Abstract
A helix-loop-helix within the group I intron has most of the canonical sequence elements of a kink turn (K-turn), yet it bends in the opposite direction. The reverse K-turn kinks toward the major rather than the minor grooves of the flanking helices. This suggests that there are two distinct subclasses of tertiary structures that a K-turn secondary structure can adopt. The final structure may be specified by external factors, such as protein binding or the tertiary structural context, rather than the intrinsic conformation of the RNA.
Keywords: reverse kink turn, RNA structure, group I intron, X-ray crystallography
INTRODUCTION
The kink turn (K-turn) is a general RNA structural motif that creates a sharp ~120° bend between two continuous helices (Klein et al. 2001). Although there is a fairly high degree of sequence variation among the broader class of K-turn motifs, there are several hallmark features that characterize the bend (Fig. 1A,B, orange). In the prototypical K-turn, a 3-nucleotide (nt) asymmetric bulge is flanked by a canonically base paired helix (C helix) and a noncanonical helix (NC helix). The NC helix usually includes two G•A mispairs immediately adjacent to the unpaired residues. The A’s in these pairs are positioned on the minor groove edge of the NC helix. They both make tertiary interactions between the helices, including a type I A-minor contact by the A in the penultimate G•A pair. Among the unpaired residues in the asymmetric loop, the first stacks on the C helix, the second stacks on the NC helix, and the third is extruded into solvent. Most K-turns are recognized by RNA binding proteins (Brodersen et al. 2002; Klein et al. 2004).
FIGURE 1.
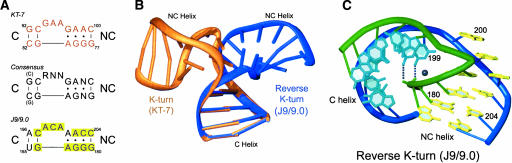
Minor groove and major groove kink turns. (A) Sequences of the consensus K-turn (middle), the archetypical K-turn KT-7 from 23S rRNA (top), and the J9/9.0 reverse K-turn. The canonical (C) and the noncanonical (NC) helices are both indicated. The nucleotide sequences of the various segments are shown. The residues in J9/9.0 that perfectly match the K-turn consensus are highlighted in yellow. The remaining three residues are all conservative substitutions based upon K-turn structures. (B) Comparison of the K-turn (KT-7, orange) and reverse K-turn (J9/9.0, blue) structures. The C helices of the KT-7 and J9/9.0 K-turns were superimposed to demonstrate the difference in NC helix placement between the minor groove and major groove K-turns. (C) Structural depiction of the reverse K-turn. The long strand is shown in blue and the short strand in green. Residues in the C helix are in cyan, while those in the NC helix are in yellow. The Mg2+ that appears to mediate the close packing of the NC phosphate backbone against the C major groove is shown as a black sphere. Hydrogen bonds between the C major groove edge and the NC backbone are shown as dashed cylinders. Panels B and C were made using Ribbons (Carson 1991).
Within the recently reported Azoarcus group I intron crystal structure there is a 90° bend at an asymmetric loop between helices P9.0 and P9 (J9/9.0, Fig. 1A,B, purple; Adams et al. 2004). The sequence elements of the J9/P9.0 bend are suggestive of a K-turn. There is a C helix (P9) that is fully base paired with canonical pairings. There is also an NC helix (P9.0) that includes purine–purine mispairs, with adenosines positioned in the minor groove, and there is a 3-nt asymmetric loop between the C and NC helices.
Although the P9-9.0 junction is very close to the K-turn consensus sequene, the J9/9.0 junction bends in the direction opposite to that expected for the K-turn motif (Fig. 1B). When the C helices of the two structures are aligned and viewed from the major groove edge, the consensus K-turn bends to the left, bringing together the minor edges of the C and NC helices (Fig. 1B, orange). In contrast, the J9/9.0 K-turn bends to the right and juxtaposes the major groove edges of the flanking helices (Fig. 1B, blue). Furthermore, the sharpest bends occur on different strands between the two folds. In the KT-7 turn, the backbone of the long strand makes a tight bend, while the short strand makes a gradual a shift in strand direction (Fig. 1B). Within the J9/9.0 turn, the long strand undergoes an incremental turn, while the short strand has a drastic kink that localizes to the purine–purine mispairs in the NC helix. The backbone conformation within this region is reminiscent of the S turn seen in the 23S sarcin-ricin loop, except the upper curve of the S is straightened by the 90° bend in the helix (Szewczak et al. 1993; Correll et al. 1998). This conformation places the phosphate backbone of the NC helix into the major groove of helix C, where the interface appears to be stabilized by a Mg2+ ion (Fig. 1C).
The tertiary structural elements that stabilize the J9/9.0 turn are different than those found in the standard K-turn. The packing of the C and NC helices also appears to be less tight (Fig. 1C), and there is no A-minor interaction, although the residue equivalent to A80 (A183) is present. Instead, the tertiary hydrogen bonding contacts are strictly between the Hoogsteen face of the unpaired nucleotides and the nonbridging phosphate oxygens of the NC helix back-bone (Fig. 1C). The retention of maximal base stacking is the most noteworthy stabilizing feature of the bend. Nucleotides in the loops are fully stacked under either the C or the NC helix. There is no bulged residue comparable to the third unpaired nucleotide in the K-turn. The bend occurs precisely between two residues on each strand, A183 and G184 in the short strand and C199 and A200 in the long strand (Fig. 1C). A200 caps the NC helix and C199 caps the C helix. The angle between these two consecutive nucleotides is particularly sharp, with C199 both perpendicular and below the plane of A200.
J9/9.0 is structurally similar to the bend between helices 54 and 55 in 23S rRNA (KT-54) (Ban et al. 2000; Duarte et al. 2003). The primary difference is the size of the asymmetric loop (five instead of three). Like the J9/9.0 region, KT-54 retains stacking interactions of all bases, bends toward the major groove, and does not bind protein. This suggests that K-turn sequences can be subdivided into two structural classes based upon the groove into which the helices are bent: (1) the standard K-turn, which bends toward the minor groove, and (2) the reverse K-turn, which bends toward the major groove.
Given the close match between the J9/9.0 sequence and the K-turn consensus sequence, what is responsible for the major groove bend? It is possible that having an A at position 97 (KT-7 numbering), instead of G, is responsible for the reversal, though this seems unlikely. In KT-7, G97 forms a sheared pair with A80. A conformationally similar mispair could also form between two adenosines.
It is more likely that structure elements external to the loop are responsible for the direction of its bend. Given the small stabilizing interface between the flanking helices, a major groove bend is unlikely to be an intrinsic feature of the asymmetric loop. In the bacterial group I intron, the end of helix P9 makes a tetraloop–tetraloop receptor (TL–TLR) that may pull the P9 helix toward the major groove (Costa and Michel 1995; Tanner and Cech 1996; Adams et al. 2004). Similarly external elements are responsible for the sharp 180° bend within J5/5a of the Tetrahymena intron P4-P6 domain (Cate et al. 1996; Matsumura et al. 2003). In that case, residues in J5/5a did not adopt an inherently bent structure in the absence of the distal tertiary contacts, including a TL–TLR (Szewczak and Cech 1997).
It is possible that any given K-turn could be in a dynamic equilibrium between an unbent conformation and the two kinked states. Fluorescence data support the inherent structural flexibility of the K-turn (Goody et al. 2004), though FRET analysis of the Mg2+-induced RNA bending might have been unable to differentiate between the major groove and minor groove variants. External stabilizing elements, such as RNA tertiary structure formation or protein binding, may be necessary to lock the RNA into one conformation or the other. Such structural heterogeneity supports the conclusion of Goody et al. (2004) that the K-turn is unlikely to be a primary organizational element of higher order RNA structure.
Acknowledgments
We thank L. Wadley for assistance with PRIMOS analysis, R. Gutell for analysis of structure alignments, and J. Cochrane for comments on the manuscript. This work was supported by NSF grant MCB-0315329.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7141504.
REFERENCES
- Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J., and Strobel, S.A. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P., and Steitz T. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Brodersen, D.E., Clemons Jr., W.M., Carter, A.P., Wimberly, B.T., and Ramakrishnan, V. 2002. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 316: 725–768. [DOI] [PubMed] [Google Scholar]
- Carson, M. 1991. Ribbons 2.0. J. Appl. Crystallogr. 24: 958–961. [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. [DOI] [PubMed] [Google Scholar]
- Correll, C.C., Munishkin, A., Chan, Y.L., Ren, Z., Wool, I.G., and Steitz, T.A. 1998. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl. Acad. Sci. 95: 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M. and Michel, F. 1995. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 14: 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte, C.M., Wadley, L.M., and Pyle, A.M. 2003. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 31: 4755–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody, T.A., Melcher, S.E., Norman, D.G., and Lilley, D.M. 2004. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA 10: 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, D.J., Moore, P.B., and Steitz, T.A. 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340: 141–177. [DOI] [PubMed] [Google Scholar]
- Matsumura, S., Ikawa, Y., and Inoue, T. 2003. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 31: 5544–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczak, A.A. and Cech, T.R. 1997. An RNA internal loop acts as a hinge to facilitate ribozyme folding and catalysis. RNA 3: 838–849. [PMC free article] [PubMed] [Google Scholar]
- Szewczak, A.A., Moore, P.B., Chang, Y.L., and Wool, I.G. 1993. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc. Natl. Acad. Sci. 90: 9581–9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, M.A. and Cech, T.R. 1996. Activity and thermostability of the small self-splicing group I intron in the pre-tRNAlle of the purple bacterium Azoarcus. RNA 2: 74–83. [PMC free article] [PubMed] [Google Scholar]