Abstract
UV-induced photochemical crosslinking is a powerful approach that can be used for the identification of specific interactions involving nucleic acid-protein and nucleic acid-nucleic acid complexes. 8-AzidoATP (8-N3ATP) is a photoaffinity-labeling agent which has been widely used to elucidate the ATP binding site of a variety of proteins. However, its true potential as a photoactivatable nucleotide analog could not be exploited due to the lack of 8-azidoadenosine phosphoramidite, a monomer used in the synthesis of RNA, and the inability of 8-N3ATP to serve as an efficient substrate for bacteriophage RNA polymerase. In this study, we explored the ability of SP6, T3, and T7 RNA polymerases and metal ion cofactors to catalyze the incorporation of 8-N3AMP into RNA. Whereas transcription buffer containing 2.0–2.5 mM Mn2+ supports T7 RNA polymerase-mediated insertion of 8-N3AMP into RNA, a mixture of 2.5 mM Mn2+ and 2.5 mM Mg2+ further improves the yield of 8-N3AMP-containing transcript. In addition, both RNA transcription and reverse transcription proceed with high fidelity for the incorporation of 8-N3AMP and complementary residue, respectively. Finally, we show that a high-affinity MS2 coat protein binding sequence, in which adenosine residues were replaced by 8-azidoadenosine, crosslinks to the coat protein of the Escherichia coli phage MS2.
Keywords: UV crosslinking, RNA–protein interaction, 8-N3ATP, modified transcript, in vitro transcription
INTRODUCTION
Biochemical machines responsible for performing diverse cellular processes such as replication of DNA and the transcription, splicing, and transport of mRNA play a crucial role in the development of a wide range of organisms (Maniatis and Reed 2002; Frouin et al. 2003). Despite significant advances in the field of cell and molecular biology, the molecular basis of these machines’ functioning is not well understood. Although biophysical tools such as X-ray crystallography and nuclear magnetic resonance (NMR) have built the knowledge base for structure–function relationships of many biologically important macromolecules, crystallization of intact cellular machinery such as the spliceo-some or its analysis by NMR remains a challenging task. In addition, the temporal nature of several protein–nucleic acid interactions within these highly dynamic cellular machines makes it even more difficult to identify interacting partners. Thus, in the absence of high-resolution structures, the development of agents that could identify key protein–nucleic acid interactions might lead to a better understanding of molecular recognition in biological systems and ultimately the functioning of biochemical machines.
UV-induced crosslinking is a powerful approach that has provided valuable information regarding the structural topography of RNA-RNA and RNA–protein assemblies (Wu and Green 1997; Shapkina et al. 2000). However, this technique requires irradiation of the biological sample with far-UV light (254 nm), which is known to cause damage to both protein and RNA. Thus, the use of photochemical agents which are activated with near-UV light (300–360 nm) and can crosslink with efficiencies greater than standard UV-induced crosslinking has become the method of choice (Favre 1990; Willis et al. 1993; Meisenheimer et al. 1996; Wang and Rana 1998; Costas et al. 2000). When inserted into RNA and irradiated, such agents not only identify interacting partners by the presence of site-specific crosslinking, but also provide detailed information about the molecular environment of ribonucleoprotein assemblies.
A variety of methods have been described for the incorporation of photocrosslinking agents into RNA (Hanna 1989; Sylvers and Wower 1993; Gaur and Krupp 1997; Yu 1999; Mundus and Wollenzien 2000). DNA template-dependent in vitro transcription in which one of the wild-type nucleoside triphosphates is replaced by a photoactivatable nucleotide analog is the most popular method for synthesizing RNAs for protein–RNA crosslinking (Milligan et al. 1987; Milligan and Uhlenbeck 1989). Alternatively, RNA containing a reactive chemical group at the 5′ or 3′ terminus is generated by in vitro transcription, followed by the coupling of photochemical probe to RNA via the reactive moiety (Burgin and Pace 1990; Fidanza et al. 1994). Additionally, a limited number of photoactivatable nucleotide analogs can also be incorporated into RNA by chemical synthesis (Shah et al. 1994; McGregor et al. 1996).
Although these approaches have proven to be useful, each suffers from some drawback. For example, p-azidophenacyl bromide, a thiol-specific photocrosslinking probe, cross-links at a location ~11 Å away from the actual site of attachment (Hixson and Hixson 1975) and thus may not necessarily describe the actual environment within the RNA–protein complex. Similarly, chemical RNA synthesis can be used to insert a photoactivatable nucleotide analog at a predetermined site; however, at present the chemical synthesis of oligoribonucleotides is limited to relatively short pieces of ~40–50 nucleotides in length (Davis 1995). Most importantly, there are very few photoactivatable nucleotide analogs that are available as phosphoramidites or function as substrates for bacteriophage RNA polymerases, and most of them are pyrimidine-based. A pyrimidine-based photo-crosslinking nucleotide analog may therefore be of limited use if: (1) an adenine base or the purine-rich region of the RNA mediates protein–RNA interaction, or (2) if the replacement of a purine base by a pyrimidine-based photo-crosslinking nucleotide abolishes the protein–RNA interaction.
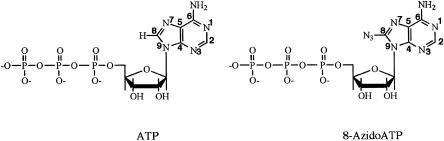
A number of adenine-based photoactivatable nucleotides have been described (MacMillan et al. 1994; Parang et al. 2002). Among them, 8-AzidoATP (8-N3ATP) has been widely used to map the active site of a variety of enzymes, and the nucleotide-binding domains of several proteins (Fig. 1 ▶; Potter and Haley 1983 and references therein). Due to the direct attachment of azide group on the base (at position 8), this nucleotide functions essentially as a zero length photoaffinity-labeling agent (Fig. 1 ▶). Moreover, it can be activated with far-UV light (~300–360 nm) that is less damaging to proteins and nucleic acids (Potter and Haley 1983; Sylvers and Wower 1993). It has been shown that 8-N3ATP functions as an elongation substrate for Escherichia coli RNA polymerase (Bowser and Hanna 1991), but appears to act as an inefficient substrate for template-dependent in vitro transcription with T7 RNA polymerase (the present study). In addition, T7 RNA polymerase-catalyzed synthesis of longer RNAs containing 8-N3AMP residues or related kinetic parameters has never been reported.
FIGURE 1.
Structures of ATP and 8-N3ATP.
In this study, we investigated the in vitro transcription conditions that would allow template-dependent insertion of 8-N3AMP into RNA. We report here that in the presence of 2.0–2.5 mM Mn2+, 8-N3ATP not only functions as a substrate for T7 RNA polymerase, but also RNAs as large as 100–300 bases can be synthesized with good yields. Additionally, both RNA transcription and reverse transcription proceeded with high fidelity for the incorporation of 8-N3AMP and its complementary residue, respectively. Finally, we show that a high-affinity MS2 coat protein binding sequence, in which adenosines were replaced by 8-azidoadenosines, crosslinked specifically to the coat protein of the E. coli phage MS2.
RESULTS AND DISCUSSION
Substitution of a pyrimidine-based photochemical nucleotide analog for a purine nucleotide has been used to analyze protein–RNA interactions (Wyatt et al. 1992). In many cases such a substitution can abolish protein–RNA interaction. For instance, replacement of the adenosine residue at the 3′ splice site AG (a highly conserved sequence element that represents the 3′ splice site of virtually all nuclear pre-mRNAs) by cytidine or uridine inhibits RNA splicing as well as the binding of a splicing factor, U2AF35 (Zhang et al. 1992; Wu et al. 1999). Thus, to identify proteins that bind specifically to an adenine base, a poly-A sequence, or a purine-rich element, a purine-based photocrosslinking nucleotide is desirable. In this study, we sought to investigate transcription conditions that may permit 8-N3ATP to function as a substrate for bacteriophage RNA polymerase. We focused our attention on 8-N3ATP because the presence of azide moiety in the imidazole ring (position 8; Fig. 1 ▶) reduces the risk of interfering with normal Watson-Crick base-pairing interactions.
8-N3ATP does not function as a substrate for bacteriophage RNA polymerase under standard transcription conditions
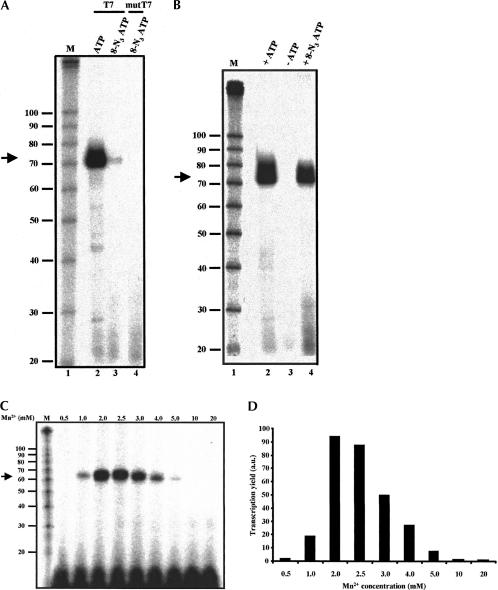
A published report indicates that RNA containing 8-N3AMP can be synthesized as T7 transcript, and can be used to probe protein–RNA interaction (Query et al. 1996). However, our attempts to synthesize 8-N3AMP-containing RNA with a variety of templates as well as by using a mutant T7 RNA polymerase (Sousa and Padilla 1995) which has been shown to incorporate modified nucleoside 5′ triphosphates resulted in inefficient incorporation of 8-N3AMP (Fig. 2A ▶). Quantitation of the data in Figure 2A ▶ suggests that replacement of ATP with 8-N3ATP led to an 86-fold decrease in the yield of full-length transcript. Additionally, transcription reactions performed in the presence of SP6 or T3 RNA polymerase failed to catalyze the insertion of 8-N3AMP into RNA (data not shown). The following reasons may account for the observed differences between our findings and those of the previous study (Query et al. 1996). First, unlike the present work, the template used in the previous study encodes a small RNA (34-mer) and contains only a single site for 8-N3AMP insertion (Query et al. 1996). Second, whereas the template used in our assay contains at least one site for the incorporation of 8-N3AMP within the first six nucleotides, Query et al. (1996) used a template in which 8-N3AMP is incorporated as the ninth nucleotide. Our results are consistent with those of earlier studies which suggest that the identity of the first six nucleotides immediately after the T7 promoter sequence plays an important role in the overall efficiency of a transcription reaction, and apparently the incorporation of a noncanonical base within the first six nucleotides of the RNA could lead to premature termination of the transcription (Milligan et al. 1987; Milligan and Uhlenbeck 1989).
FIGURE 2.
8-N3ATP does not function as a substrate for T7 or mutant T7 (T7 R&DNA) polymerase under standard in vitro transcription conditions. (A) RNAs synthesized from a BamHI-digested plasmid pPIP85.B (Query et al. 1996), which directs the synthesis of 68-mer RNA, with indicated polymerase were resolved on a 10% polyacrylamide denaturing gel. A standard in vitro transcription reaction consists of 40 mM Tris-HCl, pH 8.0, 20 mM MgCl2, 2.0 mM spermidine, 5 mM DTT, 0.01% Triton X-100, 0.4 mM CTP, UTP, GTP, and ATP, ~5 μCi [α-32P-UTP], and 25–50 units RNA polymerase. In 8-N3ATP-containing transcription reactions, ATP and DTT were replaced with 0.4 mM 8-N3ATP and 5 mM β-mercaptoethanol, respectively. M, RNA size marker (Decade Marker, Ambion). (B) In the presence of Mn2+, , 8-N3ATP serves as a substrate for T7 RNA polymerase. As in panel A, except that Mg2+ was replaced with 2.5 mM Mn2+. (C) Effect of variation of metal ion concentration on T7 RNA polymerase-catalyzed synthesis of 8-N3AMP containing RNA. As described in A, transcription reactions were performed in the presence of different concentrations of Mn2+ (0.5–20 mM), and the products of transcription reactions were resolved on a 10% polyacrylamide denaturing gel. (D) Histogram representing the yield of RNA (panel C) as a function of Mn2+ concentration. a.u., arbitrary unit.
8-N3ATP can act as a substrate for T7 RNA polymerase in the presence of Mn2+
It is well established that metal ion cofactors play an essential role in virtually every enzyme-catalyzed reaction, including the in vitro transcription with DNA-dependent RNA polymerase (Milligan et al. 1987; Conrad et al. 1995; Basu et al. 1998). Keller and coworkers (Martin et al. 1999) have shown that substitution of Mn2+ for Mg2+ could stimulate the crosslinking of 8-N3ATP into the ATP binding site of bovine and yeast poly(A) polymerases. If Mn2+ can modulate the substrate selection and catalytic properties of these polymerases, it is reasonable to expect that it may also permit the incorporation of 8-N3AMP into RNA. To test this hypothesis, and to investigate whether any other divalent metal ion might support 8-N3AMP’s incorporation into RNA, we performed T7 transcription reactions containing different metal ions (Ni2+, Co2+, Ca2+, Zn2+, and Mn2+). We found that only Mn2+ could support the insertion of 8-N3AMP into RNA (Fig. 2B ▶; data not shown).
To determine the concentration of Mn2+ that would be optimum for the incorporation of 8-N3AMP, we performed transcription reactions containing 0.5–20 mM Mn2+. We found 2.0–2.5 mM Mn2+ to be optimum for the incorpo ration of 8-N3AMP: Mn 2+ concentrations 1.0 and 3.0 mM resulted in ~80% and 45% decreases in the yield of full-length transcript, respectively (Fig. 2C,D ▶). Similar results were obtained when the BamHI-digested PIP85.B (Query et al. 1996) template was replaced by KpnI-linearized AdML (Gozani et al. 1994) template (data not shown). The finding that Mn2+-mediated transcription reaction is effective only over a narrow concentration range (2.0–2.5 mM of Mn2+ vs. 10–20 mM for Mg2+) seems to be the general property of this metal ion. Studies aimed at assessing the ability of Mn2+ ions to affect E. coli DNA polymerase I-dependent DNA synthesis or poly(A) polymerase-mediated incorporation of 8-N3AMP into poly (A) tail drew similar conclusions (el-Deiry et al. 1988; Martin et al. 1999).
Given that 8-N3ATP does not serve as an efficient substrate for T7 polymerase, and the finding that substitution of Mn2+ for Mg2+ could significantly improve its ability to act as a substrate begs explanation. Under standard reaction conditions in the presence of Mg2+ (Fig. 2A ▶), the observed poor substrate activity likely results from steric effects due to the presence of the azide substituent (Fig. 1 ▶). The large bulky azide group could conceivably force a rotational preference for the syn versus the anti conformation of the glycosidic bond to account for the poor substrate activity. However, Costas et al. (2000) showed that in the RNA, 8-N3AMP still preferentially adopts the “normal” anti conformation. Nonetheless, when bound to the enzyme in the anti conformation, the azide group could still contribute to unfavorable steric effects and limit its usefulness as a substrate, at least in the presence of the Mg2+ cofactor. Despite this poor substrate activity, 8-N3ATP clearly binds to the polymerase, as evidenced by its use as a photoaffinity probe to map the active site of T7 RNA polymerase (Knoll et al. 1992). The dramatic increase in substrate activity for 8-N3ATP in the presence of Mn 2+ suggests that the softer Mn2+ metal ion permits some conformational flexibility in the enzyme’s active site such that the steric effects induced by the azide substituent are substantially reduced. It has already been reported that the use of Mn2+ in place of Mg2+ appears to result in polymerase activity compatible for a wider range of substrates for both RNA (Conrad et al. 1995; Basu et al. 1998) and DNA (Tabor and Richardson 1989) polymerases. Other studies suggest that Mn2+ binds to polymerases with higher affinity, possibly involving two aspartic acid residues (Woody et al. 1996; Martin et al. 1999). Similar increases in binding affinity for the NTP substrates can reduce the apparent Km for the NTP substrates, which could result in tighter binding for an otherwise unfavorable NTP substrate. Indeed, in the presence of Mn2+, polyA polymerase exhibits a decreased Km value for 8-N3ATP (Martin et al. 1999). Alternatively, the ligand exchange rates for Mn2+ are roughly 100-fold faster than those measured with Mg2+ (Eigen 1963; Margerum et al. 1978) and could increase the observed 8-N3AMP incorporation by enhancing the rates of metal-mediated chemical events that require ligand exchange, or perhaps by enhancing the product release steps.
ATP is a better substrate than 8-N3ATP
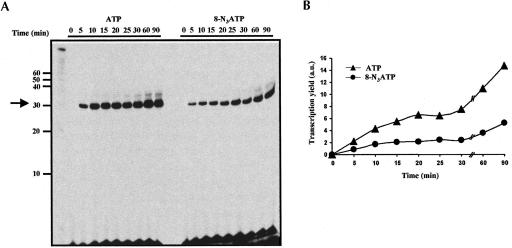
As shown in Figure 2B ▶, although Mn2+ has a stimulatory effect on the efficiency of 8-N3ATP-containing transcription reaction, the yield from ATP-containing transcription reaction is higher than that of N3ATP; substitution of 8-N3ATP for ATP resulted in ~55% decrease in the yield of full-length transcripts (Fig. 2B ▶, cf. lanes 2 and 4). It is conceivable that the addition of 8-N3AMP could have weak-ened the stability of the transcription complex formed between the polymerase and the template-RNA duplex resulting in the pausing of the elongating polymerase. This in turn could have resulted in premature termination of transcription, thus lowering the yield of full-length transcript. An alternative explanation could be that 8-N3ATP is inhibitory to T7 transcription. To determine whether incorporation of 8-N3AMP leads to premature termination of the transcription, a template that encodes a 31-mer RNA and contains only a single site for adenosine incorporation (as the last nucleotide) was constructed (see Materials and Methods). If the incorporation of 8-N3AMP promotes transcription termination, one would expect a truncated N-1 product (30-mer RNA) to be specifically produced in the transcription reaction containing 8-N3ATP. Two transcription reactions were assembled: a control transcription reaction containing all four normal NTPs, and in the second transcription reaction ATP was replaced by 8-N3ATP. Aliquots were removed at various time points, followed by the separation of RNA on a 20% denaturing polyacrylamide gel. As can be seen in Figure 3 ▶, T7 RNA polymerase was able to incorporate 8-N3AMP at a rate observed to be slower than that obtained with ATP. After a 90-min reaction period, 8-N3AMP incorporation reached ~35% of the native ATP value (Fig. 3B ▶). Significantly, however, no N-1 product was generated, suggesting that at least during transcription elongation, incorporation of 8-N3AMP did not result in the premature termination of transcription.
FIGURE 3.
Time course for the incorporation of a single AMP or 8-N3AMP residues in a 31-mer RNA. (A) Bsu36I-digested plasmid pSG30A, which encodes a 31-mer RNA and contains a single site for the incorporation of adenosine or 8-N3A, was used as a template for transcription (conditions as described in Fig. 2 ▶). The control transcription reaction contains all four normal NTPs, and in the case of 8-N3ATP transcription, 8-N3ATP replaced ATP. Aliquots were removed at various time points, followed by the separation of RNA transcripts on a 20% denaturing polyacrylamide gel. The band moving just above the full-length RNA represents N+1 product; T7 RNA polymerase has been reported to add one or more nontemplated nucleotide at the 3′ end of nascent RNA (Milligan et al. 1987). (B) Quantitation of the data from panel A. The efficiency of the transcription reaction is plotted as a function of time. (▴) ATP, (•) 8-N3ATP.
To investigate whether 8-N3ATP has an inhibitory role in the transcription, a template that encodes a 31-mer RNA but lacks any site for the incorporation of adenosine was constructed. If merely the presence of 8-N3ATP in the reaction mixture is inhibitory to the transcription reaction, then the reaction performed with only three NTPs (CTP, GTP, and UTP) should result in a higher RNA yield compared with the one containing three NTPs plus 8-N3ATP. Our results (data not shown) indicate that the presence or absence of 8-N3ATP in the transcription reaction had no effect on the yield of the transcript, suggesting that 8-N3ATP does not inhibit RNA transcription. These results suggest that the lower yield of 8-N3AMP-containing RNA is neither the result of premature termination of transcription nor the inhibition caused by 8-N3ATP, but more likely an unfavorable steric effect due to the presence of the azide moiety that apparently reduces the catalytic efficiency of 8-N3ATP as a substrate for T7 RNA polymerase (Table 1 ▶). Studies in which noncanonical NTP substrates were employed for the synthesis of modified RNAs also indicate that the introduction of an additional functional group in an otherwise wild-type substrate could adversely affect the catalytic efficiency of the modified substrate for T7 RNA polymerase (Aurup et al. 1992; Conrad et al. 1995).
TABLE 1.
Kinetic parameters for the transcription of ATP and 8-N3ATPa
| Nucleotide | Km (μM) | Vmax (pmole/h) |
| ATP | 35 | 25 |
| 8-N3ATP | 75 | 20 |
aDetermined in the presence of 2.5 mM Mn2+ and as described in Materials and Methods. The assay used to obtain values for Km and Vmax gave reproducible results, varying less than a factor of 2.
Transcription buffer containing a mixture of Mg2+ and Mn2+ improves the yield of 8-N3AMP-containing RNA
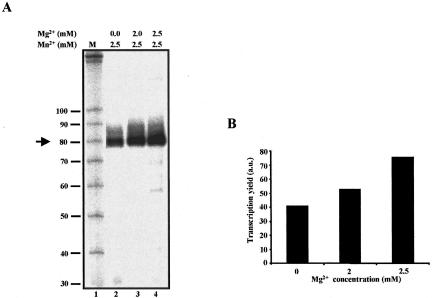
We and others have shown that transcription buffer containing a mixture of Mg2+ and Mn2+ could modulate the substrate property of modified nucleotides (Conrad et al. 1995; Gaur and Krupp 1997; Basu et al. 1998). To investigate whether a Mg2+/Mn2+ combination could further improve the yield of 8-N3AMP-containing RNA, transcription reactions containing 2.5 mM Mn2+ and increasing concentrations of Mg2+ were assembled and products were analyzed by denaturing PAGE (Fig. 4A ▶). As can be seen in Figure 4B ▶, compared with 2.5 mM Mn2+, a mixture of 2.5 mM Mg2+ and 2.5 mM Mn2+ increased the yield of the full-length transcript by ~85%. A further increase in the concentration of Mg2+ did not enhance the yield of the RNA (data not shown). These results suggest that a mixture of Mg2+ and Mn2+ enhances the yield of 8-N3AMP-containing RNA by Mn2+ likely promoting the insertion of 2+ supporting the incorporation of nor-8-N3AMP and Mg mal NTPs.
FIGURE 4.
A mixture of 2.5 Mn2+/2.5 Mg2+ improves the yield of 8-N3AMP-containing RNA. (A) As in Figure 2 ▶, except that transcription reactions were performed in the absence or with indicated concentrations of Mg2+, and KpnI- digested plasmid (pAdML) was used as a template. (B) Histogram representing the yield of RNA (from panel A) as a function of Mg2+ concentration. a.u., arbitrary unit.
Transcripts generated in the presence of 8-N3ATP are authentic and can be faithfully reverse-transcribed
Earlier reports suggest that transcripts produced by T7 RNA polymerase often contain both 5′ and 3′ termini heterogeneity (Milligan et al. 1987; Krupp 1989; Pleiss et al. 1998). It has also been shown that substitution of Mn2+ for Mg2+ could compromise the fidelity of DNA as well as RNA polymerase (el-Deiry et al. 1988; Conrad et al. 1995; Pelletier et al. 1996; Huang et al. 1997). Thus, it is important to determine: (1) whether the full-length RNA transcribed in the presence of 8-N3ATP (Fig. 2B ▶, lane 4) does in fact contain 8-N3AMP and not the result of 8-N3ATP or other NTPs being contaminated with ATP; and (2) if T7 RNA polymerase-dependent insertion of 8-N3AMP did occur in a template-dependent manner. To exclude the possibility that 8-N3ATP or any other NTP is contaminated with ATP, a transcription reaction containing only three NTPs (CTP, GTP, UTP) and a small amount of [α-32P]-UTP was performed. As illustrated in Figure 2B ▶, unlike the control transcription, exclusion of ATP from the reaction mixture failed to generate full-length transcript (cf. lanes 2 and 3), suggesting that CTP, GTP, and UTP are not contaminated with ATP.
As a more direct test to confirm the presence of 8-N3AMP in the RNA, a nearest-neighbor analysis was performed (see Materials and Methods). Briefly, full-length RNA generated in the presence of ATP or 8-N3ATP (Fig. 2B ▶, lanes 2,4) was isolated, digested to completion with RNase T2, and the resulting 3′ nucleoside monophosphates were separated by 2D-TLC (Konarska et al. 1985). To establish the identity of 32P-labeled nucleoside 3′ monophos-phates, cold yeast tRNA was included in the digestion reaction. Figure 5A ▶ shows, as expected, that the wild-type RNA yielded all four nucleoside 3′ monophosphates, confirmed by superimposition with the cold marker. However, RNA transcribed in the presence of 8-N3ATP resulted in a 3′ monophosphate whose mobility is different from adenosine 3′ monophosphate (Fig. 5B ▶, spot marked with an asterisk), confirming that 8-N3ATP is free from ATP contamination. Due to the unavailability of 8-N3Ap marker and the possibility that nitrene moiety in 8-N3Ap could have been reduced to -NH2, we have not been able to confirm whether this spot is 8-N3Ap or its reduced product, 8-NH2Ap. Nevertheless, the combined results of these experiments suggest that the transcripts generated in the presence of 8-N3ATP do contain 8-N3AMP.
FIGURE 5.
Nearest-neighbor and nucleotide analysis of [α-32P] UTP-labeled RNA. (A) Upper: Schematic representation of RNase T2 digestion. RNA transcribed in the presence of ATP was isolated and digested to completion with RNase T2 (see Materials and Methods for details), and the resulting nucleoside 3′ monophosphates were separated by 2-dimension TLC. The positions of the unlabeled nucleoside 3′ monophosphates were visualized via UV shadowing before autoradiography, and are indicated as dotted circles. (B) As described in A except that 8-N3AMP-containing RNA was used.
To determine whether the transcripts generated in the presence of 8-N3ATP are authentic and can be faithfully reverse-transcribed, a 240-mer-long AdML pre-mRNA (Gozani et al. 1994) was synthesized using a mixture of 2.5 mM Mg2+ and 2.5 mM Mn2+. A control transcription containing normal NTPs was also performed. The gelpurified RNAs were then subjected to reverse transcription and PCR. Next, dsDNAs were cloned into pCR2.1 vector using a TA cloning kit according to the instructions provided by the manufacturer (Invitrogen). Sequencing of 20 randomly selected clones (10 each from the wild-type and 8-N3AMP-substituted RNAs) revealed no misincorporation, suggesting the high fidelity of incorporation of 8-N3AMP by T7 RNA polymerase and efficient reverse transcription by MMLV reverse transcriptase (data not shown). On the basis of these results, we conclude that T7 RNA polymerase-catalyzed insertion of 8-N3AMP takes place in a template-dependent manner and 8-N3AMP-substituted transcripts can be faithfully reverse transcribed.
Steady-state kinetic parameters
Determining the kinetic parameters of a nucleotide analog is essential if an accurate comparison between the substrate property of the modified and parent nucleotide is to be made. The vector pPIP85.B digested with BamHI, used for the kinetic studies, directs the synthesis of a transcript 68 nucleotides in length. The reason for choosing this template is to avoid the incorporation of UMP within the first six nucleotides of the transcript (the sequence of the first six nucleotides of the transcript is 5′-GGGCGA-3′); it is known that incorporation of UMP residue particularly during transcription initiation leads to the generation of abortive products, which in turn could affect the determination of kinetic parameters (Martin et al. 1988; Milligan and Uhlenbeck 1989).
In the presence of 2.5 mM Mn2+ , both ATP and 8-N3ATP served as substrates for T7 RNA polymerase (Fig. 2B ▶; Table 1 ▶). Both nucleotides exhibited almost similar Vmax values; however, compared to ATP a small (approximately twofold) increase in Km for 8-N3ATP was observed. We also found that substitution of Mn2+ for Mg2+ has virtually no effect on the apparent Km of ATP, but a decrease in the Vmax value was observed. The apparent Km and Vmax values for ATP under standard transcription conditions are in the range of 20–30 μM and ~60 pmol/h, respectively (Griffiths et al. 1987), and Mn2+-dependent decrease in the apparent Vmax (25 pmol/h) for ATP is in agreement with the published data (Conrad et al. 1995). Strikingly, substitution of Mn2+ for Mg2+ leads to a dramatic increase in the affinity between T7 polymerase and 8-N3ATP (Table 1 ▶); attempts to determine the Km for Mg2+-mediated incorporation of 8-N3AMP were not successful because of the poor efficiency of the transcription reaction (Fig. 2A ▶). Although Mn2+ improves the substrate property of 8-N3ATP, the overall catalytic efficiency (Vmax/Km) of ATP is marginally better (~2.5-fold) than that of 8-N3ATP.
RNA–protein crosslinking with 8-azidoadenosine-containing RNA
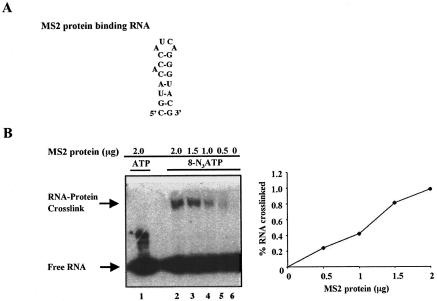
To demonstrate that 8-azidoadenosine-containing RNA can be used as a photocrosslinking probe to analyze protein–RNA complexes, a small RNA hairpin present within the genome of the bacteriophage MS2 and known to bind the dimer of the phage coat protein was transcribed in the presence of 8-N3ATP. The reason for selecting MS2 RNA-coat protein crosslinking as a model is that this system has been characterized by a cocrystal structure as well as with photocrosslinking agents (Witherell et al. 1991; Valegard et al. 1994; Stockley et al. 1995). Different concentrations (0–200 ng) of E. coli-expressed MS2 coat protein were incubated with 32P-labeled hairpin RNA (~10 fmole), and the reaction mixture was irradiated with long-wavelength UV light with a hand-held UV source. The crosslinked samples were treated with RNase A, and the mixture loaded onto a 12.5% SDS-polyacrylamide gel. As demonstrated in Figure 6 ▶, MS2 coat protein crosslinks to its cognate RNA in a dose-dependent manner. Significantly, a similar RNA in which the 8-azidoadenosine was replaced by adenosine failed to yield RNA–protein crosslink (Fig. 6 ▶, cf. lane 1 and lanes 2–5).
FIGURE 6.
MS2 protein–RNA crosslinking. (A) Sequence and proposed secondary structure of hairpin RNA known to bind MS2 protein. (B) Analysis of MS2 protein–RNA crosslinking. [α-32P]UTP-labeled RNA containing AMP (lane 1) or 8-N3AMP (lanes 2–6) was incubated with (lanes 1–5) or without MS2 protein (lane 6), and the mixture was irradiated for 15 min with long-wavelength UV light. After RNase A digestion, the crosslinked RNA was separated from free probe on a 12.5% SDS-polyacrylamide gel.
In this assay the yield of MS2 protein–RNA crosslinking is relatively low compared to the published reports where 5-bromouridine (Willis et al. 1994), 5-iodouracil (Willis et al. 1993), or 5-iodocytidine (Meisenheimer et al. 1996) substituted RNAs were used. It is conceivable that introduction of multiple 8-N3AMP residues could have lowered the affinity of MS2 protein for RNA. In order to investigate this issue, the values of Kd for the wild-type and 8-N3AMP-substituted RNAs were estimated by using gel-shift assay (Graveley and Maniatis 1998). Unlike the wild-type RNA, which binds MS2 protein with high affinity (Kd 20 nM), an ~10-fold reduction (Kd 208 nM) in the binding of 8-N3AMP RNA was observed. These results suggest that although introduction of multiple 8-N3AMP residues affects the affinity as well as the crosslinking efficiency of RNA for MS2 protein, apparently it does not greatly alter the nature of the RNA–protein interaction.
CONCLUSIONS
We have shown that under modified transcription conditions, 8-N3ATP can function as an efficient substrate for T7 RNA polymerase. The advantages of 8-N3ATP as a photoaffinity probe are many: (1) Upon UV irradiation it generates a highly reactive moiety, nitrene, that is known to react nonselectively with the chemical groups that are in the close proximity to the base; (2) the presence of the photocrosslinking moiety in the imidazole ring of 8-N3ATP does not interfere with the normal Watson-Crick hydrogen bonding; (3) RNAs with multiple incorporation of 8-N3AMP residues can be obtained in good yields; and (4) 8-azidoadenosine-containing RNA can be used to study RNA–protein interactions.
MATERIALS AND METHODS
Protein expression and purification
An overnight culture of Escherichia coli BL21 (DE3) pLysS harboring pRSET-His6MS2 (Graveley and Maniatis 1998) was diluted 1:50 in LB broth. At an OD595 of 0.5, the culture was induced with isopropyl-1-thio-β-D-galactopyranoside (1 mM). After 2 h of further growth, cells were spun down (~4 K in J-6 for 10 min) and the cell pellet was resuspended in 20 mL of buffer A (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris, pH 8.0). The cell lysate was stirred at room temperature for 30 min, followed by 30-min centrifugation at 12,000g. The resultant supernatant was mixed with 5 mL of 50% Ni2+-NTA slurry (QIAGEN) equilibrated in buffer A, and the mixture was allowed to stay at room temperature for 30 min with occasional mixing. The slurry was loaded onto a column and the resin was washed twice with buffer A and then with buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris, pH 6.3). After washing, the desired protein was eluted with 3 mL buffer B containing 100 mM EDTA, followed by 5 mL buffer B containing 100 mM EDTA. The fractions containing His-tagged-MS2 were pooled and dialyzed against buffer C (50 mM Tris, pH 8.0, 20% glycerol, 0.2 mM EDTA, 0.3 M KCl, 0.2 mM PMSF, 0.5 mM DTT) and stored at −70°C.
Template for single adenosine incorporation
The template encoding a 31-mer RNA containing a single site for adenosine residue was generated by annealing and extension of the following oligos: 5′-CCGGAATTCTAATACGACTCACTATAGGCGGTCCCTGGTGTTTG-3′ and 5′-CGCGGATCCTAAGGCGAACAGCCAAAC ACCAGGGAC-3′. The double-stranded DNA was digested with EcoRI and BamHI and subcloned into EcoRI/BamHI-digested plasmid pSP65 (Promega) to yield plasmid pSG30A. For run-off transcription, plasmid pSG30A was digested with Bsu36I.
Transcription reactions
Linearized plasmid (1 μg) was used as template for run-off transcription. A typical (10 μL) in vitro transcription reaction consisted of 40 mM Tris-HCl (pH 8.0), 2.0 mM spermidine, 5 mM β-mercaptoethanol, 0.01% Triton X-100, 0.4 mM CTP, UTP, GTP and 8-N3ATP (ICN Biochemicals), varying concentrations of MnCl2, ~5 μCi [α-32P]UTP, 25–50 units SP6, T3, T7, or T7 R&DNA polymerase (Epicentre). For the synthesis of high-specific-activity RNA, the UTP concentration was reduced to 0.1 mM and ~10 μCi [α-32P]UTP was included. All reactions involving 8-N3ATP were performed in reduced light. In the control transcription reaction, 8-N3ATP, β-mercaptoethanol, and MnCl2 were replaced by 0.4 mM ATP, 5 mM DTT, and 20 mM MgCl2, respectively. After 2 h of incubation at 37°C, the reaction was terminated by adding 10 μL stop buffer (8 M urea, 0.03% xylene cyanol/bromophenol blue), and analyzed on denaturing polyacrylamide gels. The yield of full-length RNA was estimated by Molecular Dynamics PhosphorImager scanning (ImageQuant program).
Gel mobility shift assay
The apparent dissociation constant (Kd) value, obtained from the protein concentration that gives 50% binding of the RNA, was determined with a gel mobility shift assay essentially as described (Graveley and Maniatis 1998).
Steady-state kinetic parameters
The BamHI-digested plasmid pPIP85.B (Query et al. 1996), which directs the synthesis of 68-mer RNA, was used as a template for kinetic assays. Kinetic measurements were carried out at 37°C in 500-μL Eppendorf tubes. Transcription reactions (50 μL) containing 0.5 μg DNA template, 50 units T7 RNA polymerase, 1 mM each CTP, GTP, and UTP, 8 μCi [α-32P]UTP, 2.5 mM MnCl2, and varying concentrations of ATP or 8-N3ATP were used. Reactions were initiated by the addition of T7 polymerase. Initial screenings were done in the range of 10–500 μM. Final data were obtained with 2.5 mM MnCl2 using 10, 25, 37.5, 50, 62.5, and 75 μM ATP or 50, 150, 250, 300, 350, and 400 μM 8-N3ATP. Aliquots (6 μL) were withdrawn at 0–70 min, and mixed with an equal volume of urea/dye mix and 50 mM EDTA. The time intervals were selected so that up to 10%–15% of the nucleotide is incorporated. Samples were heated at 95°C for 1 min, then chilled on ice, and transcription products were resolved on 10% denaturing polyacrylamide gels. The yields of full-length transcripts were determined by Molecular Dynamics PhosphorImager scanning. Apparent Km and Vmax values were determined as described (Conrad et al. 1995).
Nearest-neighbor analysis
Transcripts of the adenovirus MINX precursor RNA containing 8-N3AMP or AMP were prepared from BamHI-digested pRG1 plasmid (Gaur et al. 1997) as described above. The gel-purified and [α-32P]-UTP labeled RNA transcripts (1–5 pmol) were dissolved in T2 digestion buffer (50 mM sodium acetate, pH 5.2, 2.0 mM EDTA) and incubated at 37°C for 3–4 h with RNase T2 (0.4 Units; Roche Molecular Biochemicals). In order to confirm the identity of the labeled nucleoside 3′ monophosphates, yeast tRNA (40 μg) was included in the RNase T2 digestion reaction. The separation of 3′ nucleoside monophosphates was achieved by spotting the digested RNA (4 μL) onto Polygram CEL 300 cellulose plates (Macherey-Nagel), followed by the development of TLC in 2-dimensions (Konarska et al. 1985). The solvent system for the first and second dimensions were isobutyric acid/NH4OH/H2O (58/4/38) and (NH4)2SO4/1 M sodium acetate (pH 5.2)/isopropanol (80/18/2), respectively. The areas of 3′ nucleoside monophosphates were marked with UV shadowing, and the labeled 3′ nucleoside monophosphates were detected by autoradiography.
UV-induced photocrosslinking
A 20-μL reaction consisting of ~105 cpm RNA (~10–20 fmol) and MS2 protein (0.5–2.0 μg) was incubated on ice for 30 min in GS buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM EDTA, and 3 μg tRNA). Next, the reaction mixture was placed on a Parafilm-covered metal plate and irradiated with long-wavelength UV light (Spectroline; model ENF-240C) for 15 min. The UV source was at a distance of 2 cm from the sample. After UV irradiation, RNase A (6 μg; Sigma) was added and the reaction mixture was incubated at 37°C for 20 min. To the crosslinked samples, 10 μL SDS-loading buffer was added, followed by heating of the reaction mixture to 95°C for 3–5 min. As a control, the RNA transcribed with normal NTPs was treated in an identical manner. To assess protein–RNA crosslinking, the reaction mixture was loaded on a 12.5% SDS-polyacrylamide gel, and the efficiency of crosslinking was analyzed by Molecular Dynamics PhosphorImager scanning.
Reverse transcription and PCR
The reverse-transcription reaction mixture contained 50 mM Tris- HCl, pH 8.3, 50 mM KCl, 6 mM MgCl2, 5 mM β-mercaptoethanol, 0.1 mg/ml BSA, 0.25 mM each dNTPs, 50 pmole reverse primer (5′-AGGGAAAAAGAGAGAAGAAG-3′), 5 pmole RNA, and 2.5 μL MMLV reverse transcriptase. The reaction mixture was incubated at 42°C for 1 h. For PCR amplification, 20 μL of the cDNA mixture was added to 80 μL of PCR mixture containing 50 pmole of forward (5′-CGAAGATCTGGGCGAATTCGAGCTC AC) and reverse (5′-AGGGAAAAAGAGAGAAGAAG-3′) primers, 0.25 mM dNTPs, 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2) and 0.5 unit Taq polymerase (Roche Molecular Biochemicals). A total of 25 cycles with each cycle consisting of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min were performed.
Cloning and DNA sequencing
Following the PCR amplification, the amplified DNA was cloned using a TA cloning kit (Invitrogen) essentially according to the manufacturer’s instructions. Single colonies were randomly selected and plasmid was isolated using a plasmid isolation kit (QIAGEN), and the sequence was determined by ABI sequencer.
Acknowledgments
We thank Tom Maniatis (Harvard University), Robin Reed (Harvard Medical School), and Phil Sharp (MIT) for their generous gift of the plasmid pRSET-His6MS2, pAdML, and pPIP85.B, respectively; Larry McLaughlin, R-J. Lin, John Russi, Klemens Hertel, and members of the Gaur lab for helpful comments on the manuscript. We also thank Faith Osep for administrative assistance. This work was supported in part by a Department of Defense (DOD; CDMRP) grant to R.K.G. (DAMD17-03-1-0625) and from Beckman Research Institute start-up funds.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5222504.
REFERENCES
- Aurup, H., Williams, D., and Eckstein, F. 1992. 2′-Fluoro- and 2′-amino-2′-deoxynucleoside 5′-triphosphates as substrates for T7 RNA polymerase. Biochemistry 31: 9636–9641. [DOI] [PubMed] [Google Scholar]
- Basu, S., Rambo, R.P., Strauss-Soukup, J., Cate, J.H., Ferre-D’Amare, A.R., Strobel, S.A., and Doudna, J.A. 1998. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat. Struct Biol. 5: 986–992. [DOI] [PubMed] [Google Scholar]
- Bowser, C.A. and Hanna, M.M. 1991. Sigma subunit of Escherichia coli RNA polymerase loses contacts with the 3′ end of the nascent RNA after synthesis of a tetranucleotide. J. Mol. Biol. 220: 227–239. [DOI] [PubMed] [Google Scholar]
- Burgin, A.B. and Pace, N.R. 1990. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 9: 4111–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, F., Hanne, A., Gaur, R.K., and Krupp, G. 1995. Enzymatic synthesis of 2′-modified nucleic acids: Identification of important phosphate and ribose moieties in RNase P substrates. Nucleic Acids Res. 23: 1845–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas, C., Yuriev, E., Meyer, K.L., Guion, T.S., and Hanna, M.M. 2000. RNA-protein crosslinking to AMP residues at internal positions in RNA with a new photocrosslinking ATP analog. Nucleic Acids Res. 28: 1849–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R.H. 1995. Large-scale oligoribonucleotide production. Curr. Opin. Biotechnol. 6: 213–217. [DOI] [PubMed] [Google Scholar]
- Eigen, M. 1963. Fast elementary steps in chemical reaction mechanism. Pure Appl. Chem. 6: 97–115. [Google Scholar]
- el-Deiry, W.S., So, A.G., and Downey, K.M. 1988. Mechanisms of error discrimination by Escherichia coli DNA polymerase I. Biochemistry 27: 546–553. [DOI] [PubMed] [Google Scholar]
- Favre, A. 1990. 4-Thiouridine as an intrinsic photoaffinity probe of nucleic acid structure and interactions. In Bioorganic photochemistry: Photochemistry and the nucleic aids (ed. H. Morrison), pp. 379–425. Wiley, New York.
- Fidanza, J.A., Ozaki, H., and McLaughlin, L.W. 1994. Functionalization of oligonucleotides by the incorporation of thio-specific reporter groups. Methods Mol. Biol. 26: 121–143. [DOI] [PubMed] [Google Scholar]
- Frouin, I., Montecucco, A., Spadari, S., and Maga, G. 2003. DNA replication: A complex matter. EMBO Rep. 4: 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur, R.K. and Krupp, G. 1997. Chemical and enzymatic approaches to construct modified RNAs. Methods Mol. Biol. 74: 99–110. [DOI] [PubMed] [Google Scholar]
- Gaur, R.K., McLaughlin, L.W., and Green, M.R. 1997. Functional group substitutions of the branchpoint adenosine in a nuclear pre-mRNA and a group II intron. RNA 3: 861–869. [PMC free article] [PubMed] [Google Scholar]
- Gozani, O., Patton, J.G., and Reed, R. 1994. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 13: 3356–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. and Maniatis T. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1: 765–771. [DOI] [PubMed] [Google Scholar]
- Griffiths, A.D., Potter, B.V., and Eperon, I.C. 1987. Stereospecificity of nucleases towards phosphorothioate-substituted RNA: Stereochemistry of transcription by T7 RNA polymerase. Nucleic Acids Res. 15: 4145–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, M.M. 1989. Photoaffinity cross-linking methods for studying RNA-protein interactions. Methods Enzymol. 180: 383–409. [DOI] [PubMed] [Google Scholar]
- Hixson, S.H. and Hixson, S.S. 1975. P-Azidophenacyl bromide, a versatile photolabile bifunctional reagent. Reaction with glyceralde-hyde-3-phosphate dehydrogenase. Biochemistry 14: 4251–4254. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Beaudry, A., McSwiggen, J., and Sousa, R. 1997. Determinants of ribose specificity in RNA polymerization: Effects of Mn2+ and deoxynucleoside monophosphate incorporation into transcripts. Biochemistry 36: 13718–13728. [DOI] [PubMed] [Google Scholar]
- Knoll, D.A., Woody, R.W., and Woody, A.Y. 1992. Mapping of the active site of T7 RNA polymerase with 8-azidoATP. Biochim. Biophys. Acta 1121: 252–260. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M., Grabowski, P.J., Padgett, R.A., and Sharp, P.A. 1985. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature 313: 552–557. [DOI] [PubMed] [Google Scholar]
- Krupp G. 1989. Unusual promoter-independent transcription reactions with bacteriophage RNA polymerases. Nucleic Acids Res. 17: 3023–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan, A.M., Query, C.C., Allerson, C.R., Chen, S., Verdine, G.L., and Sharp, P.A. 1994. Dynamic association of proteins with the pre-mRNA branch region. Genes & Dev. 8: 3008–3020. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Margerum, D.W., Cayley, G.R., Weatherburn, D.C., and Pagenkopf, G.K. 1978. Kinetics and mechanisms of complex formation and ligand exchange. In Coordination chemistry (ed. A.E. Martell), Vol. 2, pp. 1–220, Monograph 174. American Chemical Society, Washington, D.C.
- Martin, C.T., Muller, D.K., and Coleman, J.E. 1988. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry 27: 3966–3974. [DOI] [PubMed] [Google Scholar]
- Martin, G., Jeno, P., and Keller, W. 1999. Mapping of ATP binding regions in poly(A) polymerases by photoaffinity labeling and by mutational analysis identifies a domain conserved in many nucleotidyltransferases. Protein Sci. 8: 2380–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor, A., Rao, M.V., Duckworth, G., Stockley, P.G., and Connolly, B.A. 1996. Preparation of oligoribonucleotides containing 4-thiouridine using Fpmp chemistry. Photo-crosslinking to RNA binding proteins using 350 nm irradiation. Nucleic Acids Res. 24: 3173–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenheimer, K.M., Meisenheimer, P.L., Willis, M.C., and Koch, T.H. 1996. High yield photocrosslinking of a 5-iodocytidine (IC) substituted RNA to its associated protein. Nucleic Acids Res. 24: 981–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, J.F. and Uhlenbeck, O.C. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180: 51–62. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundus, D. and Wollenzien P. 2000. Structure determination by directed photo-cross-linking in large RNA molecules with site-specific psoralen. Methods Enzymol. 318: 104–118. [DOI] [PubMed] [Google Scholar]
- Parang, K., Kohn, J.A., Saldanha, S.A., and Cole, P.A. 2002. Development of photo-crosslinking reagents for protein kinase-substrate interactions. FEBS Lett. 520: 156–160. [DOI] [PubMed] [Google Scholar]
- Pelletier, H., Sawaya, M.R., Wolfle, W., Wilson, S.H., and Kraut, J. 1996. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta. Biochemistry 35: 12762–12777. [DOI] [PubMed] [Google Scholar]
- Pleiss, J.A., Derrick, M.L., and Uhlenbeck, O.C. 1998. T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA 4: 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, R.L. and Haley, B.E. 1983. Photoaffinity labeling of nucleotide binding sites with 8-azidopurine analogs: Techniques and applications. Methods Enzymol. 91: 613–633. [DOI] [PubMed] [Google Scholar]
- Query, C.C., Strobel, S.A., and Sharp, P.A. 1996. Three recognition events at the branch-site adenine. EMBO J. 15: 1392–1402. [PMC free article] [PubMed] [Google Scholar]
- Shah, K., Wu., H., and Rana, T.M. 1994. Synthesis of uridine phosphoramidite analogs: Reagents for site-specific incorporation of photoreactive sites into RNA sequences. Bioconjug. Chem. 5: 508–512. [DOI] [PubMed] [Google Scholar]
- Shapkina, T.G., Dolan, M.A., Babin, P., and Wollenzien, P. 2000. Initiation factor 3-induced structural changes in the 30 S ribosomal subunit and in complexes containing tRNA(f)(Met) and mRNA. J. Mol. Biol. 299: 615–628. [DOI] [PubMed] [Google Scholar]
- Sousa, R. and Padilla, R. 1995. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 14: 4609–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockley, P.G., Stonehouse, N.J., Murray, J.B., Goodman, S.T., Talbot, S.J., Adams, C.J., Liljas, L., and Valegard, K. 1995. Probing sequence-specific RNA recognition by the bacteriophage MS2 coat protein. Nucleic Acids Res. 23: 2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvers, L.A. and Wower, J. 1993. Nucleic acid-incorporated azido-nucleotides: Probes for studying the interaction of RNA or DNA with proteins and other nucleic acids. Bioconjug. Chem. 4: 411–418. [DOI] [PubMed] [Google Scholar]
- Tabor, S. and Richardson, C.C. 1989. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. 86: 4076–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegard, K., Murray, J.B., Stockley, P.G., Stonehouse, N.J., and Liljas, L. 1994. Crystal structure of an RNA bacteriophage coat protein–operator complex. Nature 371: 623–626. [DOI] [PubMed] [Google Scholar]
- Wang, Z. and Rana, T.M. 1998. RNA-protein interactions in the Tat-trans-activation response element complex determined by site-specific photo-cross-linking. Biochemistry 37: 4235–4243. [DOI] [PubMed] [Google Scholar]
- Willis, M.C., Hicke, B.J., Uhlenbeck, O.C., Cech, T.R., and Koch, T.H. 1993. Photocrosslinking of 5-lodouracil-substituted RNA and DNA to proteins. Science 262: 1255–1257. [DOI] [PubMed] [Google Scholar]
- Willis, M.C., LeCuyer, K.A., Meisenheimer, K.M., Uhlenbeck, O.C., and Koch, T.H. 1994. An RNA-protein contact determined by 5-bromouridine substitution, photocrosslinking and sequencing. Nucleic Acids Res. 22: 4947–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell, G.W., Gott, J.M., and Uhlenbeck, O.C. 1991. Specific interaction between RNA phage coat proteins and RNA. Prog. Nucleic Acid Res. Mol. Biol. 40: 185–220. [DOI] [PubMed] [Google Scholar]
- Woody, A.-Y.M., Eaton, S.S., Osumi-Davis, P.A., and Woody, R.W. 1996. Asp537 and Asp812 in bacteriophage T7 RNA polymerase as metal ion-binding sites studied by EPR, flow-dialysis, and transcription. Biochemistry 35: 144–152. [DOI] [PubMed] [Google Scholar]
- Wu, S. and Green, M.R. 1997. Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J. 16: 4421–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., Romfo, C.M., Nilsen, T.W., and Green, M.R. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. [DOI] [PubMed] [Google Scholar]
- Wyatt, J.R., Sontheimer, E.J., and Steitz, J.A. 1992. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 6: 2542–2553. [DOI] [PubMed] [Google Scholar]
- Yu, Y.T. 1999. Construction of 4-thiouridine site-specifically substituted RNAs for cross-linking studies. Methods 18: 13–21. [DOI] [PubMed] [Google Scholar]
- Zhang, M., Zamore, P.D., Carmo-Fonseca, M., Lamond, A.I., and Green, M.R. 1992. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc. Natl. Acad. Sci. 89: 8769–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]