Abstract
Nonsense-mediated decay (NMD) is a eukaryotic regulatory process that degrades mRNAs with premature termination codons (PTCs). Although NMD is a translation-dependent process, there is evidence from mammalian systems that PTC recognition and mRNA degradation takes place in association with nuclei. Consistent with this notion, degradation of mammalian PTC-containing mRNAs occurs when they are bound by the cap binding complex (CBC) during a “pioneer” round of translation. Moreover, there are reports indicating that a PTC can trigger other nuclear events such as alternative splicing, abnormal 3′ end processing, and accumulation of pre-mRNA at transcription sites. To examine whether a PTC can elicit similar nuclear events in yeast, we used RNA export-defective mutants to sequester mRNAs within nuclei. The results indicate that nuclear PTC-containing yeast RNAs are NMD insensitive. We also observed by fluorescent in situ hybridization that there was no PTC effect on mRNA accumulated at the site of transcription. Finally, we show that yeast NMD occurs minimally if at all on CBC-bound transcripts, arguing against a CBC-mediated pioneer round of translation in yeast. The data taken together indicate that there are no direct consequences of a PTC within the yeast nucleus.
Keywords: NMD, nucleus, export, Rip1p, SSA4, yeast
INTRODUCTION
Nonsense-mediated decay (NMD) is a process that leads to the rapid degradation of mRNAs containing a premature termination codon (PTC). NMD exists in yeast, Caenorhabditis elegans, Drosophila, and mammals—perhaps all eukaryotes. Its function is probably to minimize the production of truncated or otherwise deleterious proteins, by reducing the concentration of incompletely or improperly processed RNAs. NMD is a translation-dependent event, which is not surprising given that NMD involves PTC recognition. As a consequence, protein synthesis inhibitors such as cycloheximide stabilize PTC-containing RNAs (Gonzalez et al. 2001). Furthermore, suppressor tRNAs that prevent stop codon recognition also reduce and in some cases eliminate NMD (Belgrader et al. 1993). NMD is therefore coupled to translation and occurs on functioning poly-ribosomes (Zhang et al. 1997)
In mammalian cells, there is also a fundamental connection between NMD and splicing, as a stop codon is recognized as premature only if there is a downstream splicing event. This is due to a requirement for the presence of an exon-junction protein complex (EJC) associated with this intron (for review, see Singh and Lykke-Andersen 2003). The EJC is deposited as a consequence of splicing 20–24 nucleotides (nt) upstream of the splice junction on mRNA and has been shown to contain factors that mediate NMD (Le Hir et al. 2000, 2001). As a consequence of this intron-EJC requirement, mammalian transcripts arising from intronless genes such as HSP70 and H4 do not undergo NMD (Maquat and Li 2001). The EJC includes the Upf3 protein, which then interacts with the perinuclear Upf2p (Kim et al. 2001; Gehring et al. 2003; Kadlec et al. 2004). If a translating ribosome encounters a PTC that is at least 50 nt upstream of an exon–exon junction, the bound Upf2p then recruits Upf1p and leads to the subsequent degradation of the transcript. Although all the Upf proteins are required for functional NMD in metazoan systems, the precise role of the individual Upf proteins in PTC recognition and subsequent RNA degradation has not been established (Culbertson and Leeds 2003).
Yeast NMD also requires Upf1p, Upf2p, and Upf3p but does not have a splicing requirement. A stop codon is thought to be recognized as premature in yeast when it precedes a downstream element, or DSE (for review, see Czaplinski et al. 1999). The DSE is a cis-element that recruits factors necessary for NMD, a role analogous to the EJC in mammalian cells. Removal of the DSE from the 3′ untranslated region (UTR) of PTC-containing mRNAs can in some cases prevent their degradation by NMD. However, there is also evidence that the distance between the polyA tail and the location of the stop codon may be a factor in premature stop codon recognition (Muhlrad and Parker 1999).
Given that bulk translation is cytoplasmic in all eukaryotic systems, an unexpected finding was that mammalian NMD occurs in association with nuclei (Cheng and Maquat 1993; Kessler and Chasin 1996; Wilkinson and Shyu 2002). In addition, other observations in mammalian systems have shown that a PTC can affect specific nuclear events, such as alternative splicing and 3 end formation (Brogna 1999; Wang et al. 2002). Muhlemann and colleagues (2001) further demonstrated by fluorescent in situ hybridization (FISH) that pre-mRNAs accumulate in larger transcription-site foci from integrated PTC-containing reporter genes, although this interpretation has been recently questioned as being due to NMD-independent events (Lytle and Steitz 2004). Taken together, the results indicate that a PTC can affect features of RNA metabolism, within or associated with mammalian nuclei.
Two models have been proposed to explain nuclear NMD. The first invokes a nuclear scanning model, which requires nuclear translation (Kessler and Chasin 1996). The second suggests that cytoplasmic translation begins during export of RNA from the nucleus to the cytoplasm. This model relies on the observations of Daneholt and colleagues, who visualized Chironimous balbiani ring mRNPs undergoing translation while still transiting the nuclear pore (Visa et al. 1996). These mRNAs might fractionate with nuclei, making it appear that NMD is a nuclear phenomenon (Ishigaki et al. 2001; McKendrick et al. 2001). Consistent with both models are recent experiments from Maquat and colleagues, showing that NMD occurs preferentially during a “pioneer” round of translation on cap binding complex (CBC)-bound RNAs (Ishigaki et al. 2001; Lejeune et al. 2002). In yeast, however, reports indicate that loss of PTC+ RNAs can occur at any stage of the translation cycle and does not necessarily occur only during the first initial round of translation (Maderazo et al. 2003; Keeling et al. 2004).
In this report, we investigate whether NMD can also occur within the yeast nucleus, a possibility that has not been rigorously investigated (Zhang et al. 1997). We utilized heat shock-inducible reporter genes and strains temperature sensitive for mRNA export to monitor mRNA during a time-course of transcriptional induction. We were therefore able to investigate the fate of newly synthesized transcripts without interference from a pre-existing equilibrium. Our results indicate that there is no detectable PTC effect on nuclear mRNA in yeast. Furthermore, we show that there are no noticeable differences in FISH signal between PTC+ and PTC− transcriptional foci. Finally, we observe no NMD on RNAs that are associated with Cbp20p, further arguing against a CBC-mediated pioneer round of translation in yeast. Our data indicate that there are little or no discernible effects of a PTC on yeast nuclear events, in contrast to higher eukaryotes.
RESULTS
NMD occurs during heat shock conditions
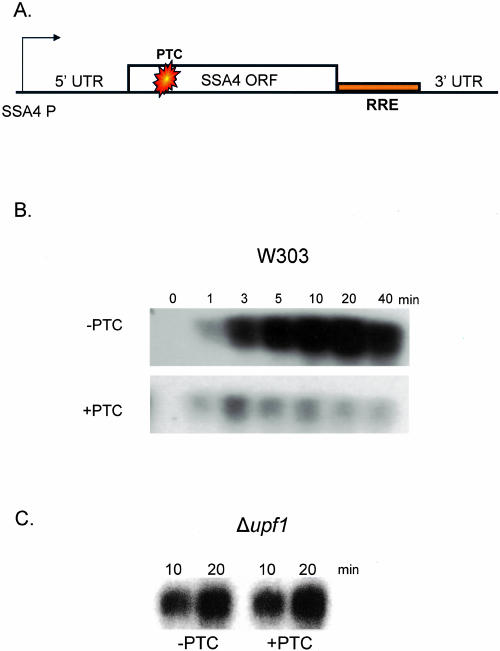
To directly assay for nuclear NMD in yeast, a PTC was introduced into the open reading frame (ORF) of a heat shock-inducible SSA4 gene (Fig. 1A). A Rev response element (RRE) was inserted between the stop codon and the 3′ UTR to distinguish the reporter RNA from endogenous SSA4 RNA (Fig. 1A). This construct allowed a rapid transcriptional induction while shifting simultaneously to the nonpermissive temperature in temperature-sensitive strains. It further allowed a monitoring of early time-points with minimal secondary effects from a prolonged heat shock.
FIGURE 1.
Analysis of the SSA4 construct. (A) Schematic of the SSA4 construct. Endogenous elements were cloned into a CEN-based plasmid. A Rev response element (RRE) was introduced to distinguish reporter from endogenous RNA. On the PTC+ construct, a PTC was introduced at position 117. (B) Wild-type cells (W303) were transformed with the two SSA4 constructs and heat shocked at 42°C, and total RNA was isolated at the indicated times. RNA was analyzed by Northern blotting using a probe against the RRE element. (C) Δupf1 cells were transformed with the two constructs and heat shocked for the indicated times. Total RNA was extracted and analyzed by Northern blot using labeled RRE.
By Northern blotting, reporter RNA was undetectable before induction (Fig. 1B), and no signal was observed with the RRE probe in strains lacking the reporter (data not shown). RNA was detected as early as 1 min after induction, with approximately equal RNA levels from the wild-type (PTC−) and PTC-containing (PTC+) reporter genes at the earliest times after the temperature shift. The PTC- reporter generated increasing levels of RNA, which were maximal after 20 min. Consistent with NMD, the PTC+ reporter RNA failed to increase between 5 and 40 min (Fig. 1B). As a control for NMD, Δupf1 yeast were examined; PTC+ containing RNA levels were restored (Fig. 1C). The lack of a PTC effect at the earliest times of induction may be due to an mRNA half-life that is too long, even with a PTC. Alternatively, these earliest transcripts may still be largely confined to the nucleus and/or not engaged in translation and therefore not yet NMD substrates.
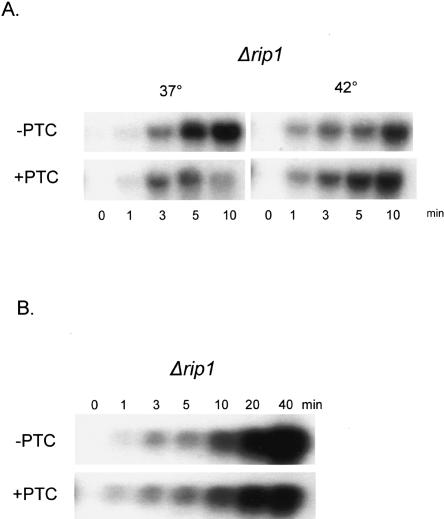
To examine directly the effect of restricting RNAs to the nucleus, we performed the same assay in a Δrip1 strain. Rip1p is a component of the nuclear pore complex, and Δrip1 cells have a tight and rapid RNA export block at 42°C (Saavedra et al. 1997). At lower temperatures such as 37°C, Δrip1 cells are permissive for export. We therefore assayed for an NMD effect at 37°C and 42°C; both temperatures induce transcription of the heat shock promoter (Fig. 2A). At the permissive temperature of 37°C, the induction time-course in the Δrip1 strain is similar to that observed for the wild-type strain, with a dramatic difference between the PTC− and PTC+ RNAs visible by 5 min after induction (Fig. 2A, left). At the nonpermissive temperature of 42°C, however, the PTC+ RNA is stabilized so that its levels resemble those of the PTC− RNA (Fig. 2A, right). The results indicate that a block to RNA export inhibits NMD. This is also true at longer times of induction (Fig. 2B), although a modest NMD effect may now be apparent (at 20 and 40 min; Fig. 2B). We interpret this to indicate some nuclear export of reporter RNA at these later times (see Discussion).
FIGURE 2.
Analysis of the SSA4 constructs in Δrip1 cells. (A) Δrip1 cells containing the SSA4 constructs were heat shocked at the permissive temperature (37°C) or the nonpermissive temperature (42°C) and total RNA was extracted at the indicated times. RNA was analyzed by Northern blot using labeled RRE. (B) Δrip1 cells were heat shocked at 42°C for the indicated times and total RNA was extracted and analyzed as in part A.
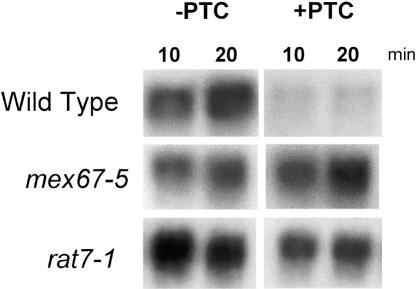
To examine more generally the effect of subcellular compartmentalization, we assayed two additional temperature-sensitive mRNA export strains, mex67-5 and rat7-1 (Gorsch et al. 1995). Mex67p is the major mRNA export receptor (Tap in mammals), and Rat7p is a nuclear pore protein (Del Priore et al. 1996; Segref et al. 1997). Both the mex67-5 and rat7-1 strains also block NMD, similar to what was observed in the Δrip1 strain (Fig. 3B). We conclude that under conditions nonpermissive for export, PTC+ RNA is stabilized to levels comparable to those in the Δupf1 strain, which is NMD deficient.
FIGURE 3.
Analysis of SSA4 constructs during general export block. Mutant yeast were transformed with the SSA4 constructs and heat shocked at 42°C for the indicated times. Total RNA was extracted and analyzed by Northern blot using labeled RRE.
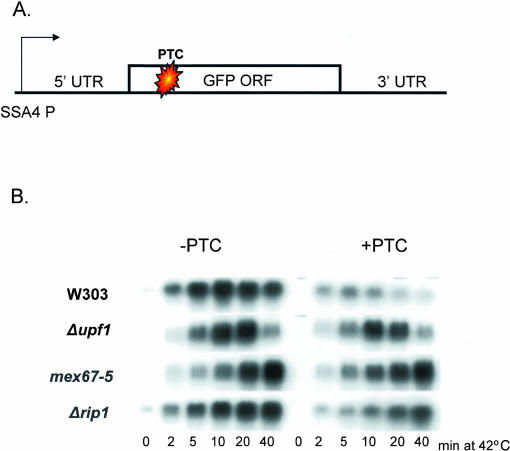
To extend the result to another reporter gene, we generated heat shock-inducible GFP constructs with and without a PTC in the open reading frame (Fig. 4A). In a wild-type strain, an NMD effect is evident by 2 min after induction (W303; cf. left and right); the effect is attenuated in a Δupf1 background. The export-deficient mex67-5 and Δrip1 strains also show an attenuation of the NMD effect, consistent with the results obtained with the SSA4 reporter (Figs. 2, 3). The results indicate that NMD is largely if not exclusively a cytoplasmic phenomenon in yeast.
FIGURE 4.
Analysis of heat shock-inducible GFP constructs. (A) Schematic of the SSA4-GFP construct. GFP was placed under control of the SSA4 promoter and contains the SSA4 3′ UTR. The PTC+ reporter contains a PTC at codon 6. (B) Wild-type and mutant yeast were transformed with the GFP constructs and heat shocked for the indicated times. Total RNA was extracted and analyzed by Northern blot analysis using a labeled GFP probe.
Visualization of RNA localization by FISH
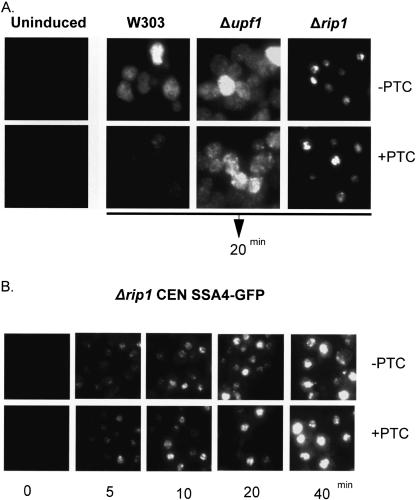
To address this issue with an independent assay, we utilized FISH with labeled oligonucleotide probes that span the GFP open reading frame (Fig. 5). Prior to heat shock, no detectable FISH signal is observed. After a 20-min heat shock at 42°C, the time at which maximal expression of the GFP reporters is observed by Northern blot, there is abundant cytoplasmic signal of PTC− RNA with a speckled appearance in the wild-type and Δupf1 strains. The PTC+ reporter shows a marked reduction in cytoplasmic signal in the wild-type strain, and this signal is substantially increased in the Δupf1 strain. In contrast to the wild-type strain, the FISH signal in the Δrip1 strain was localized to the nucleus with essentially no difference in signal intensity between the PTC− and PTC+ reporters (Fig. 5A). The similarity between the two reporters persisted over the time-course of induction (Fig. 5B). The granular appearance of the nuclear accumulation may reflect the multiple sites of transcription present with this plasmid-based reporter gene system.
FIGURE 5.
FISH analysis of heat-shock-inducible GFP reporters. (A) Wild-type (W303) and mutant yeast transformed with reporter plasmids were subjected to heat shock for 20 min. Cells were immediately fixed and analyzed by FISH. (B) Δrip1 cells containing the GFP reporter plasmids were heat shocked for the indicated times. Cells were immediately fixed and analyzed by FISH.
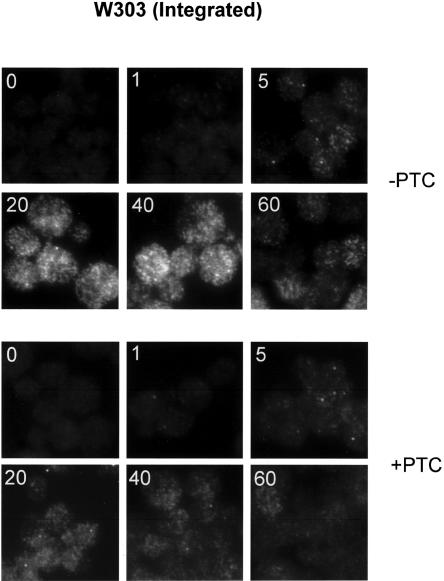
To achieve better resolution of the nuclear signal, a single copy of the heat-shock-inducible PTC− and PTC+ GFP constructs was integrated at the URA3 locus in the wild-type (W303) and Δrip1 strains. In the wild-type strain, induction of the PTC-reporter resulted in a single nuclear focus visible as early as 1 min after heat shock, presumably the site of transcription (Fig. 6, top). By 5 min after induction, the focus was brighter, and cytoplasmic signal was detectable. The cytoplasmic signal was maximal after 20 min, with a reduction in intensity evident by 60 min. A parallel reduction in the intensity of the nuclear focus took place, suggesting a decrease in transcriptional activity. A comparable nuclear focus of RNA is present with the PTC+ reporter. Although some cytoplasmic signal is evident by 20 min, this signal is clearly weaker than that observed for the PTC+ reporter. In the wild-type strain, therefore, the PTC− and PTC+ integrated reporter genes are distinguishable only in that appreciable cytoplasmic signal is seen with the PTC− RNA.
FIGURE 6.
FISH examination of integrated GFP reporters in wild-type cells. Wild-type cells containing a single copy of the heat-shock-inducible GFP reporters were heat shocked for the indicated times (inset), fixed, and analyzed by FISH.
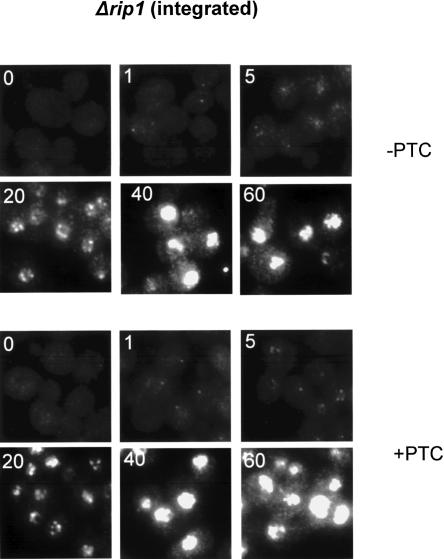
In contrast, these two reporter genes were completely indistinguishable in the Δrip1 strain; most if not all of the signal was nuclear (Fig. 7; PTC−, top; PTC+, bottom). As in the wild-type background, a single nuclear focus is detectable at 1 min of induction in the Δrip1 strain. At 5 min, however, the nuclear focus is less distinct with increasing punctate signal throughout the nucleus of the Δrip1 strain. The nuclear signal is even more intense by 20 min, with a similar punctate accumulation pattern. At later times, the signal is too intense to discern any details. The lack of a difference between the two integrated reporter genes in the Δrip1 strain recapitulates the results from the plasmid-based reporter genes (Fig. 5B). Taken together with our biochemical results, these experiments demonstrate that there are no detectable PTC effects, i.e., no NMD within yeast nuclei.
FIGURE 7.
FISH examination of integrated GFP reporters in Δrip1 cells. Conditions were the same as in Figure 6.
CBC-bound mRNAs are not subject to NMD
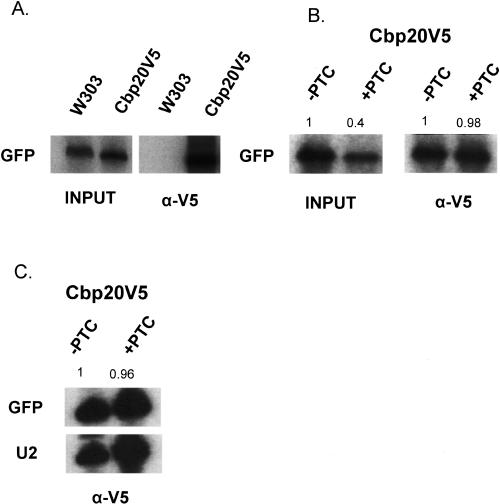
In higher eukaryotes, the PTC+ mediated reduction of RNA levels is restricted to CBC-bound RNAs, with no further reduction observed in the eIF4E-bound population (Cheng and Maquat 1993; Ishigaki et al. 2001) To examine whether these results extend to yeast, we generated a strain in which the endogenous CBP20 gene is C-terminally tagged with a V5 epitope. PTC− and PTC+ GFP reporter gene constructs were transformed into the Cbp20p-V5 strain and a control wild-type strain. In these constructs, transcription is driven by the constitutive TDH3 promoter. Immunoprecipitations from extracts were performed with an α-V5 antibody, followed by RNA extraction and primer extension analysis (IP-extension; Fig. 8).
FIGURE 8.
Analysis of GFP reporter RNAs associated with Cbp20p. (A) Wild-type (W303) yeast and yeast expressing Cbp20p-V5 were transformed with reporter plasmids expressing GFP under the constitutive TDH3 promoter. α-V5 antibodies were used to immunoprecipitate Cbp20-V5 and RNA was analyzed by primer extension (IP-primer extension). (B) Cbp20pV5-tagged yeast were transformed with plasmids containing either PTC− or PTC+ TDH3-GFP reporters and subjected to IP-primer extension. Primer extensions of total RNA (input) are shown on the left. Input RNAs are from one-hundredth of the extract used in immunoprecipitation. The numbers are the quantitation results, normalized to the GFP PTC− signal. (C) U2 snRNA was used as an internal control to quantitate the amounts of immunoprecipitated PTC− and PTC+ GFP RNAs. Numbers represent the ratio of GFP PTC+ RNA to GFP PTC− RNA, normalized to the amount of U2 snRNA.
The inputs show comparable levels of GFP RNA in the wild-type and Cbp20p-tagged strains. After immunoprecipitation with α-V5 antibody, however, only the V5-tagged extracts yielded an RNA signal (Fig. 8A). There is a clear PTC-effect when input (total) RNA is examined, with lower levels of RNA in the strain containing the PTC+ reporter gene (98% vs. 40%). However, comparable levels of PTC− and PTC+ RNAs were associated with Cbp20p (Fig. 8B). In a separate experiment, a primer complementary to U2 sn-RNA was included in the IP-extension. U2 snRNA is associated with Cbp20p, presumably as this RNA has a mono-methylated intermediate and serves as an internal control. Comparable levels of PTC− and PTC+ RNAs were associated with Cbp20p relative to U2 snRNA, despite the lower levels of PTC− total RNA (Fig. 8C). In contrast with results from higher eukaryotes, therefore, there is little or no detectable NMD in the Cbp20p-bound RNA fraction in yeast.
DISCUSSION
Although yeast NMD occurs in the cytoplasm, the possibility of nuclear NMD in yeast has never been directly examined (Maderazo et al. 2003). Using heat shock-inducible reporter genes and strains temperature sensitive for mRNA export, we show that restricting PTC-containing RNAs to the nucleus inhibits NMD. The use of a heat shock promoter allowed a rapid induction of transcription and a concomitant block to nuclear export. In this manner, we could monitor de novo transcription without interference from any RNA present prior to the heat shock and with minimal secondary effects associated with prolonged heat shock and nuclear retention. Our data indicate that there is no discernible nuclear NMD in yeast, based on biochemical as well as FISH assays.
Our results also indicate that NMD is fully functional during heat shock. It has previously been shown that mammalian NMD does not occur during heat shock on an HSP70 gene. However, the addition of an intron to this otherwise intronless gene restores NMD, indicating that the lack of an EJC rather than heat shock per se prevents NMD in mammalian cells (Maquat and Li 2001). In contrast, NMD can occur on intronless transcripts in Drosophila as well as in yeast, suggesting alternate PTC-detection mechanisms in these organisms. This may also be reflected in the fact that Drosophila NMD begins with an endonucleolytic cleavage near the PTC (Gatfield et al. 2003; Gatfield and Izaurralde 2004).
Core EJC proteins are present in mammals and flies (including eIF4IIIa, Y14, and Mago), which recruit other factors involved in RNA metabolism. These include translation factors, NMD factors, and export factors (Le Hir et al. 2001; Singh and Lykke-Andersen 2003; Nott et al. 2004; Shibuya et al. 2004). Although no evidence exists that yeast contain an EJC, some EJC proteins are conserved in yeast, such as Upf3p, Yra1p (Aly/Ref), and Sub2p (UAP56). The recruitment mechanism of these proteins to yeast mRNA is uncertain, but there is good evidence showing that at least Yra1p and Sub2p are recruited to nascent RNAs cotranscriptionally (Lei et al. 2001; Zenklusen et al. 2002; Abruzzi et al. 2004). These factors apparently mediate some of the same functions in yeast, i.e., export and NMD, but no studies have shown whether they enhance translation as in mammals (Nott et al. 2004). Given the observations that both Drosophila and yeast have no splicing requirement for functional NMD and that there is no visible nuclear effect of a PTC in yeast, NMD might also be restricted to the cytoplasm of Drosophila (Gatfield et al. 2003). On the other hand, Drosophila spliced mRNAs probably contain an EJC (Palacios et al. 2004), suggesting that nuclear NMD may occur on these mRNAs as in mammalian systems.
A comparison of the kinetics of RNA accumulation of a PTC− reporter between W303 (wild-type) and Δrip1 cells indicates that there is considerably more RNA in W303 cells (Figs.1B, 2B). This suggests that RNAs retained in the nucleus are less stable than those exported to the cytoplasm. This is consistent with observations by Das and colleagues (2003), who showed that RNAs retained in the nucleus have a higher turnover rate. Furthermore, there seems to be a modest PTC effect visible at 40 min in Δrip1 cells. We interpret this to indicate some export of RNA during the prolonged heat shock. This would then result in an accelerated destabilization of those PTC-containing transcripts that manage to access the cytoplasm.
Interestingly, the heat shock response appears to diminish over time, with decreased cytoplasmic signal and the loss of a visible transcription-site focus (e.g., Fig. 6). However, there is a marked reduction in PTC− RNA levels after 40 min of induction in the Δupf1 strain compared with the wild-type strain (Fig. 4B). This suggests that Upf1p increases cytoplasmic RNA stability during heat shock. Whether this is a heat shock-specific phenomenon remains to be determined.
Muhlemann and colleagues (2001) examined stable cell lines containing reporter constructs from the TCR-β mini gene and used FISH to examine transcription-site foci in mammalian cells. They showed that the size of the PTC+ focus was larger than that of the PTC- focus and suggested that this effect is due to PTC+ pre-mRNA accumulation at the site of transcription. However, we see no difference in focus size by FISH when examining single integrants of PTC+ and PTC− reporter genes in yeast, consistent with a PTC-independent explanation for this phenomenon (Lytle and Steitz 2004). Since there is also no observable PTC effect on nuclear-retained mRNAs by biochemical criteria, we conclude that the transcription-site foci of PTC− and PTC+ reporters are comparable. This is despite the fact that the PTC− foci were sometimes obscured by cytoplasmic signal.
It was unexpected that transcription-site focus intensity was unaffected not only by a PTC but also by a block to RNA export. This is in contrast to previous results from this and other laboratories in which foci of endogenous heat shock RNAs were visible only under conditions nonpermissive for export (Jensen et al. 2001; Thomsen et al. 2003). Furthermore, we observed that nuclear RNA accumulation from the integrated heat shock reporter genes was not restricted to a single focus in the Δrip1 strain. These discrepancies may be due to differences in chromosomal position, as the integrated reporters in this study were inserted into the URA3 locus (T.H. Jensen and K. Dower, unpubl.). Alternatively, the nature of the transcribed RNA sequences (GFP vs. endogenous heat shock sequences) may influence the fate of the nuclear RNA.
Maquat and colleagues have shown that mammalian NMD occurs preferentially during a “pioneer” round of translation on CBC-bound RNAs (Ishigaki et al. 2001; Lejeune et al. 2002). CBC-mediated translation presumably occurs within eukaryotic nuclei or in association with the nuclear membrane during or shortly after transit into the cytoplasm (Visa et al. 1996). CBC is a heterodimer of Cbp20p and Cbp80p in both yeast and mammals, and Cbp20p is the subunit that physically contacts the mRNA 5′ 7-methylguanosine cap structure. Prior to bulk translation in the cytoplasm, CBC is displaced by the translational initiation factor eIF4E (for review, see Gingras et al. 1999). Consistent with the translation of yeast mRNAs prior to Cbp20p displacement, yeast CBC can physically interact with the translational initiation factor eIF4G, suggesting that the small ribosomal subunit can be recruited to mRNA independent of eIF4E (Fortes et al. 2000). However, our experiments suggest that there is no evidence for nuclear NMD or for CBC-associated translation. This is consistent with a more recent interpretation of the CBC-eIF4G genetic interaction (Baron-Benhamou et al. 2003). The RNA that is associated with Cbp20p may therefore reflect bulk nuclear mRNA, prior to ribosome association. A prediction of our IP experiments is that the PTC-mediated decrease in RNA would be reflected in the eIF4E-bound population. Unfortunately, we have been unable to efficiently immunoprecipitate tagged eIF4E and confirm this prediction (data not shown).
In summary, we have shown that there are no observable PTC effects on yeast nuclear events. We interpret the data to indicate that there is no nuclear NMD, no nuclear translation, no translation during nuclear export, and no translation on CBC-bound mRNAs, i.e., no special pioneer round of translation in yeast.
MATERIALS AND METHODS
Plasmid construction
The wild-type SSA4 construct was generated by ligating together the following PCR fragments into pRS316: (1) the SSA4 heat shock promoter, 5′ UTR, and ORF; (2) an RRE; and (3) the SSA4 3′ UTR. The QuickChange Site-Directed Mutagenesis Kit (Stratagene) was used to generate a PTC at position 117 according to the manufacturer’s instructions. Constructions were confirmed by DNA sequence analysis.
The SSA4-GFP construct was created by cloning the SSA4 promoter and the 5′ and 3′ UTRs into pRS416. A PCR product of the GFP ORF was then inserted between the SSA4 5′ and 3′ UTRs. PTC-containing GFP was created by PCR amplification using a forward primer containing a PTC at codon 6. Constructions were confirmed by DNA sequencing.
Northern blot analysis
Cultures were grown at 30°C to an OD600 between 0.5 and 0.8 and, where necessary, an equal volume of super heated media was added to bring the cultures to 37°C or 42°C. At the indicated times, a 25-mL aliquot was immediately chilled on ice and cells were harvested by centrifugation for 5 min at 1200g. Total RNA was extracted using hot acid phenol extraction (Ausubel et al. 1994). Ten micrograms of total RNA was then separated on a formaldehyde agarose gel and analyzed by Northern blotting using radioactively labeled RRE or GFP probes generated by random primed labeling of PCR fragments (Stratagene). Methylene blue staining of membranes was performed to monitor for equal loading of RNA.
IP-extension
IP-extensions were performed according to the procedure from Vilardell et al. (2000) with some modifications. Briefly, 100-mL cultures were harvested and washed in lysis buffer (10 mM HEPES-KOH, pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, and 1 mM PMSF). Cells were resuspended in 1 mL of lysis buffer and lysed by bead beating for four 1-min pulses. After lysis, the extract was centrifuged for 5 min at maximum speed on a standard microfuge. Two hundred microliters of extract and 1.1 mL of IP buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 0.5 mM DTT, and 0.05% Nonidet NP40) were added to prepared V5-coupled agarose beads (Sigma) and incubated for 1 h at 4°C. Beads were washed four times for 10 min in IP buffer at 4°C and RNA was isolated by phenol/chloroform extraction and ethanol precipitation. Precipitated RNA was then subjected to IP-extension using GFP- and U2-specific oligonucleotides as described previously (Dower and Rosbash 2002). Beads were prepared by washing in IP buffer containing 100 ng/μL of BSA and 100 ng/μL yeast tRNA, followed by washing in IP buffer with 100 ng/μL of yeast tRNA.
Fluorescent in situ hybridization
FISH was performed using four Cy3-conjugated GFP oligonucleotides as described previously with some modifications (Vainberg et al. 2000). Cells were fixed for 10 min in 4% paraformaldehyde (EMS) and 10% acetic acid and washed twice in Sorb/KPi (1.2 M sorbitol, 0.1 M potassium phosphate, pH 6.5). Cells were allowed to adhere onto multiwell slides and were spheroplasted using oxalyticase (Enzyogenetics). Cells were permeablized with 70% ethanol for 20 min at −20°C. Wells were allowed to air dry and cells were then hydrated with 2 × SSC/50% formamide. Thirty nanograms of each probe per well was hybridized overnight in 2 × SSC/50% formamide/10% dextran sulfate, 1 μg/uL tRNA, 1 μg/uL sperm DNA, and 0.2% BSA. Cells were washed 2× in 2 × SSC/0.1% SDS for 15 min at 37°C, 2× in 1 × SSC for 15 min at room temperature, and 2× in 1× PBS. Slides were visualized using an Olympus AX70 with a 100× objective using a rhodamine filter set.
Acknowledgments
We thank our colleagues in the Rosbash and Moore laboratories for helpful suggestions and comments on the manuscript. This work was supported in part by NIH grant GM23549.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7132504.
REFERENCES
- Abruzzi, K.C., Lacadie, S., and Rosbash, M. 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 23: 2620–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 1994. Current protocols in molecular biology. John Wiley & Sons, New York.
- Baron-Benhamou, J., Fortes, P., Inada, T., Preiss, T., and Hentze, M.W. 2003. The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast. RNA 9: 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader, P., Cheng, J., and Maquat, L.E. 1993. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc. Natl. Acad. Sci. 90: 482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna, S. 1999. Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA 5: 562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J. and Maquat, L.E. 1993. Nonsense condons can reduce the abundance of nuclear messenger RNA without affecting the abundance of premessenger RNA or the half-life of cytoplasmic messenger RNA. Mol. Cell. Biol. 13: 1892–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson, M.R. and Leeds, P.F. 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13: 207–214. [DOI] [PubMed] [Google Scholar]
- Czaplinski, K., RuisEchevarria, M.J., Gonzalez, C.I., and Peltz, S.M. 1999. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays 21: 685–696. [DOI] [PubMed] [Google Scholar]
- Das, B., Butler, J.S., and Sherman, F. 2003. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 5502–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Priore, V., Snay, C.A., Bahr, A., and Cole, C.N. 1996. The product of the Saccharomyces cerevisiae RSS1 gene, identified as a high-copy suppressor of the rat7–1 temperature-sensitive allele of the RAT7/NUP159 nucleoporin, is required for efficient mRNA export. Mol. Biol. Cell 7: 1601–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower, K. and Rosbash, M. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8: 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes, P., Inada, T., Preiss, T., Hentze, M.W., Mattaj, I.W., and Sachs, A.B. 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6: 191–196. [PubMed] [Google Scholar]
- Gatfield, D. and Izaurralde, E. 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578. [DOI] [PubMed] [Google Scholar]
- Gatfield, D., Unterholzner, L., Ciccarelli, F.D., Bork, P., and Izaurralde, E. 2003. Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO J. 22: 3960–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, N.H., Neu-Yilik, G., Schell, T., Hentze, M.W., and Kulozik, A.E. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11: 939–949. [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C.I., Bhattacharya, A., Wang, W., and Peltz, S.W. 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274: 15–25. [DOI] [PubMed] [Google Scholar]
- Gorsch, L.C., Dockendorff, T.C., and Cole, C.N. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129: 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., and Maquat, L.E. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Patricio, K., McCarthy, T., and Rosbash, M. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7: 887–898. [DOI] [PubMed] [Google Scholar]
- Kadlec, J., Izaurralde, E., and Cusack, S. 2004. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat. Struct. Mol. Biol. 11: 330–337. [DOI] [PubMed] [Google Scholar]
- Keeling, K.M., Lanier, J., Du, M., Salas-Marco, J., Gao, L., Kaenjak-Angeletti, A., and Bedwell, D.M. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, O. and Chasin, L.A. 1996. Effects of nonsense mutations on nuclear and cytoplasmic adenine phosphoribosyltransferase RNA. Mol. Cell. Biol. 16: 4426–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, V.N., Kataoka, N., and Dreyfuss, G. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293: 1832–1836. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, E.P., Krebber, H., and Silver, P.A. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 15: 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, F., Ishigaki, Y., Li, X., and Maquat, L.E. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 21: 3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle, J.R. and Steitz, J.A. 2004. Premature termination codons do not affect the rate of splicing of neighboring introns. RNA 10: 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderazo, A.B., Belk, J.P., He, F., and Jacobson, A. 2003. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol. Cell. Biol. 23: 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. and Li, X. 2001. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA 7: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKendrick, L., Thompson, E., Ferreira, J., Morley, S.J., and Lewis, J.D. 2001. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m7 guanosine cap. Mol. Cell. Biol. 21: 3632–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann, O., Mock-Casagrande, C.S., Wang, J., Li, S., Custodio, N., Carmo-Fonseca, M., Wilkinson, M.F., and Moore, M.J. 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell 8: 33–43. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D. and Parker, R. 1999. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5: 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott, A., Le Hir, H., and Moore, M.J. 2004. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes & Dev. 18: 210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, I.M., Gatfield, D., St Johnston, D., and Izaurralde, E. 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757. [DOI] [PubMed] [Google Scholar]
- Saavedra, C.A., Hammell, C.M., Heath, C.V., and Cole, C.N. 1997. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes & Dev. 11: 2845–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref, A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Luhrmann, R., and Hurt, E. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16: 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya, T., Tange, T.O., Sonenberg, N., and Moore, M.J. 2004. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 11: 346–351. [DOI] [PubMed] [Google Scholar]
- Singh, G. and Lykke-Andersen, J. 2003. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 28: 464–466. [DOI] [PubMed] [Google Scholar]
- Thomsen, R., Libri, D., Boulay, J., Rosbash, M., and Jensen, T.H. 2003. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9: 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg, I.E., Dower, K., and Rosbash, M. 2000. Nuclear export of heat shock and non-heat-shock mRNA occurs via similar pathways. Mol. Cell. Biol. 20: 3996–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell, J., Chartrand, P., Singer, R.H., and Warner, J.R. 2000. The odyssey of a regulated transcript. RNA 6: 1773–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa, N., Izaurralde, E., Ferreira, J., Daneholt, B., and Mattaj, I.W. 1996. A nuclear cap-binding complex binds balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 133: 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Chang, Y.F., Hamilton, J.I., and Wilkinson, M.F. 2002. Nonsense-associated altered splicing: A frame-dependent response distinct from nonsense-mediated decay. Mol. Cell 10: 951–957. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.F. and Shyu, A.B. 2002. RNA surveillance by nuclear scanning? Nat. Cell Biol. 4: E144–147. [DOI] [PubMed] [Google Scholar]
- Zenklusen, D., Vinciguerra, P., Wyss, J.C., and Stutz, F. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22: 8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Welch, E.M., Hogan, K., Brown, A.H., Peltz, S.W., and Jacobson, A. 1997. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA 3: 234–244. [PMC free article] [PubMed] [Google Scholar]