Abstract
Studies of Escherichia coli 30S ribosomal subunit assembly have revealed a hierarchical and cooperative association of ribosomal proteins with 16S ribosomal RNA; these results have been used to compile an in vitro 30S subunit assembly map. In single protein addition and omission studies, ribosomal protein S13 was shown to be dependent on the prior association of ribosomal protein S20 for binding to the ribonucleoprotein particle. While the overwhelming majority of interactions revealed in the assembly map are consistent with additional data, the dependency of S13 on S20 is not. Structural studies position S13 in the head of the 30S subunit > 100 Å away from S20, which resides near the bottom of the body of the 30S subunit. All of the proteins that reside in the head of the 30S subunit, except S13, have been shown to be part of the S7 assembly branch, that is, they all depend on S7 for association with the assembling 30S subunit. Given these observations, the assembly requirements for S13 were investigated using base-specific chemical footprinting and primer extension analysis. These studies reveal that S13 can bind to 16S rRNA in the presence of S7, but not S20. Additionally, interaction between S13 and other members of the S7 assembly branch have been observed. These results link S13 to the 3′ major domain family of proteins, and the S7 assembly branch, placing S13 in a new location in the 30S subunit assembly map where its position is in accordance with much biochemical and structural data.
Keywords: ribosome, 30S subunit, S13, assembly
INTRODUCTION
Assembly of the Escherichia coli 30S ribosomal subunit is an elaborate process that has been under investigation for more than three decades. In early studies it was shown that the small subunit may be reconstituted in vitro from its isolated constitutive parts, 16S ribosomal RNA (rRNA) and a mixture of the Total Proteins from the 30S subunit (TP30; S2–S21), to form a functional 30S particle (Traub and Nomura 1968). Additionally it has been shown that the individually purified proteins, both natural (Traub and Nomura 1968) and recombinant (Culver and Noller 1999), could support the in vitro reconstitution. Single protein addition and omission studies of the in vitro 30S subunit reconstitution process led to the elucidation of an ordered assembly map and the definition of distinct assembly branches (Fig. 1; Mizushima and Nomura 1970; Held et al. 1974). For example, the S7 assembly branch (see Fig. 1) contains not only S7 but also r-proteins S9, S19, S14, S10, S3, and S2. Generally, there is good agreement between the dependencies indicated in the in vitro 30S subunit assembly map and structural and biochemical studies that reveal interactions and locations of proteins within the subunit. However, there is at least one significant outlier to this correlation, the positioning of r-protein S13 within the assembly map.
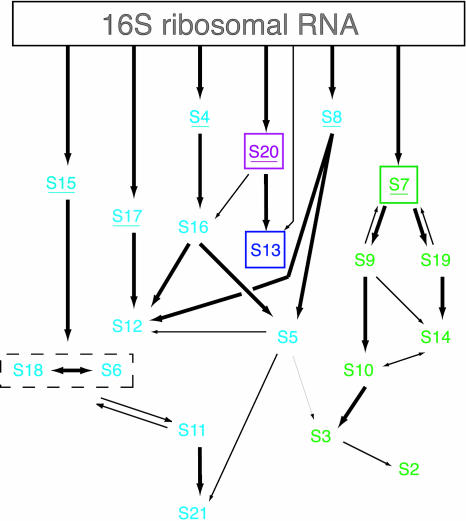
FIGURE 1.
The in vitro 30S subunit assembly map. Simplified in vitro 30S subunit assembly map (Mizushima and Nomura 1970; Held et al. 1974) highlighting the relative position of r-proteins S7 and proteins found in its assembly branch (green), S13 (blue), and S20 (magenta). The remaining r-proteins are aqua. The primary binding proteins have been underlined, and 16S rRNA is designated as a rectangle.
Experiments that led to the formulation of the assembly map (Fig. 1) indicated that S13 is dependent on S20 for association (Mizushima and Nomura 1970; Held et al. 1974); however, conflicting evidence suggested that S13 can stably assemble into a 16S-rRNA-containing RNP without the prerequisite stable association of S20 (Schaup et al. 1970; Lutter et al. 1974; Samaha et al. 1994). Studies that have focused on the positions of r-proteins within the mature 30S subunit (Lake 1975; Capel et al. 1987; Pohl and Wittmann-Liebold 1988; Wimberly et al. 2000) also suggest that S13 is misplaced in the assembly map. In the crystal structure of the Thermus thermophilus 30S subunit, S13 and S20 are located > 100 Å apart with S20 residing in the body and S13 in the head of the subunit proximal to S7 (see Fig. 4B below; Wimberly et al. 2000; Brodersen et al. 2002). Thus, the location of S13 in the head of the 30S subunit with all the components of the S7 assembly branch (Wimberly et al. 2000), and solely these components, supports inclusion of S13 within the S7 assembly branch.
FIGURE 4.
Positioning S13 in the S7 assembly branch of the in vitro 30S ribosomal subunit assembly map. (A) Revised in vitro 30S subunit assembly map. Color coding is as in Figure 1. S13 has been repositioned to show interaction with S7 and other r-proteins in this branch. (B) S13 footprinting data mapped on the tertiary structure of the 30S subunit. The enhancements are blue and protections are orange (in accordance with Figs. 2, 3). S13 is blue and S7 is green. This figure was produced with Ribbons (Carson 1997) using PDB 1J5E (Wimberly et al. 2000; Brodersen et al. 2002).
While footprinting studies can reveal assembly relationships, previous work has not dissected the requirements for S13 binding. In prior experiments, S13 interactions with 16S rRNA were probed (Heilek and Noller 1996) and footprinted (Powers et al. 1988a) in the presence of all the 1° binding proteins (S4, S7, S8, S15, S17, and S20). Therefore, to understand which 1° binding protein(s) is required for S13 assembly, chemical modification and primer extension analysis were used to examine more minimal particles containing S13. Our data support a sequential relationship between S7 and S13, not S20 and S13, resulting in a new position in the in vitro 30S subunit assembly map for S13. Additional interactions between S13 and other proteins found in the S7 assembly branch, mainly S9 and S19, are also revealed.
RESULTS AND DISCUSSION
Reconstitution of 16S-rRNA-containing RNPs
In order to determine the requirements for association of S13 with 16S rRNA, different sets of 16S rRNA r-protein-containing complexes were prepared for comparison. Initially, the appropriate recombinant primary binding protein, either S7 or S20, was added to 16S rRNA; after the binary complex was formed, S13 was added to the RNP. To investigate interactions of S13 with other proteins beyond the primary binding proteins, sequential addition of proteins, either those in the S7 assembly branch or the tertiary binding proteins, S2, S3, S10, and S14, were added to S13-containing complexes. In all cases the RNPs were used directly in probing reactions.
Footprinting S13 in the presence of S7 and S20
The reconstituted RNPs were subjected to kethoxal modification, which modifies guanine bases, and to dimethyl sulfate (DMS), which modifies both adenine and cytosine bases. Primer extension analysis was used to map the sites of 16S rRNA modification in various complexes to determine if the addition of S13 has an effect on the modification patterns. Association of S13 with the binary complexes should reveal sites of altered reactivity of 16S rRNA as compared to the modification pattern in the absence of S13.
Changes attributable to S13 binding are observed only in the presence of S7. All of the changes that are attributable to S13 footprints in the S7/S13/16S rRNA complexes are found in the 3′ major domain (Figs. 2, 3), consistent with prior structural and biochemical data (Powers et al. 1988a,b). The data representing relative changes in the modification pattern upon the addition of S13 to S7/16S rRNA have been highlighted on the secondary structure of 16S rRNA (Fig. 3) and on the 3D structure of the 30S subunit (Fig. 4B). Many changes resulting from S13 binding can be seen in the 930 to 980 region, with the most pronounced protections occurring at G944 and G945 in helix 30 and the most pronounced enhancements occurring at nucleotides 977–980 in the loop connecting helices 30 and 31 (Fig. 2A). Additional changes, many of which augment a change that is initiated by S7 binding, are observed in helices 41 and 42 (Fig. 2B). The numerous changes observed upon the addition of S13 to the S7/16S rRNA complex (Fig. 2A,B) are in sharp contrast to the total absence of changes observed upon addition of S13 to the S20/16S rRNA complex (Fig. 2C, lane 5).
FIGURE 2.
Primer extension analysis of kethoxal (A–C) and DMS (A,B,D) modified reconstituted RNPs containing 16S rRNA and different combinations of r-proteins including S13. (Lanes A,G) Dideoxy sequencing lanes; (lane 1) 16S unmodified, naked, unreacted 16S rRNA; (lane 2) 16S modified, naked 16S rRNA treated with kethoxal; (lane 8) naked 16S rRNA treated with DMS. (Lanes 3–7,9–18) RNPs that have been treated with kethoxal or DMS, respectively, and consist of a combination of naked 16S rRNA and the r-protein(s) S7, S9, S19, S13, and/or S20, as indicated at the top of the gels. (Lanes 3,9) S13 alone; (lanes 4,10) S20 alone; (lanes 5,11) S20 + S13; (lanes 6,12) S7 alone; (lanes 7,13,16) S7 + S13; (lane 14) S7 + S9; (lane 15) S7 + S19; (lane 17) S7 + S9 and S13; (lane 18) S7 + S19 and S13. The bases that have an increased reactivity, or enhancement, are noted with triangles. The bases with a decreased reactivity, or protection, are noted with circles. The DMS sequencing lanes in A have been transposed from another section of the gel. (A) Primer extension analysis using primer 945. (B) Primer extension analysis using primer 1391. (C) Primer extension analysis using primer 323. (D) Primer extension analysis using primer 945.
FIGURE 3.
S13 footprinting data mapped on the secondary structure of 16S rRNA. The 16S rRNA secondary structure (Cannone et al. 2002) with the 3′ major domain highlighted is shown, and the S13 footprinting data are mapped on this expanded domain. The bases that have an increased reactivity, or enhancement, upon the addition of S13 to the S7/16S rRNA, are noted with triangles and the bases with a decreased reactivity, or protection, are noted with circles (in accordance with the footprinting data in Fig. 2).
Since no changes are observed upon addition of S13 to a complex of S20 and 16S rRNA, it is important to verify the binding of S20 in the reconstituted RNP. In the S20/16S rRNA complex, strong kethoxal protections can be seen at nucleotides 251, 265–266, and 281 (Fig. 2C, lanes 4,5) in accordance with earlier work (Stern et al. 1988). In addition, a weak enhancement may be seen at nucleotide 278 (Fig. 4A, lanes 4,5). Thus, S20 is indeed bound in these particles, and S13 does not appear to associate with 16S rRNA solely in the presence of S20. These data support the inclusion of S13 in the S7 assembly branch rather than the S20 branch.
The relative position of S13 with regard to the other members of the S7 assembly branch (see Fig. 1) also warranted further investigation. RNPs containing various combinations of S7, S13, and other S7-dependent binding proteins were prepared and analyzed (Fig. 2D). Several increased protections are observed in the 930–960 region when S13 is added to complexes containing S7–S9 or S7–S19 particles (Fig. 2D, lanes 17,18). In these regions the addition of S13 appears to augment binding of S9 and S19 since these residues are partially protected in the absence of S13 but become more protected upon S13 addition. These results suggest a synergistic assembly relationship between S7, S13, S9, and S19. Given the complexities of the S7 assembly branch, significant further analysis will be required to dissect the role S13 plays in the association of the 3° binding proteins S2, S3, S10, and S14 that also bind to the head of the 30S subunit.
This study was designed to address the assembly relationship of S13 with the primary binding proteins S7 and S20 in the course of 30S subunit assembly. As a result, it has been shown that S13 has an assembly dependence on S7 as its binding predecessor to 16S rRNA, and that S13 binding is not affected by the presence of S20. As a consequence of these studies, S13 has been relocated in the assembly map downstream from S7 (Fig. 4A), localized within the group of proteins that bind to the 3′ major domain and form the head of the 30S subunit. Previous work suggested that S7, S9, and S19 were necessary and sufficient for S13 binding (Powers et al. 1988a). Based on the results presented here, it appears that S7 alone is necessary for S13 binding (Fig. 2), thus solidifying S13 as a secondary binding protein. Nevertheless, some synergistic effects between binding of S7, S13, S9, and S19 have been observed (Figs. 2D, 4A).
Additional sites of altered reactivity are observed when S13 is footprinted solely in the presence of S7 (Fig. 2A,B) and not a larger group of proteins (Powers et al. 1988a,b). This is likely due to the absence of the additional proteins which could mask sites of potential changes. A likely example of this is the data observed in the loop connecting helices 31 and 32. In complexes composed of S7 and 16S rRNA, nucleotides 977–980 showed enhanced reactivity (Fig. 2A; Powers et al. 1988b). Upon addition of S19 to this complex (Powers et al. 1988b), these same nucleotides become protected from reactivity. In our experiments, upon the addition of S13 to the S7/16S rRNA complexes, nucleotides 977–980 become more reactive (Fig. 2A). Thus it appears that S13 augments the effect of S7, with S19 having the opposite effect. Also, these sites are further protected upon the addition of other S7 assembly branch proteins, S2 and S3 (Powers et al. 1988a,b). These observations suggest that S13 is involved in a series of concerted and consecutive changes within the S7 assembly branch that aid 30S subunit assembly.
The majority of S13-dependent changes in reactivity are localized in the area surrounding S7 and S13 (Fig. 4B; Wimberly et al. 2000). Some of the S13 binding interactions observed correspond with direct backbone and base-specific interactions between S13 and 16S rRNA observed in the 30S subunit (Brodersen et al. 2002). Also, association of S13 with the S7/16S rRNA complex results in changes in reactivity of many nucleotides that interact with S7 within the 30S subunit (Brodersen et al. 2002). For example, in the presence of S7 minor changes in reactivity in helices 28 and 29 due to the addition of S13 are observed (Figs. 2A, 3). Various other sites show similar trends upon S13 binding. These results suggest that S13 association with the S7/16S rRNA complex enhances the association of S7 with 16S rRNA during 30S subunit assembly (see Fig. 4A). Since there are no direct contacts between S7 and S13 in the 30S subunit (Wimberly et al. 2000; Brodersen et al. 2002), the observed changes are most likely orchestrated by protein-dependent RNA conformational changes that then allow additional protein binding. Overall our data are highly consistent with the position of elements within the fully formed 30S subunit (Wimberly et al. 2000; Brodersen et al. 2002) and prescribe the inclusion of S13 in the S7 assembly branch.
MATERIALS AND METHODS
Natural 16S rRNA was extracted and purified from MRE600 as previously described (Moazed et al. 1986). Recombinant small subunit ribosomal proteins were purified as previously described (Culver and Noller 2000).
Preparation of complexes
The reconstitution was performed with 40 pmol of 16S rRNA and 280 pmol of recombinant r-proteins S7, S9, S13, S19, and S20 in appropriate combinations. 16S rRNA was heat-activated at 42°C for 15 min and cooled on ice for 20 min. The reconstitution was performed in Recon A buffer (80 mM K+-HEPES 7.6, 20 mM MgCl2, 330 mM KCl, and 0.01% Nikkol), with the final KCl concentration of 330 mM at each reconstitution stage. The first stage consists of 16S rRNA and a 7 M excess of the appropriate 1° r-protein (S7 or S20) in Recon A buffer and 1 mM β-mercapto-ethanol (BME), which was incubated at 42°C for 20 min. Next, a 7 M excess of the appropriate 2° r-protein(s) (S9, S13 and S19) is added with conditions adjusted to those of Recon A buffer, followed by a 42°C incubation for 20 min. Protein storage buffer (1 M KCl, 20 mM MgCl2, and 80 mM K+-HEPES 7.6) was added in place of the r-protein to achieve a final KCl concentration of 330 mM.
Chemical probing and primer extension analysis
The resulting RNPs were cooled on ice for 20 min, and subjected to kethoxal or dimethyl sulfate (DMS) modification on ice for 2 h (as described by Moazed et al. 1986). The rRNA was then extracted as previously described (Culver and Noller 2000). The resulting RNA-modified complexes were used for primer extension analysis with α-32P-TTP and primers (Moazed et al. 1986).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7130504.
REFERENCES
- Brodersen, C.W.J., Carter, A.P., Wimberly, B.T., and Ramakrishnan, V. 2002. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 316: 725–768. [DOI] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. 2002. The comparative RNA Web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel, M.S., Engelman, D.M., Freeborn, B.R., Kjeldgaard, M., Langer, J.A., Ramakrishnan, V., Schindler, D.G., Schneider, D.K., Schoen-born, B.P., Sillers, I.Y., et al. 1987. A complete mapping of the proteins in the small ribosomal subunit of Escherichia coli. Science 238: 1403–1406. [DOI] [PubMed] [Google Scholar]
- Carson, M. 1997. Ribbons. Methods Enzymol. 277: 493–505. [PubMed] [Google Scholar]
- Culver, G.M. and Noller, H.F. 1999. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA 5: 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol. 318: 446–460. [DOI] [PubMed] [Google Scholar]
- Heilek, G.M. and Noller, H.F. 1996. Directed hydroxyl radical probing of the rRNA neighborhood of ribosomal protein S13 using tethered Fe(II). RNA 2: 597–602. [PMC free article] [PubMed] [Google Scholar]
- Held, W.A., Ballou, B., Mizushima, S., and Nomura, M. 1974. Assembly mapping of 30S ribosomal proteins from Escherichia coli. J. Biol. Chem. 249: 3103–3111. [PubMed] [Google Scholar]
- Lake, J. 1975. Ribosomal proteins S5, S11, S13 and S19 localized by electron microscopy of antibody-labeled subunits. J. Mol. Biol. 99: 631–644. [DOI] [PubMed] [Google Scholar]
- Lutter, L., Bode, U., Kurland, C., and Stoffler, G. 1974. Ribosomal protein neighborhoods. III. Cooperativity of assembly. Mol. Gen. Genet. 129: 167–176. [DOI] [PubMed] [Google Scholar]
- Mizushima, S. and Nomura, M. 1970. Assembly mapping of 30S ri-bosomal proteins in E. coli. Nature 226: 1214–1218. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Stern, S., and Noller, H.F. 1986. Rapid chemical probing of conformations in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J. Mol. Biol. 187: 399–416. [DOI] [PubMed] [Google Scholar]
- Pohl, T. and Wittmann-Liebold, B. 1988. Identification of a cross-link in the Escherichia coli ribosomal protein pair S13–S19 at the amino acid level. J. Biol. Chem. 263: 4293–4301. [PubMed] [Google Scholar]
- Powers, T., Stern, S., Changchien, L.M., and Noller, H.F. 1988a. Probing the assembly of the 3′ major domain of 16 S rRNA. Interactions involving ribosomal proteins S2, S3, S10, S13 and S14. J. Mol. Biol. 201: 697–716. [DOI] [PubMed] [Google Scholar]
- Powers, T., Changchien, L., Craven, G., and Noller, H.F. 1988b. Probing the assembly of the 3′ major domain of 16 S ribosomal RNA. Quaternary interactions involving ribosomal proteins S7, S9 and S19. J. Mol. Biol. 200: 309–319. [DOI] [PubMed] [Google Scholar]
- Samaha, R.R., O’Brien, B., O’Brien, T.W., and Noller, H.F. 1994. Independent in vitro assembly of a ribonucleoprotein particle containing the 3′ domain of 16S rRNA. Proc. Natl. Acad. Sci. 91: 7884–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaup, H.W., Green, M., and Kurland, C.G. 1970. Molecular interactions of ribosomal components. I. Identification of RNA binding sites for individual 30S ribosomal proteins. Mol. Gen. Genet. 109: 193–205. [DOI] [PubMed] [Google Scholar]
- Stern, S., Changchien, L.M., Craven, G.R., and Noller, H.F. 1988. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J. Mol. Biol. 200: 291–299. [DOI] [PubMed] [Google Scholar]
- Traub, P. and Nomura, M. 1968. Structure and function of E. coli ribosomes, V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. 59: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons, W.M.J., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]