Abstract
Alternative splicing is a powerful means of regulating gene expression and enhancing protein diversity. In fact, the majority of metazoan genes encode pre-mRNAs that are alternatively spliced to produce anywhere from two to tens of thousands of mRNA isoforms. Thus, an important part of determining the complete proteome of an organism is developing a catalog of all mRNA isoforms. Alternatively spliced exons are typically identified by aligning EST clusters to reference mRNAs or genomic DNA. However, this approach is not useful for genomes that lack robust EST coverage, and tools that enable accurate prediction of alternatively spliced exons would be extraordinarily useful. Here, we use comparative genomics to identify, and experimentally verify, potential alternative exons based solely on their high degree of conservation between Drosophila melanogaster and D. pseudoobscura. At least 40% of the exons that fit our prediction criteria are in fact alternatively spliced. Thus, comparative genomics can be used to accurately predict certain classes of alternative exons without relying on EST data.
Keywords: alternative splicing, Drosophila, comparative genomics, bioinformatics
INTRODUCTION
Alternative splicing is a process by which a single gene can give rise to multiple mRNAs, each of which can encode proteins with distinct functions (Black 2000; Graveley 2001). It has recently been estimated that as many as 74% of human genes are alternatively spliced (Johnson et al. 2003). Moreover, some genes can generate an extraordinary number of isoforms. For instance, the Drosophila Dscam gene can potentially generate 38,016 different isoforms (Schmucker et al. 2000). As a result, alternative splicing profoundly expands the coding potential of eukaryotic genomes.
Alternative splicing also plays an important role in post-transcriptional gene regulation (Black 2000; Graveley 2001). The best characterized example of this is the sex-determination pathway in Drosophila (Forch and Valcarcel 2003). This pathway involves five genes—Sex-lethal (Sxl), transformer (tra), male-specific lethal-2 (msl-2), doublesex (dsx), and fruitless (fru)—that are each spliced differently in male and female flies. Disrupting the splicing of different genes in this pathway can cause a number of phenotypes, including male-specific lethality, transformation of the primary physical sexual traits, and alterations of male courtship behavior. Whereas alternative splicing of most of the sex-determination genes results in the production of different proteins in males and females, other alternative splicing events regulate whether or not a protein is produced. One example of this is a process called RUST (regulated unproductive splicing and translation) (Lewis et al. 2003). This process involves the alternative splicing of exons that introduce or remove premature stop codons which, in turn, control whether the mRNA is subject to nonsense-mediated decay. Thus, alternative splicing is a powerful mechanism for controlling and specifying protein production.
Current methods for identifying alternatively spliced exons involve aligning ESTs to genomic DNA or reference mRNAs (Modrek et al. 2001). These methods work well for organisms, such as human and mouse, that have extensive EST coverage. However, even when EST coverage is quite extensive, many rare alternative splicing events can still be missed (Graveley 2001). Moreover, because EST coverage is heavily biased toward the 5′ and 3′ ends of genes, many internal alternative exons are not identified by this method. These issues are even more confounding for organisms that lack extensive EST coverage. Thus, methods that facilitate the identification of alternative exons would be quite useful to assist in genome annotation. Currently, computational methods that accurately identify alternative exons do not exist. Here, we describe a comparative genomics approach that identifies alternative exons with a fairly high degree of accuracy without relying upon any EST data.
RESULTS AND DISCUSSION
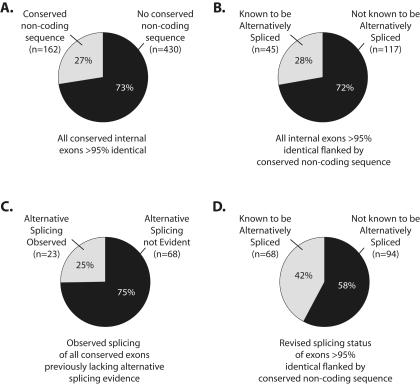
Previous studies in humans and mice have shown that alternative exons often exhibit a higher degree of sequence conservation between related species than constitutive exons (Modrek and Lee 2003; Sorek and Ast 2003; Sugnet et al. 2004). In addition, the introns flanking alternative exons, but not constitutive exons, are also highly conserved (Sorek and Ast 2003). We tested whether these criteria could be used to identify novel alternative exons by simply comparing the genomes of two related species. To do this, we analyzed the genomes of Drosophila melanogaster (Adams et al. 2000) and D. pseudoobscura (http://www.hgsc.bcm.tmc.edu/projects/drosophila/), which diverged approximately 30 millions years ago (Russo et al. 1995; Powell 1997). Consistent with the observations between humans and mice (Modrek and Lee 2003; Sorek and Ast 2003), we found that constitutively spliced exons are typically less conserved between D. melanogaster and D. pseudoobscura than known alternative exons, and that the introns flanking known alternative exons are frequently highly conserved (Fig. 1). We first identified all annotated D. melanogaster exons that are conserved in the D. pseudoobscura draft genome. The 51,432 exons common to these species are, on average, 79% identical. We next identified all D. melanogaster exons that are at least 95% identical in D. pseudoobscura (n = 1,443) and subsequently eliminated all 5′ and 3′ terminal exons from this set, leaving 592 pairs of highly conserved exons. Finally, we identified the subset of these highly conserved internal exons that are also flanked on at least one side by intronic sequence of at least 10 nucleotides (nt) that is greater than 75% identical in D. pseudoobscura (Fig. 2A). This led to a final set of 162 highly conserved exons. All available EST data indicate that 117 of the exons in this group are constitutively spliced. Interestingly, 28% (n = 45) of the exons in this final group are annotated in Release 3.2 of the D. melanogaster genome as being alternatively spliced (Misra et al. 2002; Fig. 2B). In contrast, a rough calculation suggests that less than 5% of all D. melanogaster exons are currently annotated as being alternatively spliced (Misra et al. 2002). Thus, in the current genome annotation, highly conserved exons that also contain conserved intronic sequences are at least five times more likely to be alternatively spliced than a randomly selected exon.
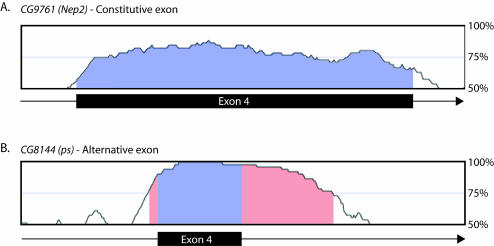
FIGURE 1.
Characteristics of alternative and constitutive exons in Drosophila. (A) A graph of the percent identity of exon 4 of CG9761 (Nep2) between D. melanogaster and D. pseudoobscura. Exon 4 is constitutively spliced and represents the average percent identity of all exons between these two Drosophila species. (B) A graph of the percent identity of exon 4 of CG8144 (ps) between D. melanogaster and D. pseudoobscura. This exon is known to be alternatively spliced, and is highly conserved between these two species. In addition, the intron sequence downstream of this exon is also highly conserved. These features are typical of known alternative exons in Drosophila. The blue shading represents the boundaries of the exons and the pink shading represents conserved noncoding sequence. The window size used for these computations is 50 nt.
FIGURE 2.
Frequency of alternative splicing among highly conserved exons. Of the 592 internal exons we identified that were at least 95% identical between D. melanogaster and D. pseudoobscura, 27% also had conserved noncoding sequence in the flanking introns (A). Of those exons, 28% were known to be alternatively spliced based on existing EST evidence (B). Experimental examination of the conserved exons that were not known to be alternatively spliced revealed that at least 25% were in fact alternatively spliced (C). When combined with the exons previously known to be alternatively spliced, at least 42% of the highly conserved exons are alternatively spliced (D).
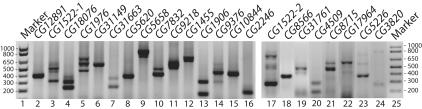
We experimentally tested whether the 117 highly conserved “constitutive” exons are actually alternatively spliced. RT-PCR was performed on a pool of RNA collected from D. melanogaster embryos, larvae, and male and female adults and the PCR products cloned and sequenced to verify their identity. Twenty-three of the 91 reactions that yielded RT-PCR products corresponding to the targeted gene exhibited some type of alternative splicing (Fig. 3). This represents the lower limit of the number of exons tested that are alternatively spliced because rare tissue-specific or developmentally regulated alternative splicing events may have been missed in our screen. Thus, these experiments revealed that at least 25% of the exons examined are alternatively spliced (Fig. 2C). To determine whether these exons were also alternatively spliced in D. pseudoobscura, RT-PCR was performed for a subset of these genes for which the D. melanogaster primers were sufficiently similar to the analogous sequence in D. pseudoobscura. Of the 13 genes tested, 11 were clearly alternatively spliced in D. pseudoobscura (data not shown). Thus, not only are the sequences of these exons conserved between the two species, but also their tendency to be alternatively spliced. When combined with the previously known alternatively spliced exons, a minimum of 42% of all highly conserved internal exons we identified in the D. melanogaster genome are alternatively spliced (Fig. 2D).
FIGURE 3.
Highly conserved Drosophila exons are alternatively spliced. Agarose gel of RT-PCR products for exons that are alternatively spliced. Each band was excised and sequenced. The identities of the alternatively spliced isoforms are shown in Figure 4.
To determine the extent to which these criteria improve the accuracy of alternative exon prediction, we tested whether 30 randomly selected exons that were not known to be alternatively spliced actually are alternatively spliced. Of these 30 exons, only one is alternatively spliced (data not shown). Interestingly, the properties of the alternative exon identified from the randomly selected group, exon 3 in CG7185, resembles the exons selected by the critera of our screen—it is 88.7% identical in D. pseudoobscura, and the sequence flanking this exon is also highly conserved. These results demonstrate that our selection criteria increase the accuracy of a priori prediction of alternative exons at least 12-fold (3.3% for randomly selected exons vs. 42% for predicted exons).
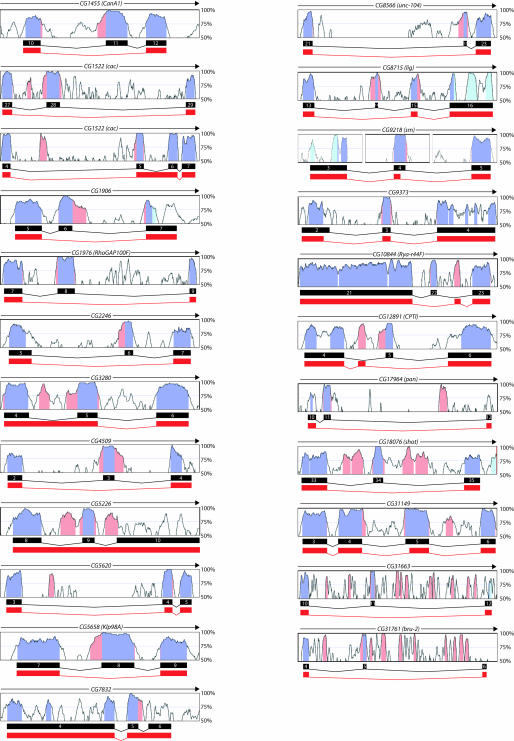
The alternative exons identified in our screen encompass nearly all varieties of alternative splicing, including alternative 5′ or 3′ splice sites, cassette exons, mutually exclusive exons, and intron retention. These newly identified alternative exons reside in genes that encode proteins with a wide variety of functions and are expressed in a broad spectrum of tissues (Table 1). In several instances, alternative splicing is expected to significantly affect the structure and/ or function of the encoded protein. For example, CG5658 (Klp98A) encodes a component of the cytoskeleton containing a kinesin motor, forkhead domain, and a PX domain (Miki et al. 2001). Exon 8 of this gene is alternatively spliced and results in a removal of the forkhead and PX domains from the protein, thereby significantly affecting the signaling properties of the molecule (Fig. 4). For two genes, CG9218 (sm) (zur Lage et al. 1997) and CG31761 (bru-2) (Delaunay et al. 2004; Fig. 4), mRNAs lacking the newly identified alternative exon would encode proteins that contain fewer RNA binding domains than the mRNAs containing the alternative exon. In each case, this could alter the spectrum of RNA sequences recognized by these proteins.
TABLE 1.
Identities and properties of newly identified alternative exons
| CG # | Name | Function | Chr # | Exon # | Exon size | % Identity | Frame | 5′ss CNSa | 3′ss CNSa | Total CNSa | Type of AS | ESTsb |
| CG12891 | CPTI | palmitoyltransferase | 2R | 5 | 60 | 96.7 | 0 | 3 | 37 | 40 | mutually exclusive | H/E/S/L/P |
| CG1522-1 | cac | volt-gated Ca2+ channel | X | 28 | 201 | 98 | 0 | 30 | 69 | 99 | cassette exon | H/S |
| CG1522-2 | cac | volt-gated Ca2+ channel | X | 5 | 101 | 100 | 2 | 10 | 46 | 56 | intron retention | H/S |
| CG18076-2 | shot | cytoskeletal binding | 2R | 34 | 66 | 100 | 0 | 34 | 6 | 40 | cassette exon | H/S/L/P/T/O |
| CG1976 | RhoGAP100F | GTPase | 3R | 8 | 258 | 98.1 | 0 | 6 | 30 | 36 | cassette exon | E/S |
| CG31149a | CG31149 | cell–cell signaling | 3R | 4 | 204 | 94.6 | 0 | 32 | 0 | 32 | alternative 5′ splice site | H |
| CG31149b | CG31149 | cell–cell signaling | 3R | 5 | 195 | 97.4 | 0 | 7 | 14 | 21 | alternative 3′ splice site | H |
| CG31663 | CG31663 | unknown | 2L | 11 | 108 | 99.1 | 0 | 17 | 44 | 61 | cassette exon | H/T |
| CG3280 | CG3280 | unknown | 3L | 5 | 105 | 98.1 | 0 | 10 | 60 | 70 | intron retention | N |
| CG5226 | CG5226 | ion transport | 2R | 9 | 69 | 97.1 | 0 | 6 | 124 | 130 | intron retention/cassette exon | H/E/T |
| CG5620 | CG5620 | unknown | 3L | 4 | 105 | 98.1 | 0 | 11 | 0 | 11 | alternative 3′ splice site | H/E/T |
| CG5658 | Klp98A | cytoskeletal component | 3R | 8 | 123 | 96.7 | 0 | 2 | 34 | 36 | cassette exon | E/T |
| CG7832 | CG7832 | unknown | 3R | 5 | 102 | 95.1 | 0 | 33 | 0 | 33 | intron retention | H/E/L/T/O |
| CG8566 | unc-104 | kinesin motor | 2R | 22 | 27 | 96.3 | 0 | 22 | 78 | 100 | cassette exon | H |
| CG9218c | sm | RNA binding | 2R | 4 | 138 | 98.6 | 0 | 22 | 0 | 22 | alternative 5′ splice site | H/E/S/L/P/T |
| CG8715a | lig | copulation | 2R | 15 | 33 | 100 | 0 | 40 | 59 | 99 | cassette exon | H/E/S/T |
| CG2246 | CG2246 | ribose phosphate kinase | 3R | 6 | 57 | 96.5 | 0 | 0 | 45 | 45 | cassette exon | H/E/L/P/O |
| CG1455e | CanA1 | phosphatase | 3R | 11 | 85 | 96.5 | 1 | 0 | 35 | 35 | cassette exon | H/E |
| CG17964 | pan | transcription factor | 4 | 11 | 166 | 96.4 | 1 | 45 | 13 | 58 | cassette exon | H/E/S/O |
| CG1906 | CG1906 | phosphatase | 3R | 5 | 55 | 100 | 1 | 39 | 6 | 45 | cassette exon | H/E/S/T |
| CG4509 | CG4509 | Ca2+ dep cell adhesion | 3R | 3 | 91 | 96.7 | 1 | 55 | 50 | 105 | cassette exon | L/P |
| CG9373 | CG9373 | RNA binding | 3R | 3 | 139 | 99.3 | 1 | 28 | 32 | 60 | alternative 5′ splice site | H/E/S/O |
| CG10844 | Rya-r44F | Ca2+ release | 2R | 22 | 113 | 95.6 | 2 | 0 | 8 | 8 | mutually exclusive | H/E/T |
| CG31761 | bru-2 | RNA binding | 2L | 5 | 83 | 96.4 | 2 | 15 | 65 | 80 | cassette exon | E/T |
a(CNS) conserved noncoding sequence.
b(H) head, (E) embryo, (S) S2 cells, (L) larvae, (P) pupal, (T) testes, (O) ovary, (N) none.
FIGURE 4.
Newly identified alternative exons. VISTA browser graphs are shown for all of the genes containing newly identified alternatively spliced exons. The blue shading represents the boundaries of the annotated exons and the pink shading represents conserved noncoding sequence. The splicing patterns shown in black are the annotated isoforms. The splicing patterns shown in red are the novel isoforms. The window size used for these computations is 50 nt.
In addition to the candidate alternative exons, we found a few novel alternative exons not predicted by our screen. For instance, exon 5 of CG12891 (CPTI) (Jackson et al. 1999), a carnitine ethyltransferase, was a candidate alternative exon that we found to be alternatively spliced. However, this exon was alternatively spliced in a mutually exclusive manner with a novel, unannotated, upstream exon (Fig. 4). The amino acid sequences encoded by these two mutually exclusive exons are 38% identical and 62% similar, and the novel exon is 92% identical in D. pseudoobscura. Similarly, the candidate exon in CG10844 (Rya-r44F) (Takeshima et al. 1994) was alternatively spliced in a mutually exclusive manner with a novel alternative exon (Fig. 4). Again, the novel exon is highly conserved (92% identity) in D. pseudoobscura, as is the flanking intron sequence. Therefore, the novel alternative exons have properties similar to our candidate alternative exons.
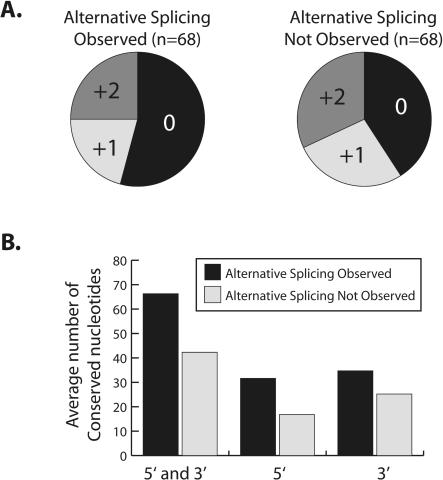
We analyzed several features of the highly conserved exons to identify properties that differ between those that we observed to be alternatively spliced and those for which alternative splicing was not observed. The group of highly conserved alternative exons we analyzed included the 23 new exons we experimentally identified as well as the 45 previously known alternative exons. Surprisingly, we found no significant differences in the relative strength or nucleotide composition of the 5′ or 3′ splice sites between the two sets of exons (data not shown). However, we identified two features that differed between these two groups of exons. First, the distribution of the exons between each of the three reading frames is different in each group. Whereas the group of exons for which alternative splicing was not observed are evenly distributed between each reading frame, the group of alternative exons is enriched in exons that maintain the reading frame (p = 0.01) (Fig. 5A). A similar preference for alternative exons to maintain the reading frame has been observed in human and mouse exons (Resch et al. 2004). The second feature that distinguishes the two groups of exons is the amount of conserved flanking intron sequence. Specifically, whereas the length of intron sequence greater than 75% identical at the 5′ and 3′ splice sites of alternative exons is an average of 65 nt, the exons for which alternative splicing was not observed are flanked by an average of 43 nt (Fig. 5B). Again, this is similar to findings in humans and mice (Sorek and Ast 2003).
FIGURE 5.
Features that distinguish exons for which alternative splicing is or is not observed. (A) Pie chart representation of the fraction of highly conserved exons of each reading frame for which alternative splicing is observed (left) or is not observed (right). (B) Graph of the average number of conserved intronic nucleotides at the 5′ or 3′ splice sites and the combination of 5′ and 3′ splice sites for exons for which alternative splicing is observed (black) or is not observed (gray).
Our results demonstrate that comparative genomics can be used to predict whether an exon is alternatively spliced with a fairly high degree of accuracy. Although the exons we tested were identified solely on the basis of their high degree of conservation, we also identified two features—higher degree of intron conservation and greater tendency to maintain the reading frame—that appear to further distinguish alternative and constitutive exons. Adding these features to the criteria of high exon and intron similarity may improve the accuracy of alternative exon prediction.
The high degree of identity used in our screen (95% exon identity, 75% intron identity) most likely exceeds the lower limits of exon and intron identity useful for accurate prediction. This is supported by the fact that the only alternative exon identified in the group of randomly selected exons we tested was 88% identical in the exon and was flanked by conserved intron sequences. Thus, further experiments will be necessary to determine the lower limits of identity that can be used to accurately predict alternative exons. This will obviously depend on the amount of divergence between the species being compared. For example, analysis of whole genome shotgun traces of five additional Drosophila species (D. simulans, D. yakuba, D. ananassae, D. mojavensis, and D. virilis) indicates that the percent identity of these conserved exons differs between species. For example, while exon 28 of CG1522 (cac) is 98% identical between D. melanogaster and D. pseudoobscura, the same exon is only 89% identical between D. melanogaster and D. virilis (data not shown). Determining these limits for each pair of species will be important since they will significantly increase the number of alternative exons that can be identified by this means.
Although this approach will be useful for identifying potential alternative exons, there are at least two classes of alternative exons that will not be identified using these criteria. The first class is small alternative exons, which will be difficult to identify based on percent identity alone. The second class of exons that will be missed by comparative genomics are those that are species specific (Modrek and Lee 2003). Recent studies in mammals have shown that a surprisingly large number of alternative exons are species specific. Additionally, there are some alternatively spliced exons that are specific to D. melanogaster or D. pseudoobscura (Graveley et al. 2004). Nonetheless, there are numerous alternative exons that are highly conserved between related species. Moreover, the finding of novel, unannotated alternative exons that are highly conserved suggests that many conserved noncoding sequences may in fact prove to be novel alternative exons. Thus, using comparative genomics to identify potential alternative exons should significantly advance our ability to accurately assess the amount of alternative splicing that occurs in any organism, thereby bringing us closer to understanding how organisms develop and function.
MATERIALS AND METHODS
Computational analysis
Percent identity data of the entire Drosophila melanogaster and Drosophila pseudoobscura genomes were downloaded from http://lbl.pipeline.gov/pseudo. All exons between 95% and 100% identical (using a window size of 50 bp) were analyzed using the VISTA browser (Mayor et al. 2000) to identify those that are flanked on one or both splice sites by intron sequence greater than 75% identical. Primers flanking all exons identified using this method were designed and the sequences are available at http://penguin.uchc.edu/~intron/philipps/oligos.html.
Experimental analysis of alternative splicing
Total RNA was isolated using Trizol (Invitrogen) from both D. melanogaster and D. pseudoobscura embryos, larvae, and adult females and males. cDNA was synthesized from 5 μg of a pool of total RNA from each developmental stage using Superscript II (Invitrogen) reverse transcriptase in a 20 μL reaction. PCR was performed using gene-specific primers and Taq DNA polymerase (Invitrogen). The reactions were incubated for 35 cycles of 94°C for 30 sec, 55°C for 15 sec, and 72°C for 1 min. PCR products were resolved by agarose gel electrophoresis. Each PCR product was excised from the gel, cloned into the pCRII-TOPO vector (Invitrogen), and sequenced.
Acknowledgments
We thank members of the Graveley laboratory and Rob Reenan for discussions and comments on the manuscript. This work was supported by an NIH grant (GM62516) to B.R.G.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7136104.
REFERENCES
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Black, D.L. 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103: 367–370. [DOI] [PubMed] [Google Scholar]
- Delaunay, J., Le Mee, G., Ezzeddine, N., Labesse, G., Terzian, C., Capri, M., and Ait-Ahmed, O. 2004. The Drosophila Bruno paralogue Bru-3 specifically binds the EDEN translational repression element. Nucleic Acids Res. 32: 3070–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forch, P. and Valcarcel, J. 2003. Splicing regulation in Drosophila sex determination. Prog. Mol. Subcell. Biol. 31: 127–151. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17: 100–107. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R., Kaur, A., Gunning, D., Zipursky, S.L., Rowen, L., and Clemens, J.C. 2004. The organization and evolution of the Dipteran and Hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. RNA 10: 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, V.N., Cameron, J.M., Zammit, V.A., and Price, N.T. 1999. Sequencing and functional expression of the malonyl-CoA-sensitive carnitine palmitoyltransferase from Drosophila melanogaster. Biochem. J. 341: 483–489. [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., and Shoemaker, D.D. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Green, R.E., and Brenner, S.E. 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. 100: 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047. [DOI] [PubMed] [Google Scholar]
- Miki, H., Setou, M., Kaneshiro, K., and Hirokawa, N. 2001. All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. 98: 7004–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, S., Crosby, M.A., Mungall, C.J., Matthews, B.B., Campbell, K.S., Hradecky, P., Huang, Y., Kaminker, J.S., Millburn, G.H., Prochnik, S.E., et al. 2002. Annotation of the Drosophila melanogaster eu-chromatic genome: A systematic review. Genome Biol. 3: RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C.J. 2003. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 34: 177–180. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29: 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J.R. 1997. Progress and prospects in evolutionary biology: The Drosophila model. Oxford University Press, NY.
- Resch, A., Xing, Y., Alekseyenko, A., Modrek, B., and Lee C. 2004. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 32: 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, C.A., Takezaki, N., and Nei, M. 1995. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12: 391–404. [DOI] [PubMed] [Google Scholar]
- Schmucker, D., Clemens, J.C., Shu, H., Worby, C.A., Xiao, J., Muda, M., Dixon, J.E., and Zipursky, S.L. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101: 671–684. [DOI] [PubMed] [Google Scholar]
- Sorek, R. and Ast, G. 2003. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 13: 1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet, C.W., Kent, W.J., Ares Jr., M., and Haussler, D. 2004. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 66–77. [DOI] [PubMed]
- Takeshima, H., Nishi, M., Iwabe, N., Miyata, T., Hosoya, T., Masai, I., and Hotta, Y. 1994. Isolation and characterization of a gene for a ryanodine receptor/calcium release channel in Drosophila melanogaster. FEBS Lett. 337: 81–87. [DOI] [PubMed] [Google Scholar]
- zur Lage, P., Shrimpton, A.D., Flavell, A.J., Mackay, T.F., and Brown, A.J. 1997. Genetic and molecular analysis of smooth, a quantitative trait locus affecting bristle number in Drosophila melanogaster. Genetics 146: 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]