them to determine its subcellular distribution by immunohistochemical methods (Fig. 6A ▶). The fluorescence staining indicated that the endogenous LeishIF4E-1 was distributed mainly in the cytoplasm (Fig. 6Aa,b ▶). The nuclei were identified by DAPI staining (Fig. 6Ac,d ▶), and the overlay of both pictures is shown in Figure 6A ▶, panels e and f.
FIGURE 6.
Subcellular localization of LeishIF4E-1. (A) Localization by immunostaining. Leishmania parasites were attached to coverslips coated with poly-lysine, fixed with methanol, and reacted with rabbit antiserum raised against LeishIF4E-1 (1:4000). The fixed cells were then incubated with goat anti-rabbit antibodies conjugated with Cy3. After washing away the second antibody, the cells were stained with DAPI and mounted over glass slides. The slides were visualized in a fluorescence microscope for Cy3 (a,b) and DAPI (c,d) staining. Pictures of similar fields were overlapped to highlight the position of nuclei (e,f). (B) Localization by Western analysis of fractionated cells. Extracts were prepared from whole cells (T) and from subcellular fractions containing the cytoplasm (S) and nucleo-cytoskeletal fractions (P) and separated over 15% SDS-PAGE. Western blots were reacted with antibodies raised against recombinant LeishIF4E-1. Control blots containing equal loads of the same fractions were reacted with a monoclonal antibody against Hsp70 and α-tubulin, representing the cytoplasmic and the nonsoluble nucleo-cytoskeletal fractions, respectively.
Similar findings were obtained from Western analysis of fractionated parasite cells (Fig. 6B ▶). Western blots were prepared from whole-cell extracts and from subcellular fractions containing the cytoplasm and the nonsoluble fraction that consisted of nuclear and cytoskeletal components. Staining by antibodies raised against LeishIF4E-1 verified that the majority of the protein was found in the cytoplasmic fraction of disrupted cells. Control blots that contained similar protein loads were reacted with a monoclonal antibody directed against Hsp70, mostly found in the cytoplasm of nonstressed cells (Velazquez and Lindquist 1984), and with antibodies against α-tubulin, representing the nucleo-cytoskeletal fraction.
LeishIF4E-1 cannot substitute the mammalian eIF4E-1 in an in vitro translation system
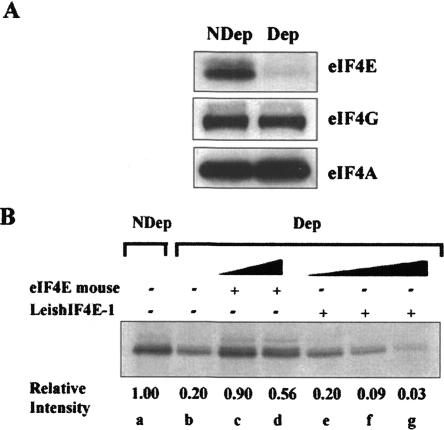
The function of LeishIF4E-1 is not yet established, and it is not clear yet whether it fulfills the functions of the traditional translation factor in Leishmania. In the absence of in vitro translation systems for Leishmania, we examined whether LeishIF4E-1 could substitute the mammalian eIF4E in a rabbit reticulocyte system. Reticulocyte lysates were selectively depleted from their endogenous eIF4E by incubation with the mammalian 4E-BP1 that competes with the endogenous eIF4G on binding to eIF4E, and excludes the participation of the eIF4E in the translation initiation complex (McKendrick et al. 2001). Addition of m7GTP-Sepha-rose beads selectively removed the complex between eIF4E and 4E-BP1. This strategy allowed maintaining the endogenous eIF4G and eIF4A that are part of the eIF4F initiation complex, as shown in Figure 7A ▶.
FIGURE 7.
LeishIF4E-1 cannot replace the mammalian eIF4E in an in vitro translation system. (A) eIF4E was selectively depleted from a rabbit reticulocyte system by addition of 4E-BP1, followed by m7GTP-Sepharose beads, which were used to remove the complex between eIF4E and the 4E-BP1. The selective depletion of eIF4E and not of other translation factors, was verified by Western analysis of the depleted (Dep) and nondepleted (NDep) extracts, using antibodies directed against the mammalian eIF4E, eIF4GI, and eIF4A. (B) Control nondepleted (NDep, lane a) lysates were used for translation of a capped and polyadenylated GFP transcript, in the absence (lane b) and in the presence of recombinant mouse eIF4E (2 μM, lane c; 10 μM, lane d) or recombinant LeishIF4E-1 (2 μM, lane e; 10 μM, lane f; 150 μM, lane g). The relative incorporation of [35S]methionine, measured by PhosphorImaging and compared with the nondepleted control, is given below the autoradiogram.
Depletion of the endogenous eIF4E from the rabbit re-ticulocyte lysate reduced the translation to 20% of the level that was measured in nondepleted lysates. Addition of the recombinant mouse eIF4E(28–217) protein (2 μM) reconstituted the translation to its original level, whereas addition of the same amount of the recombinant LeishIF4E-1 had no such effect. In view of the relatively low binding constants measured for LeishIF4E-1, increasing amounts of the external protein were added; however, the function of the depleted eIF4E was not complemented. In both cases, adding increased amounts of the external protein even had an inhibitory effect, which could originate from impurities added with the protein (Fig. 7B ▶).
DISCUSSION
In an attempt to investigate the cap-binding proteins in Leishmania, we cloned and analyzed an eIF4E homolog. A computerized structure prediction of the Leishmania protein, based on the crystal structure of the mouse eIF4E (Marcotrigiano et al. 1997), indicated that the main features that mark the cap-binding pocket in the mouse protein are conserved. These include stacking of the base between two conserved tryptophan residues, corresponding to positions 56 and 102 in the murine protein, as well as the formation of hydrogen bonds and van der Waals contacts between the N7-methyl group from the cap structure and a third conserved tryptophan, which parallels murine Trp 166. Additional conserved residues are a glutamic acid that corresponds to position 103 in the murine protein and several basic amino acids, predicted to interact with the phosphate backbone (Marcotrigiano et al. 1997; Matsuo et al. 1997; Tomoo et al. 2003). It should be noted that an additional eIF4E homolog (CAB94111), bearing 42% similarity to the mouse eIF4E and 41% homology to LeishIF4E-1, was found in the Leishmania genome database; however, this protein failed to bind either m7GTP or TMG (Y. Yoffe, A. Lerer, J. Zuberek, M. Lewdorowicz, M. Jankowska-Anyszka, E. Darzynkiewicz, and M. Shapira, in prep.).
Proteins with a broad binding specificity to both m7GTP and TMG were reported in Caenorhabditis elegans, an organism that uses both cis- and trans-splicing for processing of its mRNAs. The SL RNA of C. elegans carries a TMG structure at its 5 ′-end; thus, trans- and cis-splicing generate transcripts that are capped by TMG or m7GTP, respectively (Thomas et al. 1988; Blumenthal 1995). Five isoforms of cap-binding proteins were found in C. elegans. These display either dual binding specificities, to both m7GTP and TMG (IF4E-1, -2, and -5), or exclusive binding to m7GTP (IF4E-3 and -4) (Keiper et al. 2000; Miyoshi et al. 2002). Affinity chromatography experiments performed in this study indicated that LeishIF4E-1 bound m7GTP, allowing its purification to almost complete homogeneity on m7GTP-Sepharose. However, recombinant LeishIF4E-1 did not bind TMG, a cap structure that is typical of U snRNAs, such as U2 (Mattaj 1986; Gunzl et al. 2000). Fluorescence measurements also indicated that LeishIF4E-1 did not bind TMG. A concrete Kas value could not be calculated because of its high intrinsic fluorescence at the high concentrations required for evaluation of low affinity binding (data not shown). Thus, LeishIF4E-1 is most probably not involved in binding to snRNAs that possess a TMG cap.
Despite the great importance of structure determination of proteins, their binding affinity with different ligands cannot be extracted from the structural data, and must be obtained using alternative approaches. Measurements of fluorescence quenching provide a powerful tool to determine the affinity of binding between proteins from various species with different cap ligands (Carberry et al. 1989; McCubbin et al. 1989). The fluorescence assays in this study revealed an apparent difference between the parasite and mouse proteins. Whereas the mouse protein distinguishes between the different cap structures and binds the parasite cap with a fivefold lower affinity than m7GTP, the parasite protein, LeishIF4E-1, binds both ligands with comparable affinities, with preference toward extended cap analogs, namely, cap-0 and cap-4.
To prevent variations that could originate from truncation of the N terminus in the mouse protein eIF4E(28–217), the Kas values were measured for LeishIF4E-1 that was similarly truncated [LeishIF4E-1(12–214)], with both proteins initiating at parallel positions. Removal of the N terminus in the mouse protein did not interfere with in vitro translation activity (Marcotrigiano et al. 1997), and fluorescence studies of full-length human and truncated mouse eIF4E polypeptides showed similar affinities for m7GTP (Niedzwiecka et al. 2002). Similarly, comparable Kas values were obtained for the complete (1–214) and truncated (28–214) forms of LeishIF4E-1 (Lewdorowicz et al. 2004).
A difference of 3 orders of magnitude was observed between the Kas values for binding of LeishIF4E-1 and the mouse protein to m7GTP. In this study, different protocols were used for the purification of each protein. LeishIF4E-1 was affinity-purified from the bacterial soluble fraction over m7GTP-Sepharose, whereas the mouse protein (found mainly in inclusion bodies) was purified by refolding of the denatured protein and was not exposed to the cap ligand during the purification procedure. It should be noted that an apparent difference in the Kas was reported for human eIF4E, purified by different methods (see Table 1 ▶ in Niedzwiecka et al. 2002). However, preliminary exposure of LeishIF4E-1 to the cap ligand during its purification did not seem to affect its binding properties, because a His-tagged protein, which was purified over Ni-NTA Agarose, gave Kas values that were similar to those measured for the protein after its purification over m7GTP-Sepharose (data not shown). It is difficult to say whether the vast difference between the binding affinities observed for the mouse and parasite proteins in our in vitro assays, is valid also in vivo, and we cannot conclude if this reflects on a different mode of recognition and initiation of translation in Leishmania. Assembly of the translation initiation complex in eukaryotes involves multiple protein factors, and these could further stabilize the interaction with the 5 ′-cap. Because of the difference between the binding affinities of the two proteins, which cannot be fully explained at this stage, we concentrated on how the different modifications affected the binding to each of these two individual polypeptides.
Alignment of LeishIF4E-1 with the murine and yeast proteins shows that LeishIF4E-1 lacks sequences that are homologous to the C terminus of these two proteins. This could account for the different affinity constants measured for each of the two proteins with the different cap analogs. Indeed, truncation of the C-terminal loop in the mouse protein decreased the binding affinity to m7GTP. It also appears that the binding parameters of the murine protein truncated in its C terminus were more similar to those of the Leishmania cap-binding protein. It was less sensitive to the presence of additional nucleotides downstream to m7GTP (m7GpppApApCpU), as well as to modifications on these nucleotides, as in cap-4. The C terminus of the murine eIF4E contains amino acids that participate in cap binding (Tomoo et al. 2003), such as Ser 209, which is implicated in eIF4E phosphorylation and in cap binding (Scheper et al. 2002; Zuberek et al. 2003). Thus, because the cap-binding pocket of LeishIF4E-1 is very similar to its murine counterpart, it appears that the adaptation for binding the complex cap-4 most probably does not originate from this region. Based on the data presented in this study, we hypothesize that the lack of a region that is parallel to the murine C terminus stabilizes the interactions between LeishIF4E-1 and cap-4. However, a further structural analysis is required for mapping the nature of the exact interactions with the modified first four nucleotides in cap-4.
In trypanosomatids, cap-4 is donated to all mRNA molecules by their joining to the capped SL RNA during trans-splicing. Cap-4 biogenesis occurs gradually, it initiates with the formation of m7GTP and is followed by modification of the first four transcribed nucleotides. A cotranscriptional model of the SL RNA cap-4 methylation in T. brucei was suggested, based on the observation that modifications on the first two adenosines were detected on prematurely terminated SL RNA transcripts of 56–67 nt. The additional modifications creating the cap-4 structure occurred progressively thereafter, and the complete cap-4 was detected only after transcription of the Sm-binding site (Mair et al. 2000). Recently it was shown that modification of the first two nucleotides occurs cotranscriptionally in the nucleus, whereas modification of the fourth transcribed nucleotide (U) most probably takes place in the cytoplasm (Mandelboim et al. 2003). It was proposed that this fourth modification could signal the transport of the SL RNA between the nucleus and the cytoplasm. Thus, Leishmania and trypanosomes probably contain a variety of proteins that can differentiate between the different stages of cap-4 biogenesis. These could be involved either in the actual synthesis process or in transport of snRNPs between the nucleus and the cytoplasm, or possibly in the assembly of the translation complex on mRNAs containing the mature cap-4.
In this study we show, using immuno-histochemical assays combined with biochemical subfractionation, that LeishIF4E-1 is distributed mainly in the cytoplasm. However, at this stage, we cannot conclude yet whether LeishIF4E-1 serves as a translation factor, or whether it has a different role. LeishIF4E-1 failed to substitute the mammalian eIF4E in a rabbit reticulocyte in vitro translation system. However, because Leishmania is an early eukaryote, and in view of the absence of a better candidate for the translation initiation factor, the possibility that LeishIF4E-1 fulfills this function cannot be excluded yet. If LeishIF4E-1 is associated with translation initiation of complex, it should contain conserved regions that could interact with eIF4G (Marcotrigiano et al. 1999; Gross et al. 2003). A search of LeishIF4E-1 for conserved residues that are known to promote its interaction with eIF4G revealed that most of them are, indeed, conserved. This includes His 16 and Leu 18 in LeishIF4E-1, which correspond to His 37 and Leu 39 in the mouse and yeast proteins. Also, Val 50 in LeishIF4E-1 is conserved with Val 71 in the yeast protein, and a Phe residue at position 54 in LeishIF4E-1 parallels the yeast Trp 75. This conservation could support possible interactions with the parasite eIF4G; however, the identity of this protein in Leishmania is not clear yet. Although a straightforward BLAST search did not find a homolog for eIF4G, advanced bioinformatics tools revealed the presence of several ORFs containing domains that are characteristic of eIF4G proteins. Validation of these proteins as eIF4G homologs must first be achieved, prior to examining potential 4E–4G interactions in Leishmania.
In summary, despite the phylogenetic conservation of the cap-binding pocket, proteins that bind the unusual cap-4 structure in trypanosomatids possess structural features that distinguish them from their host counterparts. The interactions between cap-binding proteins and their corresponding ligands could serve as a potential target for drug development in the future, once the characterization of these proteins is further advanced.
MATERIALS AND METHODS
Cell cultures
Leishmania major (Friedlin) was cultured in Schneider’s medium supplemented with 10% fetal calf serum (FCS), 4 mM L-glutamine, and 25 μg/mL gentamycin.
Sequence alignment and homology modeling of Leishmania eIF4E-1
The open reading frame of eIF4E-1 was deduced from the Leish-mania genome database (LeishDB; http://www.sanger.ac.uk/Projects/L_major), accession number CAB77676. The amino acid sequence was aligned against the mouse (P20415) and yeast (P07260) proteins, with the CLUSTAL X algorithm. Homology modeling was performed using SwissPdbViewer and was based on the structure of the mouse protein (pdb 1EJ1a). The model with the lowest Z-score of the overall structure was visualized.
Cloning and expression of the Leishmania eIF4E-1 in bacteria for protein purification
The eIF4E-1 isoform of eIF4E in Leishmania was amplified by PCR, using L. major genomic DNA as a template. Forward primers corresponded to positions 1–21, 5 ′-GGAATTCCATATGTCAGC CCCGTCTTCAGTT-3 ′, and 34–54, 5 ′-GGAATTCCATATGGCG AATTTGCACAAGCTG-3 ′. The reverse primer corresponded to positions 625–645, 5 ′-CGCGGATCCCTAAGACGCCTCGCCGT GCTT-3 ′. All primers contained anchor sequences that incorporated the NdeI and BamHI restriction sites at the N termini (forward primers) and C termini (reverse primers), respectively (the restriction sites are underlined). The amplified fragments were digested with NdeI and BamHI and further cloned into the corresponding site of the pHis-parallel vector (Sheffield et al. 1999), resulting in plasmids pLeishIF4E-1 and pLeishIF4E-1(12–214). This cloning strategy eliminated the His-tag and generated a recombinant protein that initiated from its own first methionine. Two recombinant proteins were expressed, initiating at the me-thionine residue located either at position 1 or at position 12. The nucleotide sequences of the amplified fragments in pLeishIF4E-1 and pLeishIF4E-1(12–214) were compared with the genomic sequence obtained from the L. major genome project.
The recombinant pLeishIF4E-1 was introduced into Escherichia coli BL21 cells, and expression was induced at 20°C in cultures grown to OD660 0.5 for 3 h, by addition of 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG). Cells were washed once in sonication buffer (20 mM HEPES at pH 7.6, 1 mM DTT, and 2 mM EDTA, 5% glycerol), harvested, and disrupted by sonication during 2 min on ice, using pulses of 20 sec, interspaced by intervals of 20 sec. The cell extract was clarified by centrifugation during 30 min at 20,000g, and the supernatant was loaded on m7GTP-Sepha-rose, equilibrated in sonication buffer. The protein was eluted by high salt (200 mM NaCl). A similar protocol was applied for TMG-Sepharose, with elution performed with high salt, as described.
Expression and purification of the mouse eIF4E
E. coli BL21(DE3) (Novagen) cells, transformed with the vector pET3b containing a cDNA fragment of the truncated mouse eIF4E(28–217), were grown in LB medium at 37°C to OD600nm ~1 (Marcotrigiano et al. 1997). Expression was induced by addition of 0.5 mM IPTG for 3 h at 37°C. The cells were harvested by centrifugation; resuspended in a buffer solution that contained 20 mM HEPES/KOH (pH 7.2), 100 mM KCl, 5 mM EDTA, 10% glycerol, and 2 mM DTT; and disrupted by sonication. Because the majority of the protein was found in inclusion bodies, pellets of the lysed cells after centrifugation were washed three times with 1 M guanidinium hydrochloride and then solubilized in 6 M gua-nidinium hydrochloride. The protein was refolded by a single-step dialysis against 50 mM HEPES/NaOH (pH 7.2), 100 mM NaCl, 0.5 mM EDTA, and 2 mM DTT (Marcotrigiano et al. 1997) and was further purified by ion-exchange chromatography on a Hitrap SP column that was eluted by a linear gradient of NaCl (0.1–1 M NaCl, during 60 min).
The truncated fragment of eIF4E(28–204) was expressed as a fusion protein with the intein MxeGryA—a chitin-binding domain (CBD), and purified as described previously (Zuberek et al. 2003), with the exception of using 100 mM DTT instead of ethanethiol for the cleavage reaction. After purification, the lyophilized protein was dissolved in a buffer containing 4 M guanidinium hydrochloride, 50 mM HEPES/KOH, 10% glycerol, and 2 mM DTT, re-folded by a single-step dialysis and purified by ion exchange chromatography on a Hitrap SP column, as described for eIF4E(28–217). During the purification process, the proteins did not have any contact with cap analogs.
Southern analysis
Genomic DNA was extracted from L. major cultures, digested with different restriction enzymes and separated on a 0.8 % agarose gel. The DNA was blotted onto a nylon membrane and hybridized with a DIG-labeled probe consisting of the 645 nt that corresponded to the complete coding gene of LeishIF4E-1. Hybridization was carried out at 50°C, with washes containing 2× SSC/0.1% SDS for 5 min at 50°C, 0.5× SSC/0.1% SDS for 15 min at 65°C, 0.2× SSC/0.1% SDS for 15 min at 65°C, and 0.1× SSC/0.1% SDS for 15 min at 65°C. Hybridization with the DIG-labeled probe was detected by incubation with anti-DIG antibodies conjugated with alkaline-phosphatase, developed with an ECL reagent, following the manufacturer’s instructions (Roche).
Cap analogs
m7GTP, either in its free form or immobilized onto Sepharose beads, was commercially obtained (Sigma, Amersham, respectively). m32,2,7GTP, either in its free form or immobilized onto Sepharose beads, and m7GpppA were chemically synthesized as described before (Jankowska et al. 1993 and 1996, respectively). Synthesis of m7 Gpppm36,6,2′ Apm2′ Apm2′3, Cpm22′ U comprising the Leishmania cap-4 structure and m7 Gpppm36,6,2′A is described elsewhere (Lewdorowicz et al. 2004). Synthesis of m7GpppApApCpU was carried out by coupling of the nonmethylated tetranucleotide pApApCpU (Trilink Biotechnologies) with 7-methylguanosine 5′-diphosphate imidazolide [Im(m7GDP)], in a water solution and MnCl2 as catalyst, following previously published protocols (Sawai et al. 1999). m7GpppApApCpU was obtained as an ammonium salt with a calculated molecular weight of 1726.0 g/mole for its free acid, which was verified by Mass Spectrometry.
Fluorescence measurements and data analysis
The affinity of cap-binding proteins for the different cap analogs was determined by monitoring the quenching of fluorescence emission by intrinsic Trp residues in the presence of increasing amounts of the cap ligand (Carberry et al. 1989; Wieczorek et al. 1999). Freshly prepared protein samples were used for fluorescence assays, after removal of the high salt by centrifugation over 4 mL Biomax 5K NMWL filters (Millipore). Protein concentrations were determined spectrophotometrically, assuming the mouse eIF4E(28–217), eIF4E(28–204), and ε280 = 53,400 M−1 cm−1 for ε280 = 47,500 M−1 cm−1 for LeishIF4E-1(12–214), which were calculated from the amino acid composition (Gill and von Hippel 1989). Fluorescence measurements were performed on an LS-50B Spectrofluorometer (Perkin Elmer Co.) in a quartz semimicro cuvette (Hellma) with optical lengths 4 mm and 10 mm for absorption and emission, respectively. Fluorescence time-synchronized titrations (TST) (Niedzwiecka et al. 2002) were performed in 50 mM HEPES/NaOH (pH 7.2), 100 mM NaCl, 1 mM EDTA, and 1 mM DTT at 20°C by adding 1-μL aliquots of the cap analog solution to 1.4 mL of 0.1 or 0.3 μM protein solution. Each titration consisted of 40–65 data points. Fluorescence intensities (excited at 280 nm or 295 nm and observed at 337 nm or 345 nm, respectively) were corrected for sample dilution (>5%) and for the inner filter effect using an empirical calibration curve (Lakowicz 1999).
The theoretical curve for the fluorescence intensity (F) as a function of [L] was fitted to the experimental data points according to the equation:
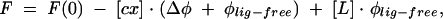 |
where the equilibrium concentration of the cap-eIF4E complex [cx] is given by
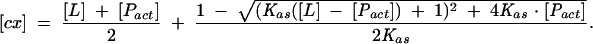 |
The parameters to be extracted from the fit were as follows: Kas, the association constant; [Pact], the concentration of the active protein; Δφ = φP-act-free − *phis;cx, the difference between the fluorescence efficiencies of the aporotein and the complex; φlig-free, the fluorescence efficiency of the free cap analog in the solution; and F(0), the initial fluorescence intensity (Niedzwiecka et al. 2002). The final Kas was calculated as a weighted average from two to five independent titration series. Regressions were performed by means of a nonlinear, least-squares method, using ORIGIN 6.0 (Microcal Software Inc.).
Antibodies
Recombinant LeishIF4E-1 was affinity-purified over m7GTP-Sepharose, emulsified with Complete Freund’s Adjuvant and injected subcutaneously into New Zealand rabbits (500 μg per rabbit). Two boosts of the protein emulsified in Incomplete Freund’s Adjuvant were given in 2-wk intervals. Serum of immunized animals was obtained 10–20 d after the boost. Antibodies against α-tubulin were commercially obtained (Yeda), and the monoclonal antibodies against Hsp70 (5A5) were a generous gift of R. Morimoto, Northwestern University. Antibodies directed against the mammalian (mouse) eIF4E, eIF4GI, and eIF4A were a generous gift of N. Sonenberg (McGill University, Canada).
Subcellular fractionation and Western analysis
For total protein extracts, Leishmania parasites (108 cells) were harvested, washed twice in phosphate-buffered saline (PBS), and extracted in SDS-PAGE sample buffer (Laemmli 1970). Protein concentrations were determined, by the BCA reagent kit (Pierce). The cells were fractionated into the cytoplasmic and nucleocyto-skeletal fractions following published protocols (Boucher et al. 2002). The cells were washed twice, and the pellet was frozen in liquid nitrogen and resuspended in Dignam buffer (10 mM HEPES at pH 7.6, 1.5 mM MgAc2, 10 mM KAc, 2 mM idodo-acetamide, 0.5 mM DTT) containing a cocktail of protease inhibitors (Sigma). The mixture was incubated for 15 min on ice, Nonidet P-40 was added to a final concentration of 0.8%, and the mixture was vortexed for 10 sec and centrifuged at 12,000g for 15 min at 4°C. The pellet was resuspended in Dignam buffer, and the protein concentrations of the cytoplasmic fraction and the resus-pended pellet were determined. Samples of whole-cell extracts and of the subcellular fractions containing equal protein loads were analyzed over 15% SDS-PAGE.
The protein blots were reacted with antibodies raised against LeishIF4E (1:8000), α-tubulin (1:400; Yeda), and Hsp70 (1:1000). The washed blots were incubated with secondary antibodies and developed with an ECL reagent (Pierce).
Indirect immunofluorescence microscopy
Wild-type L. major cells were attached to poly-lysine-coated coverslips during 10 min and fixed with methanol for 10 min at −20°C. The cells were then washed three times during 1 min in PBS, permeabilized, and blocked by incubation with 1% Triton X-100 and 10% preimmune goat serum in PBS for 1 h at room temperature. The fixed cells were incubated overnight with the antibody raised against LeishIF4E-1 (1:4000) diluted in 0.1% Triton X-100/PBS, washed three times in PBS at room temperature, and further incubated with goat anti-rabbit antibodies conjugated to Cy3 (1:500; Molecular Probes). The cells were then washed, stained with 4′,6-diamidino-2-phenylindol (DAPI) at a final concentration of 0.5 μg/mL for 30 min at room temperature, washed again in PBS, and mounted on glass slides using Elvanol. The slides were visualized in a Zeiss Axiovert 200M Fluorescence microscope, using a CCD SensiCam (PCO) camera.
In vitro transcription
A transcription vector encoding GFP, pRAT-GFP-A40, was kindly provided by N. Zilberberg (Ben-Gurion University of the Negev). The plasmid was linearized using NotI, and RNA was transcribed with T7 RNA polymerase. The resulting transcript was capped using the cap analog m7GpppG.
In vitro translation assays
4E-BP1 fused to GST was expressed from pGEX-6p1-h4E-BP1 that was kindly provided by N. Sonenberg. The protein was expressed in E. coli BL21 cells and purified as described previously (Gingras et al. 1999a). LeishIF4E-1 and the mouse eIF4E (28–217) proteins were expressed and purified over m7GTP-Sepharose. Both proteins were eluted in the presence of 200 μM m7GTP. All the proteins were dialyzed against the translation buffer (20 mM MOPS-KOH at pH 7.2, 25 mM KCl, 10 mM NaCl, 1 mM MgCl2, 7 mM β-mercaptoethanol).
In vitro translation of the capped GFP RNA (5 μg/mL) was performed in a Rabbit Reticulocyte system (Promega) following the manufacturer’s instructions. The translated proteins were labeled during 90 min at 30°C by [35S]methionine-cysteine (20 μCi; PRO-MIX, Amersham), and samples (2 μL) were analyzed on 15% SDS-PAGE and quantified by PhosphorImaging. Depletion of eIF4E from the reticulocyte lysate was carried out as described previously (McKendrick et al. 2001). Essentially, 1 μM GST-4E-BP1 was incubated with the unsupplemented lysate for 10 min at 30°C. m7GTP-Sepharose was then added, and the lysate was incubated for 5 min at 4°C. The beads were removed by centrifugation, and depletion was verified by Western analysis of samples (3 μL) that were removed from the depleted and nondepleted lysates. Lysates depleted from eIF4E were reconstituted by the murine- and Leishmania-derived proteins.
Acknowledgments
This research was supported by grants from the German-Israeli Foundation (GIF, 728-23.2/2002) and the Israel Ministry of Health (5440) provided to M.S., and by grants from the Polish Committee for Scientific Research (PBZ-KBN 059/T09/10 and KBN 3 P04A 021 25) provided to E.D. We thank Nahum Sonenberg from Mc-Gill University, Canada, for plasmids encoding the mouse eIF4E and 4E-BP1, and for antibodies against the mammalian translation factors eIF4E, eIF4GI, and eIF4A. We thank Richard Morimoto from Northwestern University at Evanston, USA, for the monoclonal antibody against hsp70. We thank Noam Zilberberg from Ben-Gurion University, Israel, for the pRAT-GFP-A40 plasmid, and Simon Morley from the University of Sussex, UK, for helpful discussions.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7520404.
REFERENCES
- Bangs, J.D., Crain, P.F., Hashizume, T., McCloskey, J.A., and Boothroyd, J.C. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267: 9805–9815. [PubMed] [Google Scholar]
- Blumenthal, T. 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11: 132–136. [DOI] [PubMed] [Google Scholar]
- Boucher, N., Wu, Y., Dumas, C., Dube, M., Sereno, D., Breton, M., and Papadopoulou, B. 2002. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 277: 19511–19520. [DOI] [PubMed] [Google Scholar]
- Carberry, S.E., Rhoads, R.E., and Goss, D.J. 1989. A spectroscopic study of the binding of m7GTP and m7GpppG to human protein synthesis initiation factor 4E. Biochemistry 28: 8078–8083. [DOI] [PubMed] [Google Scholar]
- Gill, S.C. and von Hippel, P.H. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182: 319–326. [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Gygi, S.P., Raught, B., Polakiewicz, R.D., Abraham, R.T., Hoekstra, M.F., Aebersold, R., and Sonenberg, N. 1999a. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes & Dev. 13: 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., and Sonenberg, N. 1999b. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Gorlich, D. and Mattaj, I.W. 1996. Nucleocytoplasmic transport. Science 271: 1513–1518. [DOI] [PubMed] [Google Scholar]
- Gross, J.D., Moerke, N.J., von der Haar, T., Lugovskoy, A.A., Sachs, A.B., McCarthy, J.E., and Wagner, G. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750. [DOI] [PubMed] [Google Scholar]
- Gunzl, A., Bindereif, A., Ullu, E., and Tschudi, C. 2000. Determinants for cap trimethylation of the U2 small nuclear RNA are not conserved between Trypanosoma brucei and higher eukaryotic organisms. Nucleic Acids Res. 28: 3702–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat, A., Mader, S., Pause, A., and Sonenberg, N. 1995. Repression of cap-dependent translation by 4E-binding protein 1: Competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14: 5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde, E., Lewis, J., Gamberi, C., Jarmolowski, A., McGuigan, C., and Mattaj, I.W. 1995. A cap-binding protein complex mediating U snRNA export. Nature 376: 709–712. [DOI] [PubMed] [Google Scholar]
- Jankowska, M., Temeriusz, A., Stolarski, R., and Darzynkiewicz, E. 1993. Synthesis of m2,7GTP- and m2,2,7GTP-Sepharose 4B: New affinity resins for isolation of cap-binding proteins. Collect. Czech. Chem. Commun. 58: 132–137. [Google Scholar]
- Jankowska, M., Stepinski, J., Stolarski, R., Wieczorek, Z., Temeriusz, A., Haber, D., and Darzynkiewicz, E. 1996. 1H NMR and fluorescence studies of new mRNA 5′-cap analogues. Collect. Czech. Chem. Commun. 61: S197–S202. [Google Scholar]
- Joshi, B., Cameron, A., and Jagus, R. 2004. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 271: 2189–2203. [DOI] [PubMed] [Google Scholar]
- Keiper, B.D., Lamphear, B.J., Deshpande, A.M., Jankowska-Anyszka, M., Aamodt, E.J., Blumenthal, T., and Rhoads, R.E. 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 275: 10590–10596. [DOI] [PubMed] [Google Scholar]
- Kooter, J.M., De Lange, T., and Borst, P. 1984. Discontinuous synthesis of mRNA in trypanosomes. EMBO J. 3: 2387–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond.) 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lakowicz, J.R. 1999. Instrumentation for fluorescence spectroscopy. In Principles of fluorescence spectroscopy, 2nd ed., pp. 25–61. Kluwer Academic/Plenum, New York.
- Lamond, A.L. 1990. The trimethyl-guanosine cap is a nuclear targeting signal for snRNPs. Trends Biochem. Sci. 12: 451–452. [DOI] [PubMed] [Google Scholar]
- LeBowitz, J.H., Smith, H., and Beverley, S.M. 1993. Coupling of polyadenylation site selection and trans-splicing in Leishmania. Genes & Dev. 7: 996–1007. [DOI] [PubMed] [Google Scholar]
- Lejbkowicz, F., Goyer, C., Darveau, A., Neron, S., Lemieux, R., and Sonenberg, N. 1992. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. 89: 9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewdorowicz, M., Yoffe, Y., Zuberek, J., Jemielity, J., Stepinski, J., Kierzek, R., Stolarski, R., Shapira, M., and Darzynkiewicz, E. 2004. Chemical synthesis and binding activity of the trypanosomatid cap-4 structure. RNA 10: 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair, G., Ullu, E., and Tschudi, C. 2000. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275: 28994–28999. [DOI] [PubMed] [Google Scholar]
- Mandelboim, M., Barth, S., Biton, M., Liang, X.H., and Michaeli, S. 2003. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 278: 51469–51478. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.C., Sonenberg, N., and Burley, S.K. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 6: 707–716. [DOI] [PubMed] [Google Scholar]
- Matsuo, H., Li, H., McGuire, A.M., Fletcher, C.M., Gingras, A.C., Sonenberg, N., and Wagner, G. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 9: 717–724. [DOI] [PubMed] [Google Scholar]
- Mattaj, I.W. 1986. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46: 905–911. [DOI] [PubMed] [Google Scholar]
- Matthews, K.R., Tschudi, C., and Ullu, E. 1994. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes & Dev. 8:491–501. [DOI] [PubMed] [Google Scholar]
- McCubbin, W.D., Edery, I., Altmann, M., Sonenberg, N., and Kay, C.M. 1989. Circular dichroism and fluorescence studies on five mutant forms of protein synthesis initiation factor eIF-4E, from the yeast Saccharomyces cerevisiae. FEBS Lett. 245: 261–266. [DOI] [PubMed] [Google Scholar]
- McKendrick, L., Morley, S.J., Pain, V.M., Jagus, R., and Joshi, B. 2001. Phosphorylation of eukaryotic initiation factor 4E (eIF4E) at Ser209 is not required for protein synthesis in vitro and in vivo. Eur. J. Biochem. 268: 5375–5385. [DOI] [PubMed] [Google Scholar]
- Miyoshi, H., Dwyer, D.S., Keiper, B.D., Jankowska-Anyszka, M., Dar-zynkiewicz, E., and Rhoads, R.E. 2002. Discrimination between mono- and trimethylated cap structures by two isoforms of Caenorhabditis elegans eIF4E. EMBO J. 21: 4680–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka, A., Marcotrigiano, J., Stepinski, J., Jankowska-Anyszka, M., Wyslouch-Cieszynska, A., Dadlez, M., Gingras, A.C., Mak, P., Darzynkiewicz, E., Sonenberg, N., et al. 2002. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319: 615–635. [DOI] [PubMed] [Google Scholar]
- Robalino, J., Joshi, B., Fahrenkrug, S.C., and Jagus, R. 2004. Two zebrafish eIF4E family members are differentially expressed and functionally divergent. J. Biol. Chem. 279: 10532–10541. [DOI] [PubMed] [Google Scholar]
- Rodriguez, C.M., Freire, M.A., Camilleri, C., and Robaglia, C. 1998. The Arabidopsis thaliana cDNAs coding for eIF4E and eIF(iso)4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J. 13: 465–473. [DOI] [PubMed] [Google Scholar]
- Sawai, H., Wakai, H., and Nakamura-Ozaki, A. 1999. Synthesis and reactions of nucleoside 5′-diphosphate imidazolide: A nonenzymatic capping agent for 5′-monophosphorylated oligoribonucleo-tides in aqueous solution. J. Org. Chem. 64: 5836–5840. [Google Scholar]
- Scheper, G.C., van Kollenburg, B., Hu, J., Luo, Y., Goss, D.J., and Proud, C.G. 2002. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem. 277: 3303–3309. [DOI] [PubMed] [Google Scholar]
- Shatkin, A.J. 1976. Capping of eucaryotic mRNAs. Cell 9: 645–653. [DOI] [PubMed] [Google Scholar]
- Sheffield, P., Garrard, S., and Deresenda, Z. 1999. Overcoming expression and purification problems of RhoGDI using a family of™ Parallel° expression vectors. Prot. Exp. Purif. 15: 34–39. [DOI] [PubMed] [Google Scholar]
- Sutton, R.E. and Boothroyd, J.C. 1986. Evidence for trans splicing in trypanosomes. Cell 47: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.D., Conrad, R.C., and Blumenthal, T. 1988. The C. elegans trans-spliced leader RNA is bound to sm and has a trimethylguanosine cap. Cell 54: 533–539. [DOI] [PubMed] [Google Scholar]
- Tomoo, K., Shen, X., Okabe, K., Nozoe, Y., Fukuhara, S., Morino, S., Sasaki, M., Taniguchi, T., Miyagawa, H., Kitamura, K., et al. 2003. Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J. Mol. Biol. 328: 365–383. [DOI] [PubMed] [Google Scholar]
- Tucker, M. and Parker, R. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69: 571–595. [DOI] [PubMed] [Google Scholar]
- Ullu, E. and Tschudi, C. 1995. Accurate modification of the trypanosome spliced leader cap structure in a homologous cell-free system. J. Biol. Chem. 270: 20365–20369. [DOI] [PubMed] [Google Scholar]
- Velazquez, J.M. and Lindquist, S. 1984. hsp70: Nuclear concentration during environmental stress and cytoplasmic storage during recovery. Cell 36: 655–662. [DOI] [PubMed] [Google Scholar]
- Wieczorek, Z., Niedzwiecka-Kornas, A., Chlebicka, L., Jankowska, M., Kiraga, K., Stepinski, J., Dadlez, M., Drabent, R., Darzynkiewicz, E., and Stolarski, R. 1999. Fluorescence studies on association of human translation initiation factor eIF4E with mRNA cap-analogues. Z. Naturforsch. 54: 278–284. [DOI] [PubMed] [Google Scholar]
- Zuberek, J., Wyslouch-Cieszynska, A., Niedzwiecka, A., Dadlez, M., Stepinski, J., Augustyniak, W., Gingras, A.C., Zhang, Z., Burley, S.K., Sonenberg, N., et al. 2003. Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogs by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA 9: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]