Abstract
It is theoretically possible to obtain a catalytic site of an artificial ribozyme from a random sequence consisting of a limited numbers of nucleotides. However, this strategy has been inadequately explored. Here, we report an in vitro selection technique that exploits modular construction of a structurally constrained RNA to acquire a catalytic site for RNA ligation from a short random sequence. To practice the selection, a sequence of 30 nucleotides was located close to the putative reaction site in a derivative of a naturally occurring self-folding RNA whose crystal structure is known. RNAs whose activity depended on the starting three-dimensional structure were selected with 3′–5′ ligation specificity, indicating that the strategy can be used to acquire a variety of catalytic sites and other functional RNA modules.
Keywords: catalytic RNA, in vitro selection, ligation
INTRODUCTION
Three-dimensional structures of RNA structural units in such naturally occurring RNAs as the Tetrahymena ribozyme and RNA selection technique have enabled the construction of artificial RNAs with defined three-dimensional (3D) structures (Jaeger et al. 2001; Ikawa et al. 2002) and the acquisition of new functional RNA units (Williams et al. 1994; Ohuchi et al. 2002), respectively. The self-folding P4–P6 domain of the Tetrahymena ribozyme, which consists of 160 nucleotides, is nonessential for catalytic activity (Ikawa et al. 2001, and references therein). It serves as a scaffold that stabilizes the correctly folded ribozyme and is tightly connected to the rest of the RNA (Golden et al. 1998), which consists of about 250 nucleotides containing the reaction and catalytic sites (Fig. 1A, left). The domain folds into a rigid hairpin-shaped three-dimensional structure by itself (Murphy and Cech 1993) and is composed of two subdomains, P5abc and the stacked helices composed of P4 and P6 stems. The two subdomains are associated via intramolecular long-range interactions between their structural elements.
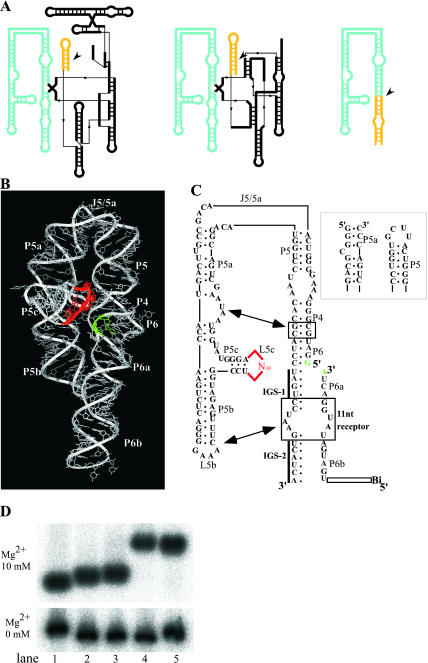
FIGURE 1.
(A) Secondary structure of Tetrahymena group I intron (left), the class hc ligase ribozyme (Jaeger et al. 1999) (middle), and P4–P6 ligase ribozyme (right). The P4–P6 domain and the helices containing the reaction site are highlighted in blue and yellow, respectively. Reaction sites are indicated by arrowheads. (B) 3D structure of P4–P6 RNA (Protein Data Bank 1GID) with highlighted L5c (red) and J6/6a (green). (C) Secondary structure of RNA with a pool (red), a reaction site in J6/6a (green), and substituted nucleotides (gray). Long-range interactions are indicated with arrows. Sequences and secondary structures of P5 and P5a regions in the bimolecular system (see last paragraph of Results) are shown in the right box. (D) Nondenaturing PAGE analysis of the P4–P6 RNA (lane 1), its derivative with termini at P6 (lane 2), and another derivative having an additional substitution at internal-loop composed of C217 – A219 and G254 –A256 between P6 and P6a with three consecutive base pairs as follows: 5′-CG, UA and AU-3′ (lane 3), a mutant RNA (this RNA is not a ligase, but a derivative of P4–P6, which has no enzymatic activity) containing L5b consisting of UUCG (L5b UUCG mutant, lane 4), and a derivative of the L5b UUCG mutant with additional substitutions described for lane 3 (lane 5). The conditions were adapted from those reported previously (Ikawa et al. 2002).
The P4–P6 domain has previously been used as a scaffold of an artificial ligase ribozyme (class hc ligase) (Jaeger et al. 1999; Fig. 1A, middle). The ribozyme was acquired by in vitro selection from a combinatorial library (RNA pool) containing three random sequences of 85 nucleotides located outside of the P4–P6 domain. The fundamental structural organization of the Tetrahymena ribozyme was succeeded to the ligase (Fig. 1A, cf. left and middle); the regions responsible for RNA ligation activity replaced the corresponding regions responsible for phosphoester transfer activity of the Tetrahymena ribozyme.
In this study, we report a new class of ligase ribozyme derived from P4–P6 RNA by using an in vitro selection technique to exploit the modular construction of RNA. The substrate binding site and a random sequence comprised of only 30 nucleotides were placed on two closely located structural elements on the scaffold (Fig. 1A, right, and B). The random sequence for constructing a catalytic unit was set directly on the specific site of the crystal structure of the P4–P6 RNA to compose the unit fixed by the RNA scaffold. As expected, the ribozyme was selected from a library of RNAs containing a short random sequence.
The advantages of the above strategy are as follows. (1) Further engineering of the selected ribozyme can be performed easier, because the anatomy of the selected ribozyme is obvious due to the modular organization of the selected molecule. Accordingly, the ribozyme can be converted to a bimolecular form by cutting it into catalytic and substrate molecules for further development. (2) The RNA library can be readily usable for selecting a variety of ribozymes with simple modifications. For the selection, two functional groups that form a covalent bond by chemical reaction can be readily set at the binding site on the scaffold.
RESULTS
Design of selection library
The putative catalytic unit was set proximal to the putative reaction site on the P4–P6 crystal structure lacking catalytic activity as follows. The P4–P6 RNA dissected from the whole intron RNA rapidly folds into a specific form consisting of two stacked helical components, P5-P4-P6-P6a-P6b and P5a-P5b, which are linked by a hinge region, J5/5a (Fig. 1B). P5c branches between P5a and P5b, and its terminal loop, L5c, is closely located to J6/6a in the 3D structure (Cate et al. 1996). The putative catalytic domain and reaction site were set at L5c and J6/6a, respectively (red and green in Fig. 1, B and C), because the L5c loop plays no role in the isolated P4–P6 RNA. As a combinatorial library from which a ligase ribozyme was to be isolated, a randomized sequence was inserted at the L5c region in place of the original UGCAA pentaloop. The size of the sequence was limited to 30 nucleotides because we hoped that a modular unit with only one function could be constructed from the relatively small numbers of nucleotides. The J6/6a internal loop was replaced with base pairs to set the reaction site within the context of a Watson–Crick duplex. Additional modifications at termini were performed to promote the isolation of ligase ribozyme by the selection procedure. The original termini of the P4–P6 RNA (positions U107 and U258 of the Tetrahymena ribozyme) were linked covalently, and new termini were generated at L6b, because circular permutation is known to maintain the 3D structure of P4–P6 (Murphy et al. 1994). At J6/6a, the resulting permuted P4–P6 was divided into two pieces, ribozyme and substrate fragments (Fig. 1C). The sequence of the 3′ region of the ribozyme fragment was engineered to serve as internal guide sequences complementary to the substrate fragments (Fig. 1C). A cytidine bulging out from P5 was deleted to stabilize the tertiary structure of the P4–P6 RNA (Juneau and Cech 1999). The ligated RNAs designed to have biotins at their 5′ ends were recovered by using streptavidine-beads.
The conservation of the overall structure of the modified P4–P6 RNA was supported by nondenaturing PAGE analysis (Fig. 1D). In the presence of magnesium ions, the RNA used as the scaffold (Fig. 1D, lane 3) and the original P4–P6 (Fig. 1D, lane 1) exhibited similar mobility, while the mutant RNAs missing the interaction at L5b (L5b UUCG mutant, Fig. 1D, lanes 4,5) showed slower mobility in comparison, suggesting that the scaffold RNA maintains the original structure.
In vitro selection experiment
A pool containing an average of 15 copies each of 2.1 × 1014 different RNA sequences was constructed and subjected to an in vitro selection procedure based on previously reported systems (see Materials and Methods, below). Two sets of substrate RNA and internal guide sequences were alternately used as selection cycles to select ribozymes without sequence specificity. The reaction time for the ligation reaction was reduced stepwise from 16 h for the first two rounds to 1 h for the 9th, 10th, and 11th rounds. In the 9th and 11th rounds, 0.08% and 0.1% of the molecules were ligated, respectively.
The ligated products from the 11th round pool were cloned, and 32 individual clones were sequenced. All clones were confirmed to have the designed ligation junctions (Materials and Methods, below) and belonged to one family (Fig. 2). A total of 87 point mutations were identified in the constant regions of 32 clones; 24 of those mutations (shown in italics in Fig. 2) were identical to those previously reported in the stable variants of the P4–P6 RNA (Juneau and Cech 1999; Juneau et al. 2001). None of the mutations in the constant region was essential for catalytic activity, although the restoration of all of the mutations of the clones reduced their ligation activity by ~30% (data not shown). Clone I with substrate I, which exhibited the highest activity in the pilot assay, was subjected to further assays. Ligation activity of clone I with substrate II was ~10% of substrate I (data not shown).
FIGURE 2.
Sequences of randomized region in selected clones. Mutations in constant regions are indicated at right; T27C and G45A (italics) were reported previously in the stable mutants of P4–P6 RNA (Juneau and Cech 1999).
Characterization of clone I
To see whether the ligation activity of the newly obtained ribozymes was dependent on the bent structure of the scaffold, two known mutations were introduced to disrupt the bent structure. The activity was diminished by J5/5a stem mutation (Fig. 3A, lane 5), which rearranges the overall structure to an extended form (Murphy et al. 1994), or by L5bGGAA mutation (Fig. 3A, lane 2), which disrupts the tertiary interaction that determines the bent structure (Young and Silverman 2002). A substitution of the original receptor with that for a GGAA-loop resulted in an active ribozyme for the L5bGGAA mutant (Fig. 3A, lane 4), indicating that the reaction depends on the correctly folded P4–P6 [Note: GAAA tetraloop is known to interact weakly with GGAA receptor (Fig. 3A, lane 3)].
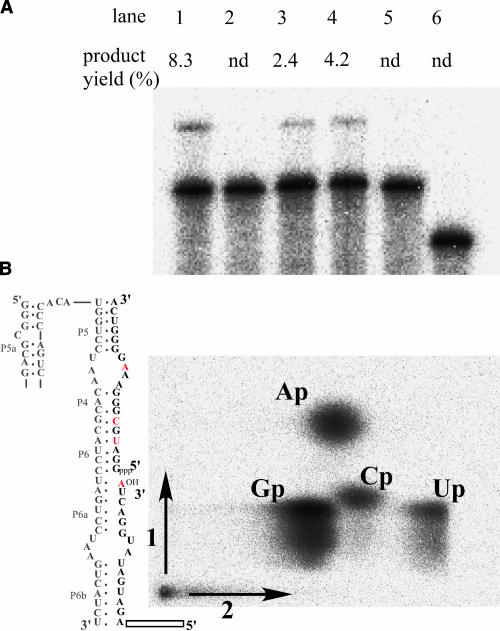
FIGURE 3.
Characterization of clone I. (A) Ligation reactions of clone I (lane 1), its derivative containing a modified L5b, which is substituted with GGAA (lane 2), a modified receptor for GGAA (GGAA-receptor) (lane 3), a GGAA at L5b and a GGAA-receptor (lane 4), J5/5a substituted with a stem structure (lane 5), and L5c with original wild-type sequence of UGCAA (lane 6). Conditions: 80 mM of MgCl2, 50 mM of KCl, 30 mM of Tris-HCl (pH 8.5), 0.5 μM of substrate RNA, and 50 nM of the ribozyme at 37°C for 1 h. (B) Analysis of the ligated phosphodiester bond. Guanylate residues in 3′-substrate was labeled with 32P. Residues containing 32P-labeled phosphate at the 3′ side are indicated in red. Substrate RNAs ligated with a modified clone I (gray) were digested with RNase T2. Resulting products were analyzed by two-dimensional denaturing gel electrophoresis.
Another mutation was introduced to see whether ligation activity actually depends on the selected sequence at L5c. Replacement of the L5c region, which consists of 30 nucleotides, with the original UGCAA pentaloop (L5cWT mutant, Fig. 3A, lane 6) completely eliminated the ligation activity, demonstrating that catalytic activity depends on the selected nucleotides at L5c.
The regiospecificity of the ligation reaction was determined with an RNase T2 digestion assay. Clone I RNA was engineered to operate as a true enzyme that joins two separate RNA substrates (Fig. 3B). The 3′ substrate RNA, which had 32P-labeled guanylate residues and was ligated to the 5′ substrate RNA by engineered clone I (represented in gray), was completely digested with RNase T2, which specifically cleaves 3′, 5′- linkages, and then analyzed by thin-layer choromatography. The ratio of 32P-labeled Ap:ApGp: Cp:Up is 2:0:1:1 if the ligation junction is composed of a 3′, 5′- linkage, whereas it is 1:1:1:1 if composed of a 2′, 5′-linkage (see residues indicated in red in Fig. 3B). The ratios of the spot intensity of Cp and Up to Ap were 0.49 and 0.42, respectively, demonstrating that the majority of the ligation junction consists of a 3′, 5′-linkage. The presence of a 3′, 5′-linkage was also confirmed by alkaline digestion (data not shown), which preferentially cleaves the 2′, 5′-phosphodiester linkage.
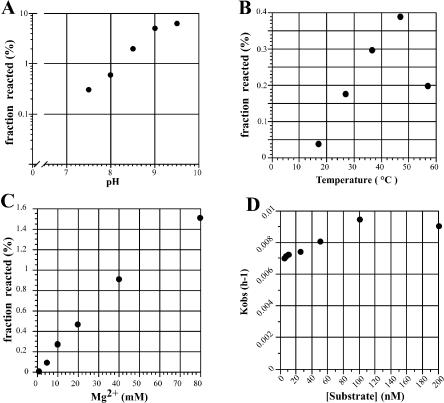
The ligation activity of clone I was examined under different conditions and enhanced with an increase of pH (up to pH 9, Fig. 4A) and magnesium ion concentration (up to 80 mM, Fig. 4C). The optimum temperature was 47°C under the conditions tested (Fig. 4B). The rate enhancement was about 104 for clone I compared with template-directed RNA ligation (Rohatgi et al. 1996). The observed rate constants were determined for clone I with various concentrations of substrate RNA (Fig. 4D). Relatively low concentrations of substrate RNA was enough for clone I to exhibit its full activity, presumably due to the long internal guide sequence. The observed rate constant and the final yield of the ligation reaction with clone I was 1.6 × 10−1 h−1 and 57% (data not shown), respectively, under optimal conditions (80 mM Mg2+, 50 mM K+, 30 mM Tris-Cl (pH 8.5) at 37°C).
FIGURE 4.
Ligation reaction of clone I under different conditions. Standard for the assay: 10 mM of MgCl2, 50 mM of KCl, 30 mM of Tris-HCl (pH 7.5), 1 μM of the substrate RNA, 0.5 μM of the ribozyme at 37°C for 1 h. Reacted fractions of substrate RNA were plotted in the range of pH 7.5–9.5 (A), 37–52°C (B), or 10–80 mM of Mg2+ (C). A plot of the observed rate constant of ligation reaction as a function of substrate concentration with 50 mM MgCl2, 50 mM KCl, and 30 mM Tris-HCl (pH 7.5) at 37°C (D).
Secondary structure of the selected sequence
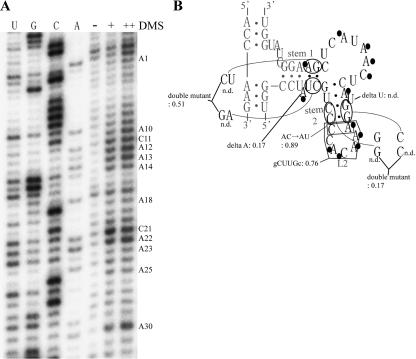
The secondary structure of clone I was predicted by using an Mfold program (Mathews et al. 1999; Zuker 2003; (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi). A dimethyl sulfate (DMS) modification experiment was performed to identify single-stranded regions and bulged-out bases in the clone, because DMS specifically modifies adenines and cytosines uninvolved in base-pairings (Fig. 5A). The predicted single-stranded regions and bulges, together with their proximal bases, were modified, indicating that the RNA actually possesses the predicted structure. This was further examined by attempting site-specific mutagenesis, because ligation activity was lost after disruption of the predicted stems, but restored by compensatory mutations (Fig. 5B). The role of L2 loop and two bulged-out nucleotides at stems 1 and 2 was assessed by mutagenesis experiments. A mutant RNA without a bulge in stem 1 retained the activity. In contrast, another mutant without a bulge in stem 2 exhibited no activity. Furthermore, a substitution of L2 loop (CCACAA) with a gCUUGc stem–loop did not eliminate the activity. These results demonstrate that L2 and a bulged-out A at stem 1 are nonessential for the activity, indicating that a catalytic site resides in the internal loop between stems 1 and 2, although it is still undeniable that a residue outside of the region is also involved with the reaction.
FIGURE 5.
Secondary structure analysis and site-directed mutations of the selected sequence in clone I RNA. (A) Autoradiogram of the DMS modification of clone I RNA. Modified bases are indicated at right. (B) Secondary structure model of clone I. Bases modified with DMS are indicated with black circles. The nucleotides derived from the randomized sequence or the constant region, are in black or gray, respectively. Relative activity is indicated for each mutant by using clone I as the standard (1.00). (n.d.) Nondetectable low activity. Conditions: 80 mM of MgCl2, 50 mM of KCl, 30 mM of Tris-HCl (pH 9.0), 0.5 μM of substrate RNA, and 50 nM of the ribozyme at 37°C for 1 h.
Evaluation of the RNA design
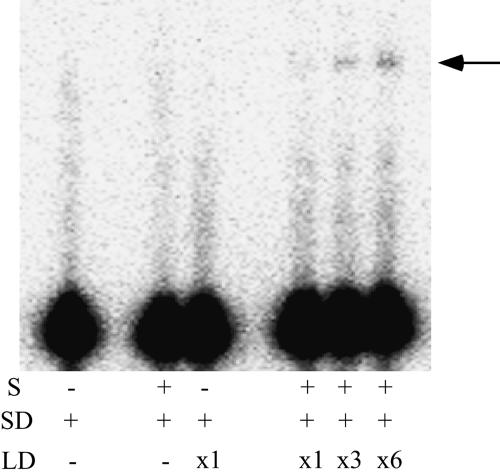
To determine whether the original modular organization is maintained in the selected ribozyme, clone I RNA was dissected physically for a reconstitution experiment (Fig. 1C) into two RNAs: substrate domain (SD RNA) and ligase domain (LD RNA). The SD RNA containing a terminal loop at the end of its P5 has an internal guide sequence for recognizing substrate RNA. The LD RNA, containing two inverted terminal base pairs for enhancing its transcription efficiency, possesses the selected catalytic core and A-rich bulge. Substrate RNA was ligated to the SD RNA in the presence of LD RNA (Fig. 6), demonstrating that the ligated product was produced by the reconstitution. The relatively weak activity of the modified ribozyme could be attributed to the weak affinity between SD and LD RNAs (Naito et al. 1998).
FIGURE 6.
Dissection and reconstruction of clone I RNA. (S) Substrate RNA. (SD and LD) Domains including IGS and random region, respectively (see Fig. 1C) (×1, ×3, ×6) The relative concentration of LD RNA (×1 corresponds to 1 μM). The bands corresponding to ligated products are indicated with an arrow.
DISCUSSION
Two strategies are available for selecting a ribozyme from a library of RNA in vitro. One is the selection from structurally unconstrained RNA containing a long random sequence, and the other is selection from a short random sequence for producing the catalytic site in the structurally constrained RNA. The number of RNA molecules in a library is practically restricted to ~1016. So, the rate of coverage for the sequence space is limited for those with a long random sequence, although their components of the selected ribozyme can be composed of any part of the RNA (Sabeti et al. 1997). In contrast, a sequence space consisting of a short random sequence can be fully or mostly covered, although its structural organization is tightly controlled by its scaffold. The latter strategy, which is suitable for in-depth exploration of a small sequence space (Hamm 1996; Davis and Szostak 2002), has rarely been studied for constructing ribozymes.
This study used the latter strategy to acquire a new class of RNA ligase ribozyme from the derivatives of self-folding P4–P6 RNA. The selection from a short random sequence for constructing a catalytic site produced a ligase ribozyme whose activity is dependent on the 3D structure of the P4–P6 RNA scaffold. Compared with the selections of ribozymes from a pool containing a long random sequence, this selection solely depends on the relative orientations of the reaction site and the selected catalytic site. In this aspect, a random sequence consisting of 30 nucleotides and its location on the P4–P6 RNA were probably reasonable choices, even though it is unclear whether they were close to the optimum.
In the reaction, the substrate oligonucleotides complementary to the internal guide sequences were ligated regio-specifically with the 3′–5′ linkage. The rate-limiting step is likely at the catalytic step, presumably due to deprotonation of the hydroxyl group at the 3′ terminal of the substrate. The rate enhancement of the selected ribozyme (104 rate enhancement) is comparable to those of previously selected ligase ribozymes without fine tuning (103–104 rate enhancement) (Bartel and Szostak 1993; Jaeger et al. 1999), although it was rather modest compared with those selected ribozymes modified by further fine tuning (106–109 rate enhancement). In many instances, rate enhancement of the selected ribozyme has been successfully achieved by additional selections for tuning the RNA conformation. For example, the rigidity of the scaffold of the ribozyme acquired in this study might have reduced the potential to fold into the most appropriate form for achieving its highest activity. Thus, further rate enhancement may be possible by performing the second selection and/or other modifications.
The elucidation of higher order structures is time consuming for the ribozymes selected from the libraries consisting of long random sequences. Even the prediction of their structural units is complicated, due to the dispersion of the nucleotides that comprise one structural unit in the primary sequences. However, the structural organization is obvious in our ligase RNA because of its modular organization. Because P4–P6 RNA and its derivatives have been successfully crystallized, the structural analysis of the selected ligase may also be possible by X-ray crystallography (Cate et al. 1996; Juneau et al. 2001).
MATERIALS AND METHODS
Construction of the RNA Library
A pool of 2.1 × 1014 different RNAs (15 copies each) was constructed, starting from the three fragments of synthetic DNA. These fragments were amplified separately by PCR (for the fragments constituting the constant regions) or primer extension (for the fragment constituting the randomized region), digested with BsaI and ligated to form DNA templates for the RNA pool. BsaI cuts outside of its recognition sequence, which enabled ligation to occur in a directional manner (Atsumi et al. 2003). 3.4 × 102 pmol of ligated DNAs were transcribed to produce 5.1 × 103 pmol of RNAs.
In vitro selection
After in vitro transcription with T7 RNA polymerase, the DNA templates were digested with RQ I DNase (Promega), and the resulting RNAs were purified by denaturing PAGE and then precipitated with ethanol. The purified RNAs and the substrate RNAs were dissolved in a buffer solution containing 1 mM Tris-Cl (pH 7.5) and 0.01 mM EDTA, denatured by incubating at 80°C for 3 min, and then preincubated at 37°C for 1 min. The mixture was subjected to folding initiation and ligation reaction by adding a fivefold concentrated reaction buffer at 37°C. The final concentrations of the pool RNA, the substrate RNA, MgCl2, KCl, and Tris-Cl (pH 7.5), were 0.5 μM, 1 μM, 10 mM, 50 mM, and 30 mM, respectively. Two substrate RNAs, 5′-(biotinU)GC UGAGCCUGCGAUUGGAGAUGAUAUGGACUA-3′ (Substrate I) and 5′-(biotinU)ACGUAACAGCACAGGAAUCGACACUUAU GGCUCG-3′ (Substrate II) have complementary sequences to the internal guide sequences as indicated by the underlining. The ligation reaction was stopped by ethanol precipitation. The products were captured on streptavidine paramagnetic particles (Pro-mega) for hybridizing with a DNA primer (5′-GGATGCGTGTTA GGACCATGT-3′) complementary to the 3′ region of the pool RNA, and reverse transcribed with RT High (TOYOBO). The resulting cDNAs were eluted from the particles by degrading the RNA with 150 mM KOH, followed by neutralization with 150 mM HCl. The cDNAs were selectively amplified by PCR using the primer for reverse transcription and selective primers complementary to the sequences of the substrate RNAs. Regenerating and type-switching PCR were carried out with the primer containing the sequence of the T7 RNA polymerase promoter and the primer having the sequence (5′-AACACTCTTAGGCTCGGGATGCGT GTTAGGACCATG-3′ or 5′-AGATGACTTAGGACTAGGATGC GTGTTAGGACCATG-3′) (The internal guide sequences are underlined). The resulting DNAs were used as templates for transcribing the pool of RNA for the next round.
Ligated RNAs from the 11th round were isolated by 6% polyacrylamide denaturing gel electrophoresis, reverse-transcribed, amplified by PCR, and then cloned into pUC119 at the HincII site. Individual clones were sequenced by using a BcaBEST Dideoxy Sequencing Kit (Takara) with an automated DNA sequencer (ALF express II, Amersham Biosciences).
Ligation assays
Uniformly, 32P-labeled RNAs were ligated to the substrate RNAs. In the reconstitution experiment, SD was 32P labeled. The standard conditions were as follows. The reaction time was 60 min, temperature was 37°C, MgCl2 concentration was 10 mM, KCl concentration was 50 mM, and Tris-HCl (pH 7.5) concentration was 30 mM. Concentrations of ribozyme and substrate RNA were identical to those in the in vitro selection, or 50 and 500 nM for the time-course assay (Fig. 4D). The ligation reaction was stopped by ethanol precipitation. Products were separated on 6% polyacrylamide denaturing gels and quantified with a BioImaging Analyzer (BAS2500; Fuji Film).
Characterization of ligation products
The identity of the nucleotides at the ligation junction was confirmed by sequencing individual clones and also by dideoxy sequencing of ligated clone I RNA with reverse transcriptase.
For the RNase T2 digestion assay, the 3′ substrate (5′-pppGGA UGCGGGAAAGGGGUCA-3′) was uniformly 32P-labeled by transcription with [α-32P]GTP, and ligated ribozymatically to substrate I. One pmol of the ligated substrate RNAs purified by denaturing PAGE was completely digested with 0.5 U of RNase T2 in a buffer containing 10 mM NaOAc (pH 4.5) and 2 mM EDTA for 2 h at 37°C. The products of RNase T2 digestion were separated by two-dimensional thin-layer chromatography, as described previously (Keith 1995).
Mutants
The template DNA for the transcription of a mutant having a UGCAA pentaloop at L5c in the structural context of clone I (mutations in the constant regions were restored) was constructed by PCR with appropriate primers and pTZIVSU (plasmid containing the whole sequence of the Tetrahymena ribozyme). The J5/5a stem mutant was constructed by PCR with appropriate primers and plasmid containing the sequence of clone I. The L5bGGAA mutants were constructed by appropriate primers and the plasmid containing clone I. The L5bUUCG mutant was constructed by PCR with appropriate primers and pTZIVSU. SD having a UUCG terminal loop at the end of P5 instead of J5/5a was constructed by PCR with three synthetic oligonucleotides. LD was constructed by PCR with plasmid containing the sequence of clone I and appropriate primers. The first two base pairs of LD were inverted for efficient transcription.
For the compensation assay of the loop-receptor of P5b × J6 (Fig. 3A, lanes 1–4), a plasmid containing the sequence of P5b GGAA mutant was constructed by PCR and ligation with the plasmid having the sequence of clone I, and confirmed to have the correct sequence. The receptor for the GGAA tetraloop was C7.34 (Costa and Michel 1997). The template for the transcription of a mutant having a 3′ sequence of C7.34 receptor (5′-CCCCACGC) instead of a 11-nt receptor was constructed by PCR with appropriate primers and the plasmid containing the clone I sequence. The sequence of substrate RNA having the 5′ sequence of C7.34 receptor was 5′-GCUGAGCCUGCGAUUGGAGAUGAGAAGGG ACUA-3′ (C7.34 sequence is underlined).
DMS modification
A reaction mixture (100 μL) containing 50 pmol of RNA, 30 mM of Tris-HCl (pH 7.5), and 0 or 50 mM of MgCl2 was incubated at 80°C for 3 min, cooled, and incubated at 37°C for 10 min. A total of 0.5 μL of 10% DMS (Sigma) in ethanol was added to the mixture and incubated at 37°C for 10 min. The reaction was stopped with 50 μL of 2-mercaptoethanol (1 M). After ethanol precipitation, the resulting RNA was reverse transcribed with 5′-prime-labeled primer, and then analyzed by performing PAGE.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7170304.
REFERENCES
- Atsumi, S., Ikawa, Y., Shiraishi, H., and Inoue, T. 2003. Selections for constituting new RNA-protein interactions in catalytic RNP. Nucleic Acids Res. 31: 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. and Szostak, J.W. 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.l., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. [DOI] [PubMed] [Google Scholar]
- Costa, M. and Michel, F. 1997. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J. 16: 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J.H. and Szostak, J.W. 2002. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl. Acad Sci. 99: 11616–11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, B.L., Gooding, A.R., Podell, E.R., and Cech, T.R. 1998. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science 282: 259–264. [DOI] [PubMed] [Google Scholar]
- Hamm, J. 1996. Characterization of antibody-binding RNAs selected from structurally constrained libraries. Nucleic Acids Res. 24: 2220–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa, Y., Yoshioka, W., Ohki, Y., Shiraishi, H., and Inoue, T. 2001. Self-splicing of the Tetrahymena group I ribozyme without conserved base-triples. Genes Cells 6: 411–420. [DOI] [PubMed] [Google Scholar]
- Ikawa, Y., Fukada, K., Watanabe, S., Shiraishi, H., and Inoue, T. 2002. Design, construction, and analysis of a novel class of self-folding RNA. Structure 10: 527–534. [DOI] [PubMed] [Google Scholar]
- Jaeger, L., Wright, M.C., and Joyce, G.F. 1999. A complex ligase ribozyme evolved in vitro from a group I ribozyme domain. Proc. Natl. Acad. Sci. 96: 14712–14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger, L., Westhof, E., and Leontis, N.B. 2001. TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 29: 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau, K. and Cech, T.R. 1999. In vitro selection of RNAs with increased tertiary structure stability. RNA 5: 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau, K., Podell, E., Harrington, D.J., and Cech, T.R. 2001. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure 9: 221–231. [DOI] [PubMed] [Google Scholar]
- Keith, G. 1995. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie 77: 142–144. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Murphy, F.L. and Cech, T.R. 1993. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry 32: 5291–5300. [DOI] [PubMed] [Google Scholar]
- Murphy, F.L., Wang, Y.H., Griffith, J.D., and Cech, T.R. 1994. Coaxially stacked RNA helices in the catalytic center of the Tetrahymena ribozyme. Science 265: 1709–1712. [DOI] [PubMed] [Google Scholar]
- Naito, Y., Shiraishi, H., and Inoue, T. 1998. P5abc of the Tetrahymena ribozyme consists of three functionally independent elements. RNA 4: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi, S.J., Ikawa, Y., Shiraishi, H., and Inoue, T. 2002. Modular engineering of a Group I intron ribozyme. Nucleic Acids Res. 30: 3473–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi, R., Bartel, D.P., and Szostak, J.W. 1996. Kinetic and mechanistic analysis of nonenzymatic, template-directed oligoribo-nucleotide ligation. J. Am. Chem. Soc. 118: 3332–3339. [DOI] [PubMed] [Google Scholar]
- Sabeti, P.C., Unrau, P.J., and Bartel, D.P. 1997. Accessing rare activities from random RNA sequences: The importance of the length of molecules in the starting pool. Chem. Biol. 4: 767–774. [DOI] [PubMed] [Google Scholar]
- Williams, K.P., Imahori, H., Fujimoto, D.N., and Inoue, T. 1994. Selection of novel forms of a functional domain within the Tetrahymena ribozyme. Nucleic Acids Res. 22: 2003–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, B.T. and Silverman, S.K. 2002. The GAAA tetraloop-receptor interaction contributes differentially to folding thermodynamics and kinetics for the P4-P6 RNA domain. Biochemistry 41: 12271–12276. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]