Abstract
Translation is a sensitive regulatory step during cellular stress and the apoptosis response. Under such conditions, cap-dependent translation is reduced and internal ribosome entry site (IRES)-dependent translation plays a major role. However, many aspects of how mRNAs are translated under stress remain to be elucidated. Here we report that reaper mRNA, a pro-apoptotic gene from Drosophila melanogaster, is translated in a cap-independent manner. In Drosophila mutant embryos devoid of the eukaryotic initiation factor 4E (eIF4E), reaper transcription is induced and apoptosis proceeds. In vitro translation experiments using wild-type and eIF4E mutant embryonic extracts show that reporter mRNA bearing reaper 5′ untranslated region (UTR) is effectively translated in a cap-independent manner. The 5′UTR of reaper exhibits a high degree of similarity with that of Drosophila heat shock protein 70 mRNA, and both display IRES activity. Studies of mRNA association to polysomes in embryos indicate that both reaper and heat shock protein 70 mRNAs are recruited to polysomes under apoptosis or thermal stress. Our data suggest that heat shock protein 70 and reaper, two antagonizing factors in apoptosis, use a similar mechanism for protein synthesis.
Keywords: reaper, hsp70, Drosophila, 1(3)67Af, eIF4E, IRES-dependent translation
INTRODUCTION
Apoptosis has been related in different ways to the control of protein synthesis (Clemens et al. 2000; Holcik et al. 2000). Recent evidence suggests that at least two pro-apoptotic genes, reaper (rpr) and grim, down-regulate inhibitors of apoptosis (IAPs) by inhibition of general protein synthesis and, in addition, by protein degradation (Holley et al. 2002; Yoo et al. 2002). Apoptosis and other stress-related processes result in a cellular environment of degraded translation factors and impaired protein synthesis. Under these conditions, it has been demonstrated that some cellular mRNAs are translated largely by internal ribosome entry site (IRES)-mediated translation (Holcik et al. 2000).
Translation of the majority of eukaryotic mRNAs requires the recognition of the 5′ cap structure (m7GpppN) by translation initiation factor 4E (eIF4E; Gingras et al. 1999). eIF4E activity is regulated by phosphorylation and eIF4E binding proteins in a process that depends on signaling mediated by a plethora of factors, such as growth factors, hormones, mitogens, and cytokines (Morley 1997; Gingras et al. 1999). Several studies have suggested a role for eIF4E on proliferation processes, cell growth, and apoptosis. The overexpression of eIF4E in cell cycle-sensitized mammalian cells results in overgrowth and malignant transformation (Sonenberg and Gingras 1998; De Benedetti and Harris 1999). Inhibition of eIF4E activity in serum-starved cells in culture induces apoptosis (Herbert et al. 2000; Polunovsky et al. 2000). In this context, translation during apoptosis would not require cap structure. However, no direct evidence of such a process has yet been described for normal cells in the context of a whole organism.
Apoptosis is a cellular process required for cell selection and for organ and tissue development, but it also figures as a reaction to eliminate cells under stress or malfunctioning ones. Apoptosis has been studied in detail in Drosophila melanogaster (Abrams 1999), and the pattern of apoptotic cells during development has been established (Abrams et al. 1993). In Drosophila, the eIF4E-1/2 gene encodes two isoforms, namely, eIF4E1 and eIF4E2 (Lavoie et al. 1996; Hernández et al. 1997). eIF4E1 is the only isoform that occurs in embryos, suggesting that it might play a major role in protein synthesis during embryogenesis (Maroto and Sierra 1989; Zapata et al. 1994; Hernández and Sierra 1995). Here we show that a null allele of the gene encoding the isoforms eIF4E1 and eIF4E2 (eIF4E-1/2) exhibits increased apoptosis during embryogenesis in correlation with up-regulated rpr transcription. We show in vitro that the 5′ untranslated region (UTR) of rpr mRNA promotes translation of reporter RNA in vitro and in vivo, either in the absence of cap, in the presence of cap competitors, in the absence of eIF4E, or in extracts derived from heat-shocked embryos, indicating that cap recognition is not required in rpr mRNA for translation. We also show that the rpr mRNA 5′UTR resembles that of Drosophila heat shock protein 70 mRNA, an antagonist of apoptosis, and that both are able to conduct IRES-mediated translation.
RESULTS
Induction of apoptosis in the absence of eIF4E
The gene eIF4E-1/2 has been mapped in the region 67A2-B1 of the Drosophila genome (Hernández et al. 1997), where several lethal alleles are known (Leicht and Bonner 1988). We sequenced the gene eIF4E-1/2 from individual homozygous embryos from all the mutants available in the region 67A2-B1. Among them, homozygous embryos corresponding to the mutant l(3)67Af exhibited a point mutation in the splicing acceptor site of the second intron of the gene eIF4E-1/2 (AG to AA; Fig. 1A ▶), which would result in a nonsense reading of the ORF of both eIF4E1 and eIF4E2 (Fig. 1A ▶; GenBank accession no. AJ11964). This mutation was absent in the other mutants analyzed, suggesting that l(3)67Af might be a bona fide allele of eIF4E-1/2. We then studied the mRNA and protein levels of eIF4E-1/2 in homozygous l(3)67Af embryos. RT-PCR analysis revealed an absence of proper splicing (Fig. 1B ▶). Western blot showed a dramatic reduction of eIF4E-1/2 levels in late embryos (Fig. 1C ▶). The embryos die during embryogenesis (Leicht and Bonner 1988; our own observations), although we observed some individuals that survived up to the state of early first instar larvae. We attribute the delayed lethality phase to the persistence of the maternal contribution of eIF4E. The removal of the maternal contribution by flipase recombination targets (FRT)-mediated recombination resulted in sterile females with no germ cell development (data not shown). Thus, we conclude that l(3)67Af is a null allele of eIF4E-1/2.
FIGURE 1.
Molecular characterization of a mutant of the Drosophila eIF4E-1/2 gene and induction of apoptosis in embryos devoid of eIF4E-1/2. (A) Schematic representation of part of the eIF4E-1/2 (Hernández et al. 1997) gene and the alternative splicing events that generate the isoforms eIF4E1 and eIF4E2. The molecular defect (a mutation AG to AA) in the splicing acceptor site of the second intron on l(3)67Af mutant is indicated, as well as the truncated proteins that are theoretically translated from unspliced mRNAs (X indicates amino acids encoded by the intron). (B) RT-PCR from wild-type (+/+), heterozygous (l(3)67Af/+), and homozygous mutant embryos (l(3)67Af/l(3)67Af) shows the absence of the spliced form in homozygous mutants. Schemes on the left represent the spliced (81-nt long) and unspliced (98-nt long) mRNAs. (C) Western blot of protein extracts derived from wild-type (+/+) and mutant (l(3)67Af/l(3)67Af) embryos. The arrow indicates the position of eIF4E1. Although always remarkable, the reduction of protein levels varied in different experiments, which we attribute to the remaining of the maternal contribution and the difficulty in properly staging mutant embryos. (D) TUNEL in wild-type embryo (stage 13) showing the pattern of cell death in the head and nervous system. (E) Apoptosis is extended in mutant embryos lacking eIF4E-1/2. Staging of mutant embryos is difficult because of developmental defects. (F) Whole-mount in situ hybridization using an antisense rpr RNA probe in wild-type embryos (stage 11). The pattern of rpr mRNA expression correlates with the pattern of apoptosis. (G) rpr expression is largely extended in embryos devoid of eIF4E-1/2. In all cases, embryos are displayed with the anterior pole to the left, dorsal side up.
Reduction of eIF4E levels has been related to apoptosis (Herbert et al. 2000; Polunovsky et al. 2000). Therefore, we analyzed the pattern of programmed cell death in l(3)67Af embryos by TUNEL technique. When compared to wild-type embryos (Fig. 1D ▶), homozygous l(3)67Af embryos showed widespread DNA fragmentation from stage 10 to 12 on Figure 1E ▶. This observation indicates that the absence of eIF4E results in the induction of apoptosis in the context of a whole developing organism, where no depletion of nutrients occurs and sensitized cells are not especially affected, requirements that have been reported as necessary in cell culture. The absence of apoptosis in early embryonic stages also correlates with the contribution of maternal eIF4E up to stage 10–12 (data not shown). One of the early pro-apoptotic genes in Drosophila is rpr (White et al. 1994), which induces apoptosis when it is expressed in cells or embryos. Thus, we monitored the expression of rpr by whole-mount in situ hybridization. When compared to wild-type embryos (Fig. 1F ▶), mutant embryos beyond stage 10 revealed extensive ectopic activation of rpr (Fig. 1G ▶), which is the likely cause of the observed apoptosis. The mechanism of increased rpr mRNA transcription in the absence of eIF4E remains to be established. However, our observations raised the question of how rpr mRNA is translated in the absence of eIF4E.
reaper mRNA is translated in a cap-independent manner in vitro
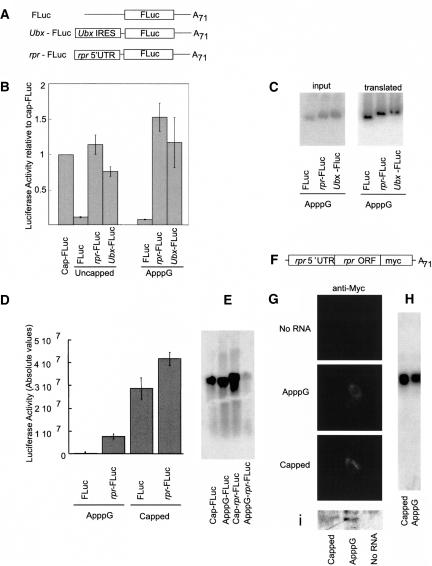
RPR is a poor antigenic protein. Despite the efforts of many laboratories, good quality antibodies against it are unavailable, and at present it is not possible to supply direct evidence for ectopic RPR protein expression in l(3)67Af embryos. For this reason, we analyzed rpr mRNA translation in a cell-free Drosophila-embryo translation system (Maroto and Sierra 1989; Gebauer et al. 1999). We used different in vitro-transcribed, mono-cistronic and polyadenylated reporter mRNAs containing several 5′UTRs either capped with m7GpppG or uncapped (Fig. 2A ▶). The mRNAs were added to the translation reaction at different concentrations and the activity of the encoded reporter gene (FLuc, firefly luciferase) measured at different time points within a linear response range (see Supplemental Figure, panel B, at www.mpibpc.gwdg.de/abteilungen/060/publications/Hernandez_RNA/Hernandez_RNA2004_SuppInfo.html). The translation of an uncapped control reporter mRNA (FLuc; Fig. 2B ▶) was 10× less efficient than the capped control mRNA (cap-FLuc; Fig. 2B ▶). In contrast, uncapped reporter mRNA containing the previously defined IRES from the Ubx mRNA (Ubx-Fluc) (Fig. 2B ▶; Hart and Bienz 1996) was translated five to eight times more efficiently than the uncapped control mRNA (FLuc). Similarly, the uncapped reporter mRNA containing the 5′UTR of rpr mRNA (rpr-Fluc; Fig. 2B ▶) was translated at levels equivalent to the capped control transcript cap-FLuc. Hence, rpr mRNA 5′UTR enhanced the translation of an uncapped reporter transcript in vitro, despite the fact that the stability of uncapped transcripts, measured by the addition of [32P]-labeled transcript to the reaction, was strongly reduced when compared to the capped reporter (see Supplemental Figure, panel C). The use of polyadenylated reporter mRNAs bearing a nonfunctional cap structure (ApppG instead of m7GpppG) also resulted in impaired cap-dependent translation (Fig. 2B ▶). In comparison with cap-FLuc, either uncapped or ApppG-FLuc transcripts were translated 10× less efficiently than the capped control. The translation efficiency of ApppG-capped rpr-FLuc and ApppG-capped Ubx-FLuc was equivalent to the capped control (cap-FLuc) and even more efficiently translated than the capped control with the presence of the cap analog. This is most likely the result of the increased stability of the reporter mRNAs (Fig. 2C ▶; Supplemental Figure, panel C). The same effect was observed in vivo after transfection of Drosophila S2 cells with m7GpppG- or ApppG- capped reporter mRNAs (Fig. 2D ▶). Although we consistently observed a decreased stability of ApppG-capped rpr-FLuc mRNA as analyzed by Northern blot (Fig. 2E ▶), we observed that the transcript bearing rpr 5′UTR was translated at least 10× better than the ApppG-capped control (Fig. 2D ▶). To rule out an effect of sequences downstream of the AUG or the rpr ORF, we designed a transcript bearing both rpr 5′UTR and the entire rpr ORF, tagged at the C terminus with a myc epitope (Fig. 2F ▶). Transfection of S2 cells with m7GpppG- or ApppG-capped mRNAs resulted in the translation of both forms as assessed by immunofluorescence (Fig. 2G ▶) and Western blot (Fig. 2I ▶) using anti-myc antibody. Northern blot analysis revealed that both transcripts are neither degraded nor cleaved in the experimental conditions used (Fig. 2H ▶). It is important to note that the translation in vivo of in vitro-transcribed ApppG-capped RNA indicates cap-independent translation and avoids the possible presence of cryptic promoters in the 5′UTRs (see below). Thus, the results confirm a cap-independent mechanism for rpr mRNA translation.
FIGURE 2.
rpr 5′UTR enhances cap-independent translation. (A) Reporter monocistronic mRNAs used in in vitro translation assays. A71 denotes a poly-A tail of 71 nt. (FLuc) Firefly luciferase. (B) Relative translation of uncapped and ApppG-capped transcripts in embryonic translation extracts relative to a 7mGpppG-capped control. (C) mRNA stability of the ApppG-capped transcripts assayed in B. (D) Reporter gene activity 6 h after transfection of S2 cells with mock-capped and capped transcripts assayed in A. The bars represent mean and standard deviation of four independent experiments using 5 ng (in vitro translation reaction) of 5 μg (cell transfection) of reporter mRNA. (E) Northern blot of transfected S2 cells of D reveals the integrity of the transcripts after 6 h of incubation. (F) rpr5′UTR-ORF-Myc-tagged construct used on in vivo assays of G. (G) Immunofluorescence detection of myc epitope in S2 cells mock-transfected (upper panel); transfected with in vitro transcribed, ApppG-capped, poly-adenylated rpr5′UTR-ORF-Myc (middle panel); or 7mGpppG-capped, polyadenylated rpr5′UTR-ORF-Myc transcript (lower panel). (H) Northern blot detection of the transcript in the cells corresponding to the transfection experiment shown in G. (I) Detection by Western blot of the translation products from the cells transfected with different forms of rpr5′UTR-ORF-Myc described in G using anti-myc antibody.
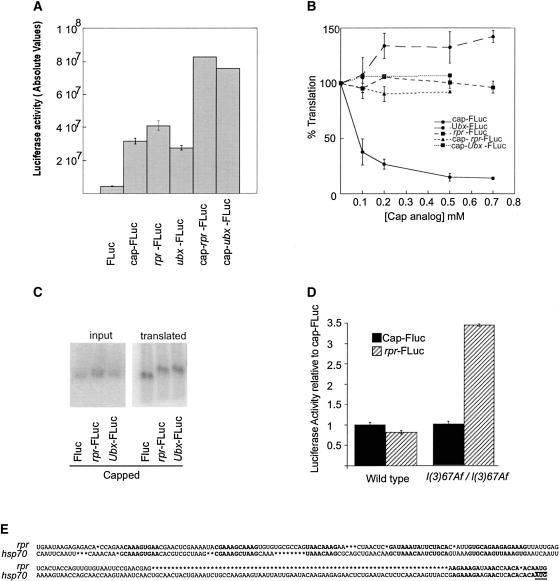
Capping either rpr-FLuc or Ubx-FLuc reporter mRNAs resulted in even higher levels of absolute translation in wild-type extracts with respect to the uncapped transcripts (Fig. 3A ▶). This may either be the consequence of increased mRNA stability or the result of the addition of cap-dependent and cap-independent translation initiation mechanisms. To distinguish between the two possibilities, we performed competition experiments by addition of the cap analog m7GpppG to the in vitro translation reaction in wild-type extracts, which mimics the lack of eIF4E in the lysate (Maroto and Sierra 1988). The addition of 0.1 mM cap analog resulted in a 60% decrease in the translation efficiency of cap-FLuc (Fig. 3B ▶, circles and solid line). Further addition of the cap analog reached a plateau at 90% inhibition, a residual activity equivalent to the remaining translation level observed when uncapped or ApppG-capped reporter transcripts were used (cf. Fig. 2B ▶). In contrast, the translation of either uncapped (circles and dashed line) or m7GpppG-capped (squares and dotted line) Ubx-FLuc and rpr-FLuc (squares and dashed line or triangles and dashed lines, respectively) was not significantly reduced by the addition of the cap analog (Fig. 3B ▶). Transcript degradation was not observed during the translation reaction (Fig. 3C ▶). The competition experiments described above rule out an additive effect of cap-dependent initiation on cap-containing reporters and suggest that the translation level observed for the capped rpr-Luc reporter mRNA is the result of improved mRNA stability. Therefore, we conclude that cap recognition is not required for the translation of the mRNA containing the rpr 5′UTR.
FIGURE 3.
Translation of capped rpr 5′UTR is not competed by free cap analog. (A) In vitro translation of capped and uncapped monocistronic reporter transcripts in translation extracts derived from wild-type embryos. (B) Competition of cap-dependent translation with increasing concentrations of free cap analog m7GpppG. The efficiency is referred to the translation of the reporter mRNA in the absence of the cap analog. The translation reactions proceed as described in Figure 2 ▶. (C) Stability analysis of the capped transcripts assayed in A and B. Radiolabeled transcripts were added to the translation mixture, purified at the end of the reaction, and separated in agarose gels. The same translation reaction was measured in parallel for expression of the reporter gene. (D) Translation of cap-FLuc and uncapped rpr-FLuc reporter transcripts in embryonic translation extracts prepared from wild-type and l(3)67Af homozygous embryos. The translation reaction was set up using an equivalent amount of embryos representing equal amounts of protein for each reaction. Note that extracts from different batches cannot be compared in absolute values, as their activity varies from lot to lot (see Materials and Methods for details). We observed a lower rate of translation in absolute values when using the mutant extracts (data not shown). However, the relative level of translation of cap-independent reporter transcripts to cap-dependent reporter is higher in extracts derived from l(3)67Af homozygous embryos. (E) Sequence comparison between the 5′UTR of Drosophila hsp70 and rpr mRNAs. Nucleotides highlighted in bold indicate motifs of similarity.
To prove that rpr can be translated in the eIF4E-1/2 mutant condition, where eIF4E is absent, we performed the same in vitro experiments using a translation extract derived from homozygous l(3)67Af embryos (Fig. 3D ▶). Equal amounts of protein were used in every assay to make relative results comparable (see Materials and Methods). We observed that the absolute values of translation in mutant extracts decreased with respect to wild-type extracts, a likely consequence of diminished overall translation activity (data not shown). As described earlier, the translation level of both cap-FLuc and uncapped rpr-FLuc was similar in wild-type extracts. However, in the mutant extracts, the relative efficiency of translation of rpr-FLuc was 3.5× higher than the cap-FLuc control mRNA (Fig. 3D ▶). This indicates that rpr-FLuc is more efficiently translated than a capped transcript in the absence of endogenous eIF4E in the extracts.
The mode of translation described here for rpr resembles that described for Drosophila heat shock protein mRNAs. Analysis of the 5′UTR of rpr shows that it is short (170 nt), has a high content of adenine (45%), and lacks an obvious secondary structure. In addition, it neither contains short ORFs nor the polypyrimidin track characteristic of some IRESs (Jackson 2000). These features are nonetheless found to be frequent in heat shock protein mRNA 5′UTR (Schneider 2000). Sequence comparison showed that rpr mRNA 5′UTR displays tracts of high similarity to the 5′UTR of heat shock protein 70 mRNA from D. melanogaster (Dm-hsp70; Fig. 3E ▶). This similarity was not observed when rpr or Dm-hsp70 5′UTRs were compared to human-hsp70. This led us to consider that rpr and Dm-hsp70 might share similarities in the translation mechanism.
rpr and Dm-hsp70 mRNA 5′UTRs display IRES activity
To evaluate the mechanism of cap-independent translation, we performed in vitro translation assays with capped dicistronic reporter transcripts, which either lack intercistronic sequences (FLuc/RLuc, negative control), bear the IRES of the Ubx mRNA (FLuc/Ubx/RLuc), or contain the rpr 5′UTR (FLuc/rpr/RLuc) in the intercistronic region (Fig. 4A ▶). As expected, the presence of the Ubx IRES increased the efficiency of translation of the second cistron (RLuc) by a factor of three (with respect to the first cistron, FLuc) when compared to the control without an IRES (Fig. 4B ▶). The presence of rpr 5′UTR resulted in a modest but statistically significant increase of 1.5-fold of the second to the first cistron when compared to the control (Fig. 4B ▶). The same effect was observed when a transcript bearing Dm-hsp70 5′UTR was used as an intercistronic region. On the contrary, the insertion of rpr 5′UTR in antisense orientation (FLuc/rpr anti/RLuc) resulted in the reduction of the second cistron activity. The integrity of the transcripts, assessed by the addition of labeled mRNA as tracer and evaluated after the translation reaction, was not affected within the level of detection of the method used, indicating that the translation of the second cistron was not the result of mRNA cleavage (Fig. 4C ▶). Competition experiments using cap analog showed that the efficiency of translation of the second cistron (IRES-dependent) increased, while the translation of the first cistron (cap-dependent) decreased (data not shown). However, we observed that the second cistron of the reporter FLuc/RLuc displayed a relatively high level of translation, likely due to ribosome readthrough (see Fig. 4B,G ▶). Therefore, we could not rule out at this point that the relatively modest, although significant, IRES activity of rpr 5′UTR and hsp70 5′UTR could have been the consequence of enhanced intercistronic readthrough. This argument is relevant if we consider that both rpr and hsp70 5′UTRs are short and do not have any potential secondary structure. To address this problem, we assayed IRES activity using the dicistronic reporter vector pFLuc/cad/RLuc, which contains the 5′UTR of the Drosophila maternal caudal (cad) mRNA downstream of the first cistron (Fig. 4D ▶). cad 5′UTR is not an IRES, possesses a significant secondary structure, and efficiently prevents readthrough in the same way as observed for the use of hairpins or inactive IRESs (P. Vázquez-Pianzola and R. Rivera-Pomar, unpubl.). The insertion of the Ubx 5′UTR in the reporter FLuc/cad/Ubx/RLuc resulted in a 12-fold relative increase of the second cistron translation with respect to the control mRNA FLuc/cad/RLuc, which is likely the consequence of the decrease in ribosomal readthrough in the control RNA (Fig. 4E ▶). The analysis of the reporter FLuc/cad/rpr/RLuc also showed that the second cistron was efficiently translated with a 14-fold increase with respect to the control FLuc/cad/RLuc and equivalent to that for FLuc/cad/Ubx/RLuc (Fig. 4E,G,H ▶). The increase of IRES activity was also evident for the insertion of hsp70 5′UTR sequence. In all of these cases, the integrity of the reporter mRNAs was not altered during the translation reaction (Fig. 4F ▶). Although we established that blocking the readthrough improves the detection of the IRES activity of rpr and Dm-hsp70 5′UTRs, we observed that in the presence of cad 5′UTR and the rpr or Dm-hsp70 5′UTR sequences, the absolute activity of the second cistron also increases. Moreover, the use of ApppG-capped dicistronic reporter transcripts showed that while the activity of the first cistron is stopped, the second cistron is still translated (Fig. 4 ▶, cf. H and G). Therefore, we could not rule out that the sequence within cad 5′UTR (and translation factors potentially binding therein) enhances the activity of rpr and hsp70 5′UTR. To exclude the effect of cad 5′UTR on the activity of the second cistron, we inserted a synthetic hairpin of high stability (see Materials and Methods; Coldwell et al. 2001) to prevent readthrough (Fig. 4I ▶). The results of the in vitro translation activity and mRNA stability experiment, including a control of the rpr 5′UTR inserted in antisense orientation, confirmed the data obtained using the cad-based reporter mRNA and strongly indicated that rpr and hsp70 5′UTR display IRES activity (Fig. 4J,K ▶).
FIGURE 4.
rpr and Dm-hsp70 5′UTR do contain an internal ribosome entry site that functions in vitro. (A) Reporter dicistronic mRNAs used in the in vitro translation assays. rpr-anti indicates a construction in which rpr 5′UTR was cloned in inverse orientation with respect to the second cistron AUG. (RLuc) Renilla luciferase. (B) Normalization of the translation efficiency of dicistronic transcripts. The graph shows the ratio of RLuc activity (translation of the second cistron) to FLuc activity (first cistron) normalized with respect to the same ratio in the FLuc/RLuc transcript. (C) Stability analysis of the dicistronic reporter mRNAs used in B. (D) Reporter dicistronic mRNAs containing the 5′UTR of caudal to prevent readthrough of ribosomes. (E) Translation efficiency of dicistronic transcripts depicted in D is normalized as in B but to the ratio derived from the use of FLuc/cad/RLuc reporter RNA. (F) Stability analysis of the dicistronic reporter mRNAs used in E. (G) Absolute values of translation corresponding to the first cistron (FLuc, white bars) and the second cistron (RLuc, black bars) of the constructs indicated in A and D using m7GpppG-capped transcript. (H) Absolute values of translation corresponding to the first cistron (FLuc, white bars) and the second cistron (RLuc, black bars) of the constructs indicated in A and D using ApppG-capped transcript. Note that the scale is one order of magnitude lower than G. Absolute values are comparable, as all of the experiments were performed with the same lot of extracts. The activity of the first cistron decreases dramatically, while the activity of the second cistron remains either constant or increases. (I) Reporter dicistronic mRNAs containing a synthetic, stable hairpin that prevents readthrough of ribosomes. (J) Translation efficiency of dicistronic transcripts depicted in I normalized to the FLuc/cad/RLuc. (K) Stability analysis of the dicistronic reporter mRNAs used in J. Values of all experiments represent at least four independent experiments.
To validate our observations, we then proceeded to study reporter mRNAs in vivo. We transfected Drosophila S2 cells with the same transcripts used in the in vitro experiment shown in Figure 4, A–C ▶. We observed that m7GpppG-capped dicistronic reporter mRNAs bearing either rpr or Dm-hsp70 5′UTRs display IRES activity as described for the in vitro translation assays (cf. Figs. 5A ▶ and 4B ▶). Northern blot experiments showed that the transcript is stable and remains intact after the transfection and incubation time (Fig. 5B ▶). We finally analyzed the translation of several dicistronic reporter plasmids in DNA transfected S2 cells. The reporter mRNAs used contain the minimal Adh promoter under control of five upstream activation sequence (UAS) sites, which are activated by Gal4, and two reporter cistrons: myc-tagged cyan fluorescent protein (myc-CFP, first cistron) and hemagglutinin-tagged yellow fluorescent protein (HA-YFP-NLS bearing a nuclear localization signal, second cistron; Fig. 5C ▶). Cotransfection of Drosophila S2 cells with pActin-Gal4 and either the dicistronic reporter gene without intercistronic sequences or bearing the rpr 5′UTR was performed (Fig. 5D ▶). We observed that CFP (first cistron) is expressed both in cells transfected with the control plasmid and in cells transfected with the rpr-containing construct, while YFP (second cistron) is only detected in the nuclei of cells that contains the rpr 5′UTR upstream of the second cistron (Fig. 5D ▶). The score of the cells that display CFP expression showed that none of them expressed YFP in the absence of rpr 5′UTR sequences, but all of them showed nuclear YFP when rpr 5′UTR sequences were inserted. Although we believe we have demonstrated the presence of an IRES transfecting dicistronic reporter RNA, we have yet to confirm that the activity determined using the plasmidic reporter was not the result of cryptic promoters within rpr 5′UTR, either in its natural state or in a Gal4-dependent manner. We cloned the rpr 5′UTR in a promoterless vector and cotransfected the cells with pActinGal4 (see Materials and Methods). None of the cells transfected with the rpr-HA-YFP-NLS plasmid expressed YFP (Fig. 5D ▶), while a control bearing the 5′UTR of an alternatively spliced isoform of Drosophila eIF4E1 mRNA, which contains a cryptic promoter (P. Vázquez-Pianzola and R. Rivera-Pomar, un-publ.), indeed expressed YFP (Fig. 5D ▶).
FIGURE 5.
In vivo analysis of rpr and Dm-hsp70 IRES. (A) Relative translation activity of dicistronic reporter gene expression in S2 cells transfected with the m7GpppG-capped transcripts depicted in Figure 4A ▶. (B) Northern blot experiments on transfected S2 cells show that the transcripts are stable after 6 h of incubation. There is no sign of degradation or cleavage. (C) Dicistronic plasmids containing the cyan fluorescent protein ORF (myc-CFP, first cistron, cytoplasmic) and the yellow fluorescent protein ORF (HA-YFP-NLS, second cistron, nuclear) under the Gal4-dependent UAS-Adh promoter. (SV40) The SV40 t-antigen 3′UTR and polyadenylation signal. (D) Drosophila S2 cells cotransfected with the plasmids depicted in C and actin-Gal4, which activates the UAS-Adh promoter. Merged figure displays digitalized colors, CFP (green) and YFP (red). (Ph. C.) Phase contrast. The panels correspond to the construct used for transfection. For details, see text.
We believe that the entire set of in vitro and in vivo experiments, designed to assess translation of a variety of reporter mRNAs, its stability, and the absence of cryptic promoters, provides clear evidence of IRES activity in the rpr and Dm-hsp70 5′UTRs.
Do rpr and hsp70 share a common mechanism of translation?
To further analyze rpr and hsp70 mRNA translation, we tested the ability of different reporter mRNAs to function in vitro in translation extracts derived from untreated and heat-shocked embryos, where cap-dependent initiation is severely impaired and translation of heat shock protein mRNAs is favored (Schneider 2000). In extracts derived from untreated embryos, ApppG-capped hsp70-FLuc and rpr-FLuc mRNAs were efficiently translated at a level comparable to m7GpppG-capped FLuc control (Fig. 6A ▶, data normalized to hsp70-FLuc). In translation extracts derived from heat-shocked embryos, translation of the capped FLuc control is dramatically reduced up to 10-fold, as we have observed in the competition experiments and by using ApppG-capped transcripts (Figs. 2B ▶, 3B ▶) when compared to hsp70-FLuc translation (Fig. 6B ▶). At the same time, the translation of ApppG-capped rpr-FLuc reporter increases, likely because of the availability of idle translational machinery. These results are in agreement with the data presented above, which show the ability of rpr-FLuc to be translated in a cap-independent manner. We next assayed the ability of the different 5′UTRs to support translation of the second cistron in dicistronic reporter mRNAs containing either a functional cap (Fig. 6C ▶) or the nonfunctional cap analog ApppG (Fig. 6D ▶) in heat-shocked extracts. The absolute values for the first and second cistron for the different dicistronic mRNAs used are summarized in Figure 6E and F ▶. These data demonstrate that the 5′UTR of Dm-hsp70 mRNA supports IRES-dependent translation, as it does rpr 5′UTR also in conditions of heat shock. When combined, these data indicate that rpr and Dm-hsp70 mRNAs are translated in a similar cap-independent fashion, and that both 5′UTRs support IRES activity.
FIGURE 6.
rpr and Dm-hsp70 5′UTR IRESs are functional during heat shock. Monocistronic mRNA reporter expression in translation extracts derived from untreated (A) and heat-shocked (B) embryos. Normalized activity of capped (C) and ApppG-capped (D) dicistronic mRNA reporters in translation extracts derived from heat-shocked embryos. In all experiments, the normalization is referred to the hsp70-containing reporter mRNA. (E) Absolute values of reporter gene expression activity of 7mGpppG-capped transcripts in heat shock extracts. Cap-dependent translation is abolished. (F) Absolute values of reporter gene expression activity of ApppG-capped transcripts in heat shock extracts.
rpr and hsp70 mRNAs are associated with polysomes after heat shock and apoptosis induction in embryos
To obtain definitive evidence of the ability of the endogenous rpr and hsp70 mRNAs to be translated in vivo, we separated ribosomal fractions from translation extracts derived from 0- to 12-h-old Drosophila embryos under normal conditions of development or after either heat shock or irradiation with X-rays, which induces the transcription of hsp70 mRNA and rpr mRNA, respectively. The heat-shocked embryos mimic the absence of eIF4E in the l(3)67Af homozygous mutant. We used them for polysome analysis because the acquisition of enough staged homozygous mutant embryos by hand sorting was unworkable. By ultracentrifugation in sucrose gradients, we isolated mRNAs associated with two well-defined ribonucleoprotein fractions: (1) initiation complexes (43S, 48S, 80S), which correspond to untranslated or newly initiated mRNAs (U in Fig. 7A ▶), and (2) the polysomal fraction that contains mRNAs that are being efficiently translated into proteins (P in Fig. 7A ▶). When compared to untreated embryos (Fig. 7A ▶, left panel), the sucrose profile of heat-shocked embryos (Fig. 7A ▶, middle panel) showed a sharp decrease of polysomes, reflecting the inhibition of translation that occurs during heat shock. A reduction in the amount of polysomes was also observed in sucrose profiles derived from X-ray-irradiated embryos (Fig. 7A ▶, right panel). The effect of irradiation is less dramatic than the effect of heat shock and might reflect a smaller fraction of cells damaged after irradiation with respect to the number of cells responding to heat shock. We then studied the presence of the transcripts corresponding to Actin5C (a cap-dependent transcript; Fig. 7B ▶, left panel), hsp70 (Fig. 7B ▶, middle panel), and rpr (Fig. 7B ▶, right panel) mRNAs in both untranslated and polysome-associated fractions derived from untreated and heat-shocked embryos. The fraction of polysome-associated mRNAs with respect to the untranslated ones provides an indication of their level of translation (P/U in Fig. 7 ▶ insets). We first observed that the combined amount of hsp70 and rpr mRNA is lower than the amount of Actin5C mRNA in the control (Fig. 7B ▶, black lines). Actin5C mRNA is actively translated under normal conditions, as indicated by a P/U ratio 2 (Fig. 7B ▶, left panel), while a ratio of 1 was observed for hsp70 mRNA (Fig. 7B ▶, middle panel) and rpr mRNA (Fig. 7B ▶, right panel). This implies that all three mRNA are being translated, although hsp70 and rpr mRNAs are present in lower amounts than Actin5C. After heat shock treatment (Fig. 7B ▶, red lines), we observed an increase of the total amount of hsp70 mRNA and a decrease in the levels of Actin5C and rpr mRNAs relative to untreated embryos (Fig. 7D ▶). However, a major change was evident: The total amount of Actin5C mRNA associated with polysomes decreased (P/U < 1), while the ratio of polysome-associated rpr mRNA remained stable and that of hsp70 mRNA increased (Fig. 7B ▶). Our data demonstrate that during heat shock, a cap-dependent mRNA decreases its translation level, while both rpr and hsp70 mRNAs are still actively translated, irrespective of their relative level of transcription.
FIGURE 7.
Recruitment of actin5C, hsp70, and rpr mRNAs to polysomes during heat shock and apoptosis in Drosophila embryos. (A) Sedimentation profiles in sucrose gradients from untreated (left), heat-shocked (middle), and X-ray-irradiated (right) Drosophila embryos. The position of ribosomal complexes 40/43/48S and 80S is indicated. (U) Fraction containing untranslated/initiated mRNAs; (P) polysomal fraction. (B) Amplification of actin5C, hsp70, and reaper mRNAs in U and P fractions by real-time quantitative-RT-PCR experiments from heat-shocked embryos. (C) Same experiment as in B, conducted with X-ray-irradiated embryos. The amplification course shows the mRNA present in the control (black lines) or treated (red lines) embryos either in the fraction P (closed circles) or U (crosses). The insets represent the P/U ratio in the control (dashed bars) or treated (grey bars) embryos for each mRNA tested. (E) Comparison of the total amounts of mRNAs from the experiments performed in B and C. The amount of mRNA is calculated as P + U and normalized to untreated embryos.
It has been shown that irradiation with X-rays induces the transcription of rpr mRNA in embryos and, in consequence, apoptosis, and that the induction of apoptosis results in impaired translation (Clemens et al. 2000; Ollmann et al. 2000). Thus, we conducted an equivalent experiment using translation extracts derived from X-ray-irradiated Drosophila embryos to provide evidence for the ability of rpr to be translated during apoptosis induction (Fig. 7C ▶). As expected, the overall transcription of rpr mRNA increased (Fig. 7D ▶), no major changes were observed in the levels of Actin5C mRNA, while transcription of hsp70 was reduced. However, the association of Actin5C mRNA with polysomes decreases (P/U = 1.5; Fig. 7C ▶, left panel), while the level of hsp70 and rpr mRNAs associated with polysomes slightly increases (P/U = 1.5; Fig. 7C ▶, middle and right panels). Although the results are less dramatic than those observed in heat-shocked embryos, perhaps due to the number of cells effectively undergoing apoptosis, cap-dependent translation decreases, whereas cap-independent initiation is maintained or slightly elevated. Our data indicate that during apoptosis, mRNAs are poorly translated, reflecting a lower initiation rate, and suggest that cap-dependent, but not cap-independent, translation is preferentially impaired. The increase in rpr mRNA association with polysomes indicates that rpr mRNA is translated in vivo under conditions of diminished cap-dependent initiation through both heat shock and irradiation. The higher proportion of hsp70 and rpr mRNAs in polysomes can be attributed to the increased availability of the basic translation machinery on inhibition of cap-dependent translation, eventually resulting in an important increase of the absolute mass of HSP70 and RPR protein synthesized during heat shock and apoptosis, respectively.
DISCUSSION
Previous studies have shown that the inhibition of eIF4E function in serum-starved, but not in serum-fed, cells in culture induces apoptosis (Herbert et al. 2000; Polunovsky et al. 2000). Our data indicate that the reduction of eIF4E promotes apoptosis in cells developing in their natural environment. Despite the important work performed on eIF4E since the early 1970s, a complete knock-out of the gene has not yet been described in a multicellular organism. In yeast, the mutant cdc33-1, which partially affects the function of eIF4E, shows a reduced cap-binding activity and, therefore, a slow growing rate (Brenner et al. 1988; Altmann and Trachsel 1989). In higher organisms, loss-of-function studies have been limited to antisense experiments in mammalian cells in culture (De Benedetti et al. 1991; Joshi-Barve et al. 1992). The use of RNAi in Caenorhabditis elegans first allowed the study of the role of eIF4E in developmental processes (Keiper et al. 2000), but gene redundancy rendered functional assessment very difficult, and no evidence for induced apoptosis has been provided to date. More recently, a hypomorphic, nonembryonic lethal allele of Drosophila eIF4E-1/2 was shown to affect cell growth and proved to be very useful to determine Ser 209 phosphorylation in vivo and its role for eIF4E function (Lachance et al. 2002). Here we showed that the mutant l(3)67Af (see also Lachance et al. 2002) is a null allele of eIF4E that lacks the isoforms 1 and 2. l(3)67Af is recessive embryonic lethal and shows widespread induction of apoptosis in homozygosis, a process likely triggered by the up-regulation of at least one pro-apoptotic gene, rpr. Activation of rpr has also been observed in other genetic backgrounds, such as crumbs (Nordstrom et al. 1996). How rpr transcriptional activation occurs under these circumstances is still unknown, and the link between a lack of eIF4E and the activation of rpr requires further investigation. Another important conclusion from our genetic analysis is that, at least in Drosophila, eIF4E is not a limiting factor in vivo, as it is now accepted for the studies carried out in rabbit reticulocyte lysates (Rau et al. 1996). In fact, heterozygous l(3)67Af flies, in which the dose of eIF4E is reduced to 50%, are viable, normal, and fertile. Finally, as in C. elegans, several eIF4E genes have been annotated in the Drosophila genome (Lasko 2000). However, the embryonic lethality of the homozygous l(3)67Af mutant indicates an absolute requirement for eIF4E1 and/or eIF4E2 in early developmental stages. This agrees with two observations: only eIF4E-1/2 (the gene analyzed here) is expressed during embryogenesis and eIF4E1 is the unique isoform identified from eIF4F complexes purified from embryos (Maroto and Sierra 1989; Zapata et al. 1994; Hernández and Sierra 1995; G. Hernández, J.M. Sierra, and R. Rivera-Pomar, unpubl.).
It has recently been shown that general inhibition of protein synthesis takes place in Drosophila, Xenopus, and rabbit reticulocyte translation extracts in vitro in the presence of RPR (Holley et al. 2002; Yoo et al. 2002). Yoo et al. consider that if RPR (as well as GRIM, another pro-apoptotic gene product) promotes apoptosis by inhibition of protein synthesis, the inhibition of translation by other mechanisms might be expected to promote apoptosis. They report, indeed, that cycloheximide treatment of Drosophila cells results in apoptosis. The mechanism of protein synthesis inhibition by RPR remains to be established. Presumably, it would be a general process that might include rpr mRNA translation inhibition as well. However, we show that the lack of an essential factor for protein synthesis induces rpr mRNA transcription, which in turn must escape translation inhibition. We demonstrate that rpr can be translated in the absence of cap-binding activity in vitro by using translation extracts derived from eIF4E mutant or heat-shocked embryos or by competition with a cap analog. We revealed that rpr and hsp70 mRNAs, two antagonizing genes induced in response to stress, are translated in a cap-independent manner in vitro and that they are associated with polysomes in vivo in conditions of diminished cap-dependent translation mediated by stress. Our experiments coincide with the observation of Joshi-Barve et al. (1992) that hsp70 but not actin mRNA is associated with polysomes when cap-binding activity is reduced in HeLa cells by antisense eIF4E RNA, which we now extend to the translation of those genes during stress and apoptosis.
IRESs have been identified in some cellular mRNAs that encode apoptotic regulators such as XIAP, p36, BiP, and Apaf-1 (Holcik et al. 2000). Thus, IRES-independent translation is a common theme during apoptosis and possibly in other stress responses (Johannes et al. 1999). The resemblance and similar behavior of rpr and Dm-hsp70 mRNA 5′UTRs correlates with the ability of both rpr and Dm-hsp70 mRNAs to be translated under heat shock and X-ray irradiation conditions, as our polysome analysis showed. A very recent finding suggests that human-hsp70 mRNA might also support IRES activity (Rubtsova et al. 2003), in contradiction to previously published evidence (Vivinus et al. 2001). Although the IRES capability of human-hsp70 5′UTR remains a matter of controversy, more definitive evidence has been provided to support that human-hsp70 mRNA uses a shunting mechanism (Yueh and Schneider 2000). The evidence presented here shows that both Dm-hsp70 and rpr mRNAs are indeed translated in an IRES-dependent manner and that their 5′UTR shows sequence similarity, although most of the homology blocks between rpr and hsp70 5′UTR seem to be dispensable for reporter mRNA translation in a cap-independent context (P. Vázquez-Pianzola and R. Rivera-Pomar, un-publ.). The mechanism of shunting demonstrated by Yueh and Schneider (2000) for the translation of human-hsp70 mRNA involves the pairing of sequences within the 5′UTR and the 18S rRNA. In agreement with the observation of Yueh and Schneider (2000) for Dm-hsp70 5′UTR, we could not find any pairing possibility between rpr or Dm-hsp70 5′UTR mRNAs and Drosophila 18S rRNA, and the similarity we observed between rpr and Dm-hsp70 does not extend to human-hsp 70 mRNA, indicating that the shunting mechanism established for human-hsp70 mRNA might not necessarily apply to the Drosophila homolog.
Discrepancy with respect to the existence and nature of cellular IRESs has recently arisen (Kozak 2001a,b; Schneider et al. 2001). A positive consequence of this debate has been that, while IRESs do indeed exist, the experimental standards required to prove their activity have increased. We consider that we have fulfilled them in this study. Indisputable IRES, namely, the 5′UTR of certain viruses, implies a complex RNA secondary structure, sometimes the presence of several short ORFs, and a polypyrimidin track. Cellular IRESs are less characterized and show enormous heterogeneity (Jackson 2000). In this context, the IRESs of rpr and Dm-hsp70 appear to be unusual, suggesting that, rather than being defined structures, IRESs may constitute the intrinsic activity of certain mRNAs. Accordingly, the definition of IRES can be very broad and implies a variety of mechanisms, including shunting. IRESs such as those described in this study may reflect the selective advantage of an unstructured 5′UTR with the ability to direct the passive recruitment of ribosomes without the requirement of other factors such as the helicase eIF4A and, possibly, without scanning. Our experiments using translation extracts from heat-shocked embryos indicate that the IRESs of rpr and Dm-hsp70 are indeed translated in an eIF4F-independent manner. This mechanism might be advantageous under stress conditions where the amount of eIF4F complex decreases and the availability of idle ribosomes increases.
Both HSP70 and RPR act, by different pathways, in response to stress (Abrams 1999; Schneider 2000). Heat shock treatment does not result in increased apoptosis, likely due to the counteracting role of HSP70, which is produced at higher rates. HSP70 blocks apoptosis by binding to Apaf-1 (Saleh et al. 2000; Beere and Green 2001) and to the apoptosis-inducing factor AIF (Ravagnan et al. 2001). Thus, a simple mechanism of translation would allow Dm-hsp70 mRNA to escape the repression of translation by caspase-mediated cleavage of translation factors, at least during the initial steps of apoptosis. It is a challenging idea that, at least in Drosophila, where rpr and other pro-apoptotic genes have been identified, early apoptosis genes may be translated in a similar manner as heat shock protein mRNAs. Studies from our laboratory on the translation of other Drosophila pro-apoptotic genes, such as head involution defect (hid) and grim support this assumption (P. Vázquez-Pianzola and R. Rivera-Pomar, unpubl.). The hypothesis that heat shock response and apoptosis evolved in a similar context using common molecular mechanisms during cellular stress has recently been put forward (Beere and Green 2001). Here, we provide evidence supporting the idea of a shared mechanism of protein synthesis in genes involved in different developmental processes, one protecting and the other eliminating cells at risk.
MATERIALS AND METHODS
Mutant analysis and fly work
Mutants from the chromosomal region 67A2-B1 (l(3)67Aa2 ri1 pp/TM3, Sb1; l(3)67Ab2 ri1 pp/TM3, Sb1; l(3)67Ac1 ri1 pp/TM3, Sb1; l(3)67Ad1 ri1 pp/TM3, Sb1; l(3)67Ae1 ri1 pp/TM3 Sb1 and l(3)67Af1 ri1 e4/TM3, Sb1) (Leicht and Bonner 1988) were obtained from Mid-America Drosophila Stock Center. To identify homozygous mutant embryos, we balanced the mutations either to TM2 Ubx130, hkb-lacZ, or TM3, Act-GFP chromosomes. Wild type, heterozygous, and homozygous embryos were hand sorted after β-ga-lactosidase activity staining or in vivo using the GFP marker. eIF4E-1/2 gene was PCR-amplified from genomic DNA of single homozygous l(3)67A* mutant embryo, using primers corresponding to the nucleotides −652 to −623 and 2737–2708 of the eIF4E-1/2 gene (Hernández et al. 1997). Amplified DNA fragments were cloned into the pCR2.1 vector (Invitrogen) and fully sequenced in both strands. RNA was isolated from 10 to 20 embryos using the Total RNA purification kit (Qiagen) and digested with RNAse-Free DNase I (Qiagen). RT-PCR reactions were carried out with RNA derived from five embryos (stage 6–12) using the Access RT-PCR system (Promega) and primers corresponding to the nucleotides 1368–1391, 1437–1463, and 1511–1534 of the eIF4E-1/2 gene (Hernández et al. 1997). They amplified either an 81-nt-long (spliced) or a 98-nt-long (nonspliced) fragment from eIF4E-1/2 mRNAs. Western blot analysis was performed by loading 10–20 homozygous embryos (stage 8–12) per lane and was revealed with rabbit anti-Drosophila/eIF4E-1/2 antibody (Maroto and Sierra 1989). Apoptosis was detected by TUNEL with the In Situ Cell Death Reaction Kit, AP (Roche Diagnostics GmbH). Whole-mount in situ hybridization was performed using antisense RNA probes as described (Klinger and Gergen 1993).
Plasmids
The plasmids used in this study were derived from the monocistronic pLUC-cassette (Gebauer et al. 1999). 5′UTR cDNA sequences of Ubx (Hart and Bienz 1996), rpr (White et al. 1994), caudal, and Dm-hsp70 (McGarry and Lindquist 1985) were cloned into the SacI-NcoI site of pLUC-cassette. The dicistronic reporter vector (pFLuc/RLuc) was generated by cloning the Renilla luciferase (RLuc) gene into the HpaI site of pLUC-cassette (Gebauer et al. 1999). To prevent translation readthrough, we generated the plasmid pFLuc/cad/RLuc by cloning the caudal (cad) 5′UTR (Mlodzik and Gehring 1987) into the BglII site of pFLuc/RLuc. The 5′UTRs to be assayed for IRES activity were cloned into the ORF-intervening BglII site of both pFLuc/RLuc and pFLuc/cad/RLuc. The 5′UTR of the gene Ultrabithorax (Ubx), an already defined IRES in vivo (Hart and Bienz 1996; Ye et al. 1997), was used as positive control. The construct rpr-myc was generated by cloning the entire reaper 5′UTR and ORF, including a C-terminal myc tag, into the EcoRV site of pBluescript KS (Invitrogen). Further plasmids preventing readthrough of the second cistron were generated by insertion of a stable synthetic hairpin containing a 62-nt palindromic sequence (−687 Kcal/mol) (Coldwell et al. 2001) upstream of the second cistron (RLuc). This hairpin was shown to significantly reduce the translation of a second cistron when placed upstream of the second cistron of a bicistronic reporter without translational enhancement of the IRES activity (Coldwell et al. 2001). The hairpin was introduced into the BamHI site of the constructs FLuc/RLuc, FLuc/hsp70/RLuc, FLuc/rpr/RLuc, and FLuc/rprantisense/RLuc to create the plasmids FLuc/hairpin/RLuc, FLuc/hairpin/hsp70/RLuc, FLuc/hairpin/rpr/RLuc, and FLuc/hairpin/rpr antisense/RLuc, respectively.
In vitro translation assays
Translation extracts were prepared from 0 to 12-hour-old Drosophila embryos as described (Tuschl et al. 1999). In vitro translation was performed as described (Maroto and Sierra 1989; Gebauer et al. 1999) for 60 min at 30°C. Detailed protocols are found in the mentioned reference. The activity of every extract lot was titrated using different amounts of reporter mRNA and time-course experiments to determine the nonsaturating linear range of the reaction (Gebauer et al. 1999; see Supplementary figure, www.mpibpc.gwdg.de/abteilungen/060/publications/Hernandez_RNA/Hernandez_RNA2004_SuppInfo.html). We observed that different batches of translation extracts vary in their absolute translation activity, but the relative activity of each mRNA remains stable. This has also been observed by others (F. Gebauer, pers. comm.). It is important to note that the comparison of absolute values for different batches of the extracts or extracts derived from different treatments is impossible. In all cases where we compared absolute values, we used the same batch of extract. The degradation of the transcripts was assessed using traces of [32P]-labeled reporter RNA in the translation reaction. After completion of the reaction, the RNA was purified and analyzed by denaturing agarose gel electrophoresis and phosphorim-aging (see Supplementary figure, www.mpibpc.gwdg.de/abteilungen/060/publications/Hernandez_RNA/Hernandez_RNA2004_SuppInfo.html). Extracts from heat-shocked embryos were prepared from pools of 0- to 12-h embryos which have been treated for 45 min at 37°C and processed without further recovery. Translation extracts from mutant embryos were prepared as described above. The mutant embryos were hand sorted and staged by using the GFP marker as indicated above. Because of the difficulties in obtaining the required amount of fresh embryos in the proper stage, different batches of extract (~50–100 embryos per batch) were prepared, frozen, and pooled for the assays as described above. Protein content on the extracts was measured by Bradford assay. Either m7GpppG or ApppG-capped transcripts (New England BioLabs) or uncapped transcripts were synthesized using T3 RNA polymerase (Ampliscribe mRNA transcription kit, Biozym Diagnostik GmbH). Reporter gene expression (Firefly and Renilla luciferases) was determined using the Dual-luciferase reporter assay system (Promega) and detected in a Monolight 2010 Luminometer (Analytical Luminescence Laboratory).
Cell transfection
Drosophila Schneider S2 cells (80% confluent) were cotransfected in 24-well plates with 150 ng of the dicistronic plasmids and 50 ng of the pActGal4 plasmid by using the Effectene transfection reagent (Qiagen). After 48 h of transfection, the fluorescence signal was acquired using a Zeiss LSM 310 confocal microscope and the appropriate filters set. For RNA transfection of S2 cells, RNA was prepared with the Ampliscribe mRNA transcription kit (Biozym Diagnostik GmbH) as described above. 1 × 106 Drosophila S2 cells were seeded in 35-mm Petri dishes and transfected with 5 μg of RNA using the Effectene reagent (Qiagen). Six hours after transfection, the cells were harvested and assayed for luciferase activity (~2.5 × 105 cells/assay) and Northern blot (1 × 105 cells/lane). The RNAs were separated in formaldehyde-agarose gels, transferred onto nylon membranes, and hybridized with [32P]-labeled anti-sense RNA probes, as described (Hernández and Sierra 1995). The expression of tagged RPR protein was detected using the mono-clonal antibodies 9E10 either by Western blot (1:10) or immunofluorescence (1:1).
Polysome analysis
D. melanogaster Oregon R embryos were collected in apple juice-agar plates and dechorionated. Embryos that were 0- to 12-h old were heat shocked for 45 min at 37°C and processed without recovery. For apoptosis induction, 0- to 8-h embryos were exposed to 4000 radians using a Torrex 150D X-ray irradiator and, after further development for 4 h at 25°C, processed. One hundred fifty milligrams of embryos were homogenized in 300 μL ice-cold buffer A (30 mM HEPES at pH 7.4, 100 mM K acetate, 2 mM Mg acetate, 5 mM DTT, 50 μ/mL RNasin, 2 mg/mL heparin, and EDTA-free protease inhibitor cocktail Complete [Roche Diagnostics GmbH]). The homogenate was clarified by centrifugation at 20,000g for 20 min. Two hundred microliters of the cleared lysate were layered onto a 10 mL 10%–50% sucrose gradient prepared in 15 mM Tris-HCl (pH 7.5), 15 mM MgCl2, 300 mM NaCl, and 1 mg/mL heparin and centrifuged in a Beckman Ti-SW41 rotor for 2.5 h at 36,000 rpm at 4°C. Ultraviolet (UV) absorbance was recorded at 254 nm and 0.5 mL fractions were collected. The fractions corresponding to preinitiation and initiation complexes (40/43/48S and 80S) and those corresponding to polysomes were pooled.
Quantitative real-time RT-PCR
Pooled fractions of preinitiation/initiation complexes or polysome-bound mRNAs were digested with proteinase K (150 μg/mL) in the presence of 1% SDS for 30 min at 37°C. The digestion was adjusted to sodium acetate 0.3 M and the RNA precipitated with ethanol. The pellet was dissolved in H2O. The resulting RNA was digested with RNase-free DNase I to prevent any contamination with genomic DNA, further purified using the RNeasy Mini Kit (Qiagen), and quantified by spectrophotometry. One hundred nanograms of RNA was used for quantitative real-time RT-PCR using the QuantiTect SYBR Green RT-PCR kit (Qiagen) in a DNA Engine Opticon System (M.J. Research Inc.). Sequence-specific 25-mer oligonucleotides for Drosophila actin5C, hsp70, and rpr mRNAs were designed to amplify 100-bp fragments.
Acknowledgments
We thank G. Vorbrueggen for the rpr probe, M. Bienz for the Ubx cDNA, F. Gebauer for the pLUC-cassette vector, S. Campuzano for her valuable advice during the initial genetic work, and S. Höppner for proofreading. This work was supported by the Max Planck Gesellschaft and the Deutsches Bundesministerium für Bil-dung und Forschung (grant 031U215B to R.R.-P.), and by grants from the Spanish Ministerio de Educación y Cultura (J.M.S.), and Fundación Ramón Areces (to the CBMSO).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7154104.
REFERENCES
- Abrams, J.M. 1999. An emerging blueprint for apoptosis in Drosophila. Trends Cell Biol. 9: 435–440. [DOI] [PubMed] [Google Scholar]
- Abrams, J.M., White, K., Fessler, L.I., and Steller, H. 1993. Programed cell death during Drosophila embryogenesis. Development 117: 29–43. [DOI] [PubMed] [Google Scholar]
- Altmann, M. and Trachsel, H. 1989. Altered mRNA cap recognition activity of initiation factor 4E in the yeast cell cycle division mutant cdc33. Nucleic Acids Res. 17: 5923–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beere, H.M. and Green, D.R. 2001. Stress management-heat shock protein-70 and the regulation of apoptosis. Trends Cell Biol. 11: 6–10. [DOI] [PubMed] [Google Scholar]
- Brenner, C., Nakayama, N., Goebl, M., Tanaka, K., Tohe, A., and Matsumoto, K. 1988. CDC33 encodes mRNA cap-biding protein eIF-4E of Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 3556–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, M.J., Bushell, M., Jeffrey, I.W., Pain, V.M., and Morley, S.J. 2000. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 7: 603–615. [DOI] [PubMed] [Google Scholar]
- Coldwell, M.J., deSchoolmeester, M.L., Fraser, G.A., Pickering, B.M., Packham, G., and Wills, A.E. 2001. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene 20: 4095–4100. [DOI] [PubMed] [Google Scholar]
- De Benedetti, A. and Harris, A.L. 1999. eIF4E expression in tumors: Its possible role in progression of malignancies. Int. J. Biochem. Cell Biol. 31: 59–72. [DOI] [PubMed] [Google Scholar]
- De Benedetti, A., Joshi-Barve, S., Rinker-Schaeffer, C., and Rhoads, R.E. 1991. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol. Cell. Biol. 11: 5435–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer, F., Corona, D.F.V., Preiss, T., Becker, P.B., and Hentze, M.W. 1999. Translational control of dosage compensation in Drosophila by Sex-lethal: Cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 18: 6146–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Hart, K. and Bienz, M. 1996. A test for cell autonomy based on a dicistronic messenger translation. Development 122: 747–751. [DOI] [PubMed] [Google Scholar]
- Herbert, T.P., Fahraeus, R., Prescott, A., Lane, D.P., and Proud, C.G. 2000. Rapid induction of apoptosis mediated by peptides that bind initiation factor eIF4E. Curr. Biol. 10: 793–796. [DOI] [PubMed] [Google Scholar]
- Hernández, G. and Sierra, J.M. 1995. Translation initiation factor eIF-4E fromDrosophila: cDNA sequence and expression of the gene. Biochim. Biophys. Acta 1261: 427–431. [DOI] [PubMed] [Google Scholar]
- Hernández, G., Diez del Corral, R., Santoyo, J., Campuzano, S., and Sierra, J.M. 1997. Localization, structure and expression of the gene for translation initiation factor eIF4E from Drosophila melanogaster. Mol. Gen. Genet. 253: 624–633. [DOI] [PubMed] [Google Scholar]
- Holcik, M., Sonenberg, N., and Korneluk, R.G. 2000. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 16: 469–473. [DOI] [PubMed] [Google Scholar]
- Holley, C.L., Olson, M.R., Colon-Ramos, D.A., and Kornbluth, S. 2002. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat. Cell Biol. 4:439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J. 2000. Comparative view of initiation site selection mechanisms. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 127–184. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Johannes, G., Carter, M.S., Eisen, M.B., Brown, P.O., and Sarnow, P. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. 96: 13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Barve, S., De Benedetti, A., and Rhoads, R.E. 1992. Preferential translation of heat shock mRNAs in HeLa cells deficient in protein synthesis initiation factors eIF-4E and eIF-4γ. J. Biol. Chem. 267: 21038–21043. [PubMed] [Google Scholar]
- Keiper, B.D., Lamphear, B.J., Deshpande, A.M., Jankowska-Anyszka, M., Aamodt, E.J., Blumenthal, T., and Rhoads, R.E. 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 275: 10590–10596. [DOI] [PubMed] [Google Scholar]
- Klinger, M. and Gergen, P. 1993. Regulation of runt transcription by Drosophila segmentation genes. Mech. Dev. 43: 3–19. [DOI] [PubMed] [Google Scholar]
- Kozak, M. 2001a. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 21: 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001b. New ways of initiating translation in eukaryotes? Author’s reply. Mol. Cell. Biol. 21: 8241–8246. [Google Scholar]
- Lachance, P.E.D., Miron, M., Raught, B., Sonenberg, N., and Lasko, P. 2002. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol. Cell. Biol. 22: 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko, P. 2000. The Drosophila melanogaster genome: Translation factor and RNA binding proteins. J. Cell Biol. 150: F51–F56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, C.A., Lachance, P., Sonenberg, N., and Lasko, P. 1996. Alternatively spliced transcripts from the Drosophila eIF4E gene produce two different cap-binding proteins. J. Biol. Chem. 271: 16393–16398. [DOI] [PubMed] [Google Scholar]
- Leicht, B.G. and Bonner, J.J. 1988. Genetic analysis of chromosomal region 67A-D of Drosophila melanogaster. Genetics 119: 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto, F.G. and Sierra, J.M. 1988. Translational control in heat-shocked Drosophila embryos. J. Biol. Chem. 263: 15720–15725. [PubMed] [Google Scholar]
- ———. 1989. Purification and characterization of mRNA cap-binding protein from Drosophila melanogaster embryos. Mol. Cell. Biol. 9: 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry, T.J. and Lindquist, S. 1985. The preferential translation of Drosophila hsp70 mRNA requires sequences in the untranslated leader. Cell 42: 903–911. [DOI] [PubMed] [Google Scholar]
- Mlodzik, M. and Gehring, W.J. 1987. Expression of caudal gene in the germ line of Drosophila: Formation of RNA and protein gradient during early embryogenesis. Cell Death Differ. 48: 464–478. [DOI] [PubMed] [Google Scholar]
- Morley, S.J. 1997. Intracellular signalling pathways regulating initiation factor eIF4E phosphorylation during the activation of cell growth. Biochem. Soc. Trans. 25: 503–509. [DOI] [PubMed] [Google Scholar]
- Nordstrom, W., Chen, H., Steller, H., and Abrams, J.M. 1996. Activation of the reaper gene during ectopic cell killing in Drosophila. Dev. Biol. 180: 213–212. [DOI] [PubMed] [Google Scholar]
- Ollmann, M., Young, L.M., Di Como, C.J., Karim, F., Belvin, M., Robertson, S., Whittaker, K., Demsky, M., Fisher, W.W., Buchman, A., et al. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101: 91–101. [DOI] [PubMed] [Google Scholar]
- Polunovsky, V.A., Gingras, A.C., Sonenberg, N., Peterson, M., Tan, A., Rubins, J.B., Manivel, J.C., and Bitterman, P.B. 2000. Translational control of the antiapoptotic function of Ras. J. Biol. Chem. 275: 24776–24780. [DOI] [PubMed] [Google Scholar]
- Rau, M., Ohlmann, T., Morley, S.J., and Pain, V.M. 1996. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J. Biol. Chem. 271: 8983–8990. [DOI] [PubMed] [Google Scholar]
- Ravagnan, L., Gurbuxani, S., Susin, S.A., Maisse, C., Daugas, E., Zamzami, N., Mak, T., Jaattela, M., Penninger, J., Garrido, C., et al. 2001. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat. Cell Biol. 3: 839–843. [DOI] [PubMed] [Google Scholar]
- Rubtsova, M.P., Sizova, D.V., Dmitriev, S.E., Ivanov, D.S., Prassolov, V.S., and Shatsky, I.N. 2003. Distinctive properties of the 5′-unstranslated region of human Hsp70 mRNA. J. Biol. Chem. 278: 22350–22356. [DOI] [PubMed] [Google Scholar]
- Saleh, A., Srinivasula, S.M., Balkir, L., Robbins, P.D., and Alnemri, E.S. 2000. Negative regulation of Apaf-1 apoptosome by Hsp70. Nat. Cell Biol. 2: 476–483. [DOI] [PubMed] [Google Scholar]
- Schneider, R,J. 2000. Translational control during heat shock. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 615–635. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schneider R.J., Agol, V.I., Andino, R., Bayard, F., Cavener, D.R., Chappell, S.A., Chen, J.J., Darlix, J.L., Dasgupta, A., Donze, O., et al. 2001. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 21: 8238–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg, N. and Gingras, A.C. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10: 268–275. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Zamore, P.D., Lehmann, R., Bartel, D.P., and Sharp, P.A. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 13: 3191–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivinus, S., Baulande, S., van Zanten, M., Campbell, F., Topley, P., Ellis, J.H., Dessen, P., and Coste, H. 2001. An element within the 5′ untranslated region of human Hsp70 mRNA which acts as a general enhancer of mRNA translation. Eur. J. Biochem. 268: 1908–1917. [DOI] [PubMed] [Google Scholar]
- White, K., Grether, M.E., Abrams, J.M., Young, L.M., Farrell, K., and Steller, H. 1994. Genetic control of programmed cell death in Drosophila. Science 264: 677–683. [DOI] [PubMed] [Google Scholar]
- Ye, X., Fong, P., Izuka, N., Choate, D., and Cavener, D.R. 1997. Ul- trabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol. Cell. Biol. 17: 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.J., Huh, J.R., Muro, I., Yu, H., Wang, L., Wang, S.L., Feldman, R.M., Clem, R.J., Muller, H.A., and Hay, B.A. 2002. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 4: 416–424. [DOI] [PubMed] [Google Scholar]
- Yueh, A. and Schneider, R.J. 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes & Dev. 14: 414–421. [PMC free article] [PubMed] [Google Scholar]
- Zapata, J.M., Martínez, M.A., and Sierra, J.M. 1994. Purification and characterization of eukaryotic polypeptide chain initiation factor 4F from Drosophila melanogaster embryos. J. Biol. Chem. 269: 18047–18052. [PubMed] [Google Scholar]