Abstract
U2 snRNA, a key player in nuclear pre-mRNA splicing, contains a 5′-terminal m3G cap and many internal modifications. The latter were shown in vertebrates to be generally required for U2 function in splicing, but precisely which residues are essential and their role in snRNP and/or spliceosome assembly is presently not clear. Here, we investigated the roles of individual modified nucleotides of HeLa U2 snRNA in pre-mRNA splicing, using a two-step in vitro reconstitution/complementation assay. We show that the three pseudouridines and five 2′O-methyl groups within the first 20 nucleotides of U2 snRNA, but not the m3G cap, are required for efficient pre-mRNA splicing. Individual pseudouridines were not essential, but had cumulative effects on U2 function. In contrast, four of five 2′O-methylations (at positions 1, 2, 12, and 19) were individually required for splicing. The in vitro assembly of 17S U2 snRNPs was not dependent on the presence of modified U2 residues. However, individual internal modifications were required for the formation of the ATP-independent early spliceosomal E complex. Our data strongly suggest that modifications within the first 20 nucleotides of U2 play an important role in facilitating the interaction of U2 with U1 snRNP and/or other factors within the E complex.
Keywords: mRNA splicing, modified nucleotides, pseudouridine, ribose methylation, E complex
INTRODUCTION
The removal of introns from pre-mRNA is achieved by a highly dynamic ribonucleoprotein complex termed the spliceosome. It is formed by the ordered association of the U1, U2, U4/U6, and U5 snRNPs (small nuclear ribonucleoproteins) and non-snRNP proteins with conserved regions of the pre-mRNA at the 5′ and 3′ splice sites and the so-called branch site (for review, see Reed and Palandijan 1997; Burge et al. 1999). The first step of spliceosome assembly includes the ATP-independent recognition of the 5′ splice site by U1 snRNP, forming the E (early) complex. In an ATP-requiring step, U2 snRNA then base-pairs with the branchsite, generating the pre-spliceosome or A complex. Subsequently, the U4/U6.U5 tri-snRNP binds, giving rise to the B complex. After major RNA–RNA rearrangements, the catalytically activated spliceosome is formed. During these rearrangements, the U4 and U6 snRNAs dissociate and the 5′ end of U2 enters into base-pairing interactions with U6, forming part of the spliceosome’s catalytic core. The activated spliceosome catalyses the first transesterification step of splicing and complex C is formed. After the second step of splicing, the mRNA is released and the post-spliceosomal complex, containing the excised intron and the U2, U5, and U6 snRNPs, disassembles and the snRNPs are then recycled for new rounds of splicing. U2 snRNA thus plays an active role during nearly all stages of the splicing process. Indeed, purified E complexes were recently shown to contain both U1 and U2 snRNPs, indicating that U2 may play an important role already during the earliest steps of intron recognition (Das et al. 2000). However, the role of U2 snRNP components in E-complex formation is largely unknown.
The protein composition of the human 17S U2 snRNP is well characterized. It contains the seven Sm proteins, common to U1, U2, U4, and U5, the stably associated U2-specific A′ and B″ polypeptides, and the splicing-essential SF3a and SF3b heteromeric protein complexes (for review, see Krämer 1996; Das and Reed 1999; Krämer et al. 1999; Will et al. 2001). SF3a and SF3b facilitate the interaction of U2 with the branch site (Gozani et al. 1996; Das et al. 1999; Will et al. 2001). Recently, a number of additional proteins, including Prp5, Prp43, SPF30/SMNrp, SPF31, and SPF45 were identified in purified 17S U2 snRNPs (Will et al. 2002); these proteins were also found in purified human pre-spliceosomes (Hartmuth et al. 2002).
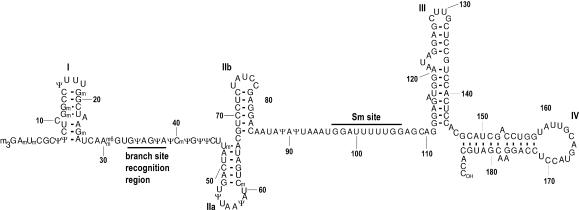
Of the U snRNAs, U2 has the most post-transcriptional internal modifications. Human U2 snRNA (Fig. 1) contains ten 2′O-methylated (2′O-Me) residues and 13 pseudouridines (5-β-D-ribofuranosyluracil). The synthesis of most of these modifications is directed by sequence-specific guide RNAs in the nucleoplasmic Cajal bodies following import of U2 from the cytoplasm (Jády et al. 2003). In U snRNAs, most modified nucleotides are found in regions of functional importance (for review, see Gu et al. 1996). For example, almost all modified bases of U2 are found in its 5′ half (Fig. 1). Additionally, spliceosomal U snRNAs, except U6, obtain cotranscriptionally at their 5′ end an m7G cap, which is converted to m3G in the cytoplasm and serves as a signal for reimport of the U snRNPs into the nucleus (Fischer and Lührmann 1990; Hamm et al. 1990).
FIGURE 1.
Primary and secondary structure of human U2 snRNA. (m3G) N2,2,7-trimethylguanosine; (m) 2′O-methyl; (Ψ) pseudouridine; (m6) N6-methyl. The Sm site and branchpoint recognition sequences are indicated. Stem loops are numbered with roman numerals.
Neither the m3G cap nor internal modifications are essential for the function of U1 (Will et al. 1996), U4 (Wersig and Bindereif 1992), U5 (Ségault et al. 1995), or U6 (Wolff and Bindereif 1992) snRNAs in splicing in vitro. In contrast, post-transcriptional modifications are essential for U2 function in Xenopus oocytes (Pan and Prives 1989) and HeLa nuclear extract (Ségault et al. 1995), but their role remains poorly understood. Modifications within the first 27 nucleotides of U2 snRNA, including the m3G cap, were shown to be required for splicing and/or U2 snRNP biogenesis in Xenopus oocytes (Yu et al. 1998). More recent data suggest a role for pseudouridines in and near the branchsite-binding sequence of U2 in splicing/U2 assembly (Zhao and Yu 2004).
Here, we investigate the roles of individual modified nucleotides of HeLa U2 snRNA in pre-mRNA splicing. We used a two-step reconstitution/complementation system that generates mature U2 snRNP particles from U2 snRNAs containing a given number of modified residues. We demonstrate that the three pseudouridines and five 2′O-methyl groups within the first 24 nucleotides of HeLa U2 snRNA are to various degrees required for pre-mRNA splicing. In contrast, the 5′ m3G cap is dispensable. Further, we show that modified bases in U2 snRNA are not required for 17S U2 snRNP assembly in vitro, but rather are essential for E-complex formation.
RESULTS
Experimental system for depletion/complementation assays
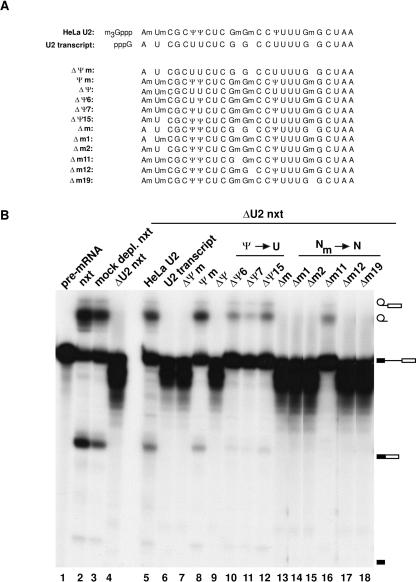
To explore the roles of individual internal modified nucleotides within the 5′ end of U2 snRNA in splicing, we chemically synthesized RNA oligonucleotides corresponding to the 5′-terminal 24 nucleotides of U2, which contained differing numbers and types of modifications (Fig. 2A). These were ligated to an unmodified in vitro transcript encompassing the remaining nucleotides of U2, resulting in chimaeric U2 snRNA molecules. A total of 17S U2 snRNPs were then reconstituted by first assembling core U2 snRNPs from RNA-free, purified Sm proteins and U2 snRNA, and subsequently adding nuclear extract depleted specifically of core U2 snRNP (Ségault et al. 1995). Splicing activity of the reconstituted U2 snRNPs was then assayed by addition of 32P-labeled pre-mRNA and incubation for 3 h at 30°C (Fig. 2B). The splicing activity of mock-depleted nuclear extract (Fig. 2B, lane 3) was comparable to untreated nuclear extract (Fig. 2B, lane 2). In contrast, U2-depleted nuclear extract was no longer active in splicing (Fig. 2B, lane 4). Reconstituted U2 snRNPs containing U2 snRNA purified from HeLa U2 snRNPs (Fig. 2B, lane 5) restored splicing to the level of mock-depleted nuclear extract. Consistent with earlier results (Ségault et al. 1995), reconstituted U2 snRNPs containing in vitro transcribed U2 failed to complement splicing (Fig. 2B, lane 6). In the absence of functionally active U2 snRNPs, the pre-mRNA was apparently more readily degraded in nuclear extract under splicing conditions (e.g., cf. Fig. 2B lanes 4,6 and 3,5).
FIGURE 2.
Internal modifications in the first 24 nucleotides of U2 snRNAs are required for splicing. (A) The sequence composition of the chemically synthesized oligonucleotides corresponding to the first 24 nucleotides of U2 snRNA. The following notation is used: ΔΨm, replacement of all pseudouridines (Ψ) by uridines (U) and deletion of all 2′O-methylations (2′O-Me); (Ψm) retention of all Ψ and 2′O-Me; (ΔΨ) replacement of all Ψs by U; (ΔΨn) replacement of Ψ at position n with U; (Δm) deletion of all 2′O-Me; (Δmn) deletion of 2′O-Me at position n. The oligonucleotides were ligated to in vitro-transcribed U2 starting at G25. For comparison, sequences of purified HeLa U2 and U2 transcript are shown. (B) Internal modifications within the first 24 nucleotides of U2 snRNA are required for efficient complementation of splicing. Splicing complementation of U2 snRNAs differing in the number and type of internal modifications was assayed in the two-step reconstitution system by monitoring splicing of 32P-labeled pre-mRNA (lane 1). Untreated (lane 2), mock-depleted (lane 3), and U2-depleted (lane 4) nuclear extract are shown as controls. (Lanes 5–18) Complementation of U2-depleted nuclear extract with U2 snRNPs reconstituted with the U2 snRNA indicated above each lane. RNA was analyzed by denaturing PAGE and visualized by autoradiography.
Internal modifications at the 5′ end of U2 snRNA, but not the m3G cap structure, are required for splicing in vitro
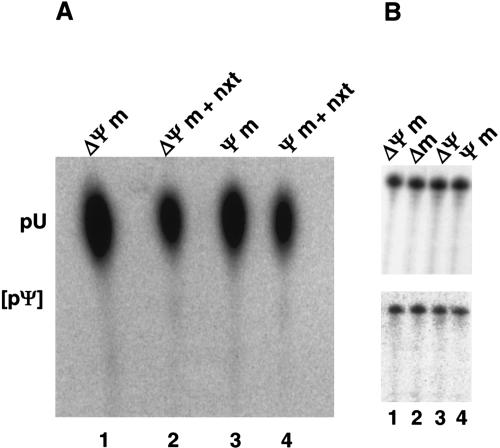
We next tested whether modifications at the 5′ end of U2 are generally required for splicing. As observed with U2 transcript (Fig. 2B, lane 6), a chimaeric U2 snRNA lacking any modification (ΔΨm-U2 snRNA) was found to be inactive in splicing (Fig. 2B, lane 7). In contrast, the Ψm-U2 snRNA, which contains all 2′O-methylated and pseudouridine residues within the first 24 nucleotides, but lacks an m3G cap, complemented splicing almost as efficiently as HeLa U2 snRNA (Fig. 2B, cf. lanes 5 and 8). Importantly, HeLa nuclear extract on its own does not support pseudouridylation of chimaeric RNAs Ψm and ΔΨm under a variety of conditions assayed (Fig. 3A). This is consistent with the previous results where pseudouridine was not detected in in vitro-transcribed U2 snRNA after in vitro reconstitution and splicing (Ségault et al. 1995). Furthermore, during reconstitution and splicing, no selective degradation of ΔΨm or three other tested U2 snRNAs was observed (Fig. 3B, lanes 1–4). Thus, internal modifications have no significant effect on the stability of U2 snRNA in nuclear extract. Taken together, we conclude that only the internal modifications present in Ψm-U2 snRNA, namely those within the first 24 nucleotides, functionally contribute to splicing complementation in our in vitro system, while the m3G cap is dispensable.
FIGURE 3.
Chimaeric U2 snRNAs are not pseudouridylated and remain stable during reconstitution and splicing in the HeLa cell nuclear extract. (A) Chimaeric U2 RNAs Ψm (lanes 1,2) and ΔΨm (lanes 3,4) bearing internal 5′[32P]U labels downstream of G25 were treated with nuclease P1 before (lanes 1,3) or after reconstitution and splicing (lanes 2,4) and analyzed by TLC on PEI plates. The position of uridine 5′-monophosphate (pU) is indicated. Pseudouridine 5′-monophos-phate (pΨ) would be expected at the position indicated by square brackets. (B) Internal RNA modifications do not differentially affect the stability of U2 snRNP in nuclear extract. Equivalent amounts of four different 3′ end-labeled chimaeric U2 snRNAs (indicated above the panels) were analyzed before (top) and after (bottom) an in vitro splicing complementation reaction, using unlabeled pre-mRNA.
The three pseudouridines within the first 24 nucleotides contribute to U2 snRNA function in splicing
We next investigated the contribution of individual pseudouridines to splicing complementation (Fig. 2B). A U2 snRNA in which all pseudouridines within the first 24 nucleotides are replaced by uridines, but the 2′O-methyl groups are retained [ΔΨ-U2 did not support splicing (Fig. 2B, lane 9)], demonstrating that one or more of these pseudouridines is required for splicing. However, when individual pseudouridines at positions 6, 7, or 15 were replaced by uridines, generating the ΔΨ6-U2, ΔΨ7-U2, and ΔΨ15-U2 snRNAs, splicing activity was significantly reduced, as compared with HeLa U2 or Ψm-U2, but not abolished (Fig. 2B, lanes 10–12). Thus, individual pseudouridines are not absolutely required for splicing complementation, but contribute to U2 function. The inactivity of the ΔΨ-U2 snRNA thus arises from the cumulative effect of the loss of individual pseudouridines at positions 6, 7, or 15 of U2 snRNA.
Four 5′-terminal 2′O-methylations are essential for the function of U2 snRNA in pre-mRNA splicing
To investigate the role of 2′O-methylations, we performed reconstitution/complementation studies with chimaeric U2 snRNAs lacking one or more 2′O-methyl groups. A U2 snRNA lacking all five 2′O-methyl groups, but containing pseudouridines (Δm-U2), was inactive in splicing (Fig. 2B, lane 13). Chimaeric U2 snRNAs lacking individual 2′O-methyl groups at positions 1, 2, 12, or 19 (i.e., Δm1-U2, Δm2-U2, Δm12-U2, and Δm19-U2) also failed to complement splicing activity (Fig. 2, lanes 14,15,17,18). In contrast, deletion of the 2′O-methyl of nucleotide G11 reduced, but did not abolish, splicing (Fig. 2, lane 16). Thus, 2′O-methyl groups at positions 1, 2, 12, and 19, but not at position 11, are individually required for the function of U2 snRNA in pre-mRNA splicing.
Internal modifications at the 5′ end of U2 snRNA are required for E-complex formation
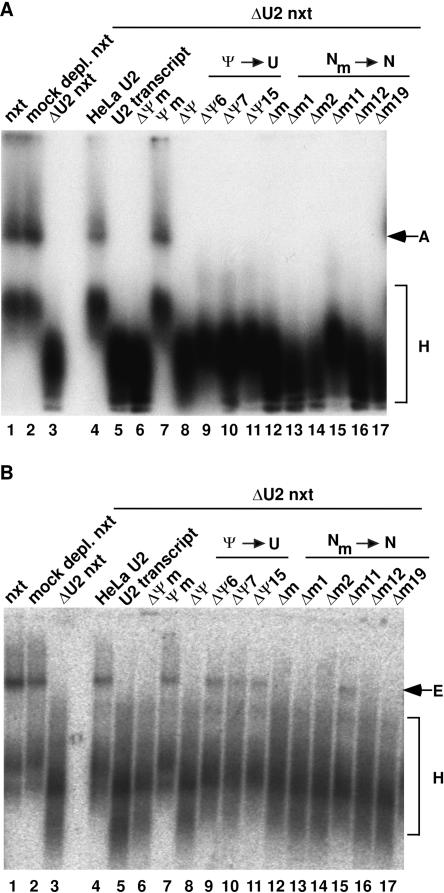
To determine whether modified U2 nucleotides act prior to the catalytic steps of splicing, we next investigated whether they affect spliceosome assembly. For this purpose, we performed splicing in the presence of ATP and analyzed A-complex formation on agarose gels (Das and Reed 1999). HeLa and Ψm-U2 snRNA supported A-complex formation as efficiently as untreated and mock-depleted nuclear extract (Fig. 4A, cf. lanes 4,7 and lanes 1,2). In contrast, no A complex could be detected with U2-depleted extract or upon complementation with any of the other U2 snRNAs (Fig. 4A, lanes 3,8–17). The apparent lack of A-complex formation with the ΔΨ6-U2, ΔΨ7-U2, ΔΨ15-U2, and Δm11-U2 snRNAs was unexpected, as low levels of splicing activity had been observed with these snRNAs (Fig. 2B). However, A complexes formed with these U2 chimera could have escaped detection either due to their inherent instability during gel electrophoresis or due to fast turnover of the low amounts formed. These data indicate that individual 2′O-methyl groups at positions 1, 2, 12, and 19, and collectively pseudouridines at positions 6, 7, and 15, are required either prior to or at the time of A-complex formation.
FIGURE 4.
Internal modifications within the first 24 nucleotides of U2 snRNA are required for efficient spliceosome assembly. Analysis of A-complex (A) and E-complex (B) formation with chimaeric U2 snRNAs. Untreated (lane 1), mock-depleted (lane 2), and U2-depleted (lane 3) nuclear extract are shown as controls. (Lanes 4–17, each panel) Complementation of the U2-depleted nuclear extract with U2 snRNPs reconstituted with the U2 snRNA indicated above each lane. Positions of the H, A, and E complexes are indicated. The ATP depletion of nuclear extract for the E-complex assay (B) was complete, as no A complex could be detected when control samples were analyzed in the presence of heparin (data not shown). Complexes were analyzed on native agarose gels and visualized by autoradiography.
To determine whether internal modifications are required at an earlier step, we next performed splicing in the absence of ATP (see Introduction) and assayed E-complex formation (Fig. 4B). In contrast to mock-depleted extract, U2-depleted nuclear extract on its own did not support E-complex assembly (Fig. 4B, lane 3), corroborating the requirement for U2 snRNP during this process (Das et al. 2000). HeLa and Ψm-U2 snRNA supported E-complex assembly as efficiently as untreated or mock-depleted nuclear extract (Fig. 4B, cf. lanes 4,7 and lanes 1,2). U2 transcript or ΔΨm-U2 snRNA (Fig. 4B, lanes 5,6) on the other hand, did not support E-complex assembly, suggesting that at least some of the modifications are required for U2 function at this assembly stage. U2 snRNPs reconstituted from the ΔΨ6-U2, ΔΨ7-U2, ΔΨ15-U2, and Δm11-U2 snRNAs formed E complex, albeit less efficiently than HeLa or Ψm-U2 (Fig. 4B, lanes 9–11,15), consistent with their reduced splicing activity (Fig. 2B). All other 2′O-methyl deletions (Fig. 4B, lanes 13,14, 16,17) abolished E-complex formation, demonstrating that these modifications are individually required for E-complex formation.
Modifications are not required for the association of 17S U2-specific proteins
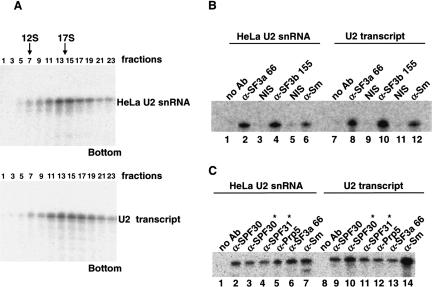
The inability of certain U2 snRNA chimaera lacking modified nucleotides to support E-complex formation could be due to inhibition of U2 snRNP assembly. We therefore investigated whether 17S U2 snRNP formation is compromised in the absence of modifications. U2 snRNPs were thus reconstituted with 32P-labeled HeLa snRNA or U2 transcript and analyzed by glycerol gradient centrifugation. U2 snRNPs assembled with either RNA peaked in the 17S region of the gradient (fractions 13–17; Fig. 5A) and no apparent difference in their distribution across the gradient could be detected. These results provided a first indication that neither internal modifications nor the m3G cap are required for 17S U2 snRNP assembly.
FIGURE 5.
17S U2 snRNP assembly is not dependent on the presence of modifications in U2 snRNA. (A) Both HeLa U2 snRNA (top) and U2 transcript (bottom) assemble into a 17S U2 snRNP in vitro. The 3′ end-labeled RNAs were used for two-step reconstitution and RNPs were analyzed on a 10%–30% glycerol gradient. The RNA content of the odd fractions was analyzed by denaturing PAGE and visualized by autoradiography. Native 17S U2 snRNP, isolated according to Will et al. (2002), exhibited an identical sedimentation behavior (data not shown). (B,C) Immunoprecipitation assays of U2 snRNPs reconstituted with 3′ end-labeled HeLa U2 snRNA (B, lanes 1–6, C, lanes 1–7) or U2 transcript (B, lanes 7–12, C, lanes 8–14). The antibody used is indicated above each lane. (NIS) Nonimmune serum of the antibody in the preceeding lane. Antibodies denoted with * were affinity purified. Major (B) and substoichiometric (C) components of U2 snRNP were analyzed.
However, the reconstituted 17S U2 snRNPs could potentially lack one or (Behrens et al. 1993; Will et al. 2002) more U2-specific protein, which would not necessarily be detectable by sedimentation analysis (cf. Will et al. 2002). Thus, U2 snRNP formation was additionally assayed by immunoprecipitation with antibodies against a subset of U2 snRNP-specific proteins. As expected, both fully modified and unmodified U2 snRNAs were precipitated with anti-Sm (Fig. 5B, lanes 6,12) and anti-U2 B″ (data not shown) antibodies, indicating that 12S U2 snRNP formation is not dependent on modifications. In addition, both RNAs were precipitated by SF3a66 (Fig. 5B, lanes 2,8) and SF3b155 (Fig. 5B, lanes 4,10) antibodies. As SF3a66 and SF3b155 are components of the highly stable heteromeric SF3a and SF3b protein complexes, respectively, it is highly likely that all subunits of SF3a and SF3b are present. Thus, consistent with the observed sedimentation behavior of reconstituted U2 snRNPs, association of these complexes with U2 snRNA does not require modifications. Further, when immunoprecipitations were performed with antibodies against SPF30/SMNrp, SPF31, and hPrp5, which are substoichiometrically present in 17S U2 snRNPs (Fig. 5C), no significant difference in precipitation efficiency was observed between HeLa U2 snRNA and U2 transcript. Taken together, we conclude that U2 snRNA modifications are not required for the formation of intact 17S U2 snRNPs.
DISCUSSION
In this report, we show that efficient pre-mRNA splicing in vitro is dependent on the three pseudouridines and the five 2′O-methyl groups within the first 24 nucleotides of U2 snRNA. In contrast, neither the m3G cap nor internal modifications downstream of A24 are required. Moreover, we could show that the internal modifications at the 5′ end of U2 snRNA are not required for 17S U2 snRNP formation in vitro (see also below). Strikingly, all internal modifications required for splicing were also shown to be essential for the ATP-independent formation of spliceosomal E complexes. Our data thus corroborate previous observations, indicating that U2 snRNP is functionally associated with the pre-mRNA already in the E complex (Das et al. 2000), and show for the first time that internal modifications within the first 24 nucleotides of U2 are required for this functional interaction.
Data presented here regarding the function of the U2 snRNA’s 5′ m3G-cap indicates that the cap does not play a differential role in the function/assembly of the major spliceosomal U snRNAs (i.e., U1, U2, U4, and U5). In all cases, the cap is dispensable for their activity in splicing and also for their assembly in vitro (see Introduction). We note that the results obtained with U2 snRNPs reconstituted in HeLa nuclear extract differ somewhat from those observed with Xenopus oocytes (Yu et al. 1998; Zhao and Yu 2004). In the latter experiments, pseudouridines, and to some extent, also the m3G cap structure, were found to be critical for the assembly in the nucleus of a stable 17S U2 snRNP particle. This could either be due to species-specific differences or to differential requirements of 17S U2 snRNP assembly in vivo versus in vitro.
Our analysis of the contribution of individual modifications to splicing allowed us to clearly distinguish between the effects of pseudouridines and 2′O-methylations. When individual pseudouridines at positions 6, 7, and 15 of U2 snRNA were replaced by uridines, residual E-complex formation and splicing was observed. However, substitution of all pseudouridines by uridines resulted in a complete loss of splicing (Fig. 2B). These results thus provide evidence for a cumulative effect of pseudouridines on E-complex formation and splicing. In contrast, four of the five 2′O-methylations within the 5′-terminal 24 nucleotides of U2 snRNA were absolutely required for E-complex formation and splicing; only deletion of the 2′O-methyl at G10 was not completely detrimental to splicing (Fig. 2B).
Our finding that as many as eight modifications within a short stretch of 24 nucleotides of U2 snRNA are either essential or contribute significantly to U2 function is quite surprising considering that post-transcriptional modifications in many RNA molecules often have no significant effect on their function, at least not when tested in vitro (for review, see Charette and Gray 2000). Some cases, where modifications have been documented to be required for RNA function, are known, however. For example, post-transcriptional modifications in the primer tRNA3Lys are required for initiation and elongation of HIV1 reverse transcription (Isel et al. 1993). Whereas it is thought that the initiation of the reverse transcriptase with the priming complex is affected (Isel et al. 1996), the contribution of individual modified residues of tRNA3Lys to the functional interaction with the reverse transcriptase remains to be investigated. Another example is the selective translational activation of stored maternal mRNAs during early maturation of Xenopus oocytes. Here, 2′O-methylation of the first two bases, that is, the synthesis of a cap II structure on dormant maternal mRNAs, was shown to be required for translational activation (Kuge and Richter 1995; Kuge et al. 1998). The exact function of the two methyl groups in this activation process is not yet understood. Finally, modified nucleotides that are clustered in the peptidyl transferase center of the 23S and 28S rRNAs of the large subunits of Escherichia coli and yeast ribosomes, respectively, also appear to be required for efficient translational activity of the ribosome (Green and Noller 1996; King et al. 2003). The effects of individual modified residues appear to be primarily cumulative ones; only a single pseudouridine residue in the peptidyltransferase center of yeast 28S rRNA was found to contribute substantially on its own to translational efficiency (King et al. 2003).
Why might pseudouridines and 2′O-methylations in the 5′-terminal region of U2 snRNA be required for U2 snRNP function in E-complex formation and splicing? One of the few well-established roles of pseudouridine is that it stabilizes the conformation of RNA by base stacking, with an approximately additive effect on stability when multiple pseudouridines are present (Meroueh et al. 2000), and by coordinating a structural water molecule between its additional hydrogen bond donor at N1-H and the phosphate backbone (Arnez and Steitz 1994). Similarly, 2′O-methylation is a powerful biological means to stabilize an RNA helix (for review, see Davies 1998). Thus, one explanation would be that both requirements are a reflection of the necessity for a stabilized U2 5′ stem loop structure during E-complex formation.
In addition to stabilizing RNA structure, modifications could also play a role in RNA–protein interactions, and thus in this way, be of functional importance for either U2 snRNP structure or E-complex assembly. However, several observations do not support a structural role for modifications in the architecture of U2 snRNP. First, modifications were not required for in vitro assembly of a 17S U2 snRNP particle, and their absence had no detectable effect on the association pattern of most U2 snRNP-specific proteins with the U2 snRNA, as evidenced by immunoprecipitation analyses (Fig. 5B,C). Second, although we observed a number of protein–RNA cross-links between U2-specific proteins and the 5′ region of U2 snRNA in purified HeLa 17S U2 snRNPs, these cross-links were not found to be dependent on the presence of modifications when U2 snRNPs were reconstituted in our two-step in vitro system (O. Dybkov, G. Dönmez, K. Hartmuth, C. Will, and R. Lührmann, unpubl.). Whereas these arguments are not decisive and we cannot rigorously exclude the possibility that U2 snRNA modifications play a role in fine tuning RNA–protein interactions within the 17S U2 snRNP, we favor the idea that modified nucleotides contribute functionally first at the time of E-complex formation. It will therefore be interesting in the future to investigate how U2 snRNP communicates with U1 snRNP and/or other essential factors in the E complex and whether 5′-terminal U2 modifications facilitate RNA–RNA or protein–RNA interactions between U2 snRNP and other E-complex components. These interactions could also involve components of the U4/U6.U5 tri-snRNP complex, as the latter has been reported to contribute to recognition of the 5′ splice site at an early step of spliceosome assembly (Maroney et al. 2000). Our demonstration in this study that functionally active U2 snRNPs can be reconstituted in vitro from chimaeric U2 snRNAs, which, in addition, may also contain reporter molecules, including those useful for cross-linking or hydroxyl radical probing, will considerably facilitate such studies.
MATERIALS AND METHODS
Materials
DNA oligonucleotides used:
K78: 5′-CCTAATACGACTCACTATAGATCGCTTCTCGGCCTTTTGGC-3′;
K79: 5′-GCCATGCTCGAGGGTGCACCGTTCCTGGAGGTAC-3′;
K80: 5′-GTATCAGATATTAAACTGATAAGAACAGATACTACACTTGATCTTAGCCAAAAGGCCGAGAAGCGAT-3′;
K82: 5′-CCTAATACGACTCACTATAGATCAAGTGTAGTATCTGTTCTTATC-3′.
RNA oligonucleotides were purchased from Dharmacon (USA) or RNA-Tec (Belgium). The bU2 biotinylated 2′O-methylated oligonucleotide, complementary to nucleotides 1–20 of U2 snRNA, (Ségault et al. 1995) was obtained from RNA-Tec. The following antibodies were used: anti-SF3a66, anti-hPrp5 (Will et al. 2002); anti-SF3b155 (Will et al. 2001); anti-Sm (Y12, Lerner and Steitz 1979), anti-U2 B″ (4G3, Habets et al. 1989), and anti-SPF30 (Meister et al. 2001). Anti-SPF31 antibodies were raised against a peptide (amino acids 1–17) of the SPF31 protein. Where appropriate, nonimmune serum (NIS) was used as a negative control. Peptide antibodies were affinity purified as described previously (Will et al. 2002).
Preparation and analysis of RNA
DNA templates for in vitro transcription of U2 snRNA constructs were prepared by PCR amplification of pMRG3U2-27 (Jacobson et al. 1993) using primers K78/K79 (U2 snRNA) or primers K82/K79 (G25-U2 snRNA, starting at nucleotide G25), and cleaved with XhoI. Large-scale T7 RNA polymerase transcription was performed as previously described (Milligan and Uhlenbeck 1989). All RNAs were purified on 9.6% polyacrylamide-8.3 M urea gels. HeLa U2 snRNA was prepared by gel fractionation of a HeLa 5S RNA fraction prepared from HeLa cell nuclear extract by phenol extraction and fractionation. The 3′-end labeling of RNA was performed as described (England and Uhlenbeck 1978). The G25-U2 snRNA transcript was first dephosphorylated, then 5′-phosphorylated with polynucleotide kinase and subsequently 3′ end-labeled. Chimaeric U2 snRNAs were prepared by mixing the respective RNA oligonucleotide with the G25-U2 snRNA and K80 splint oligonucleotide, and then ligating with T4 DNA ligase (Moore and Sharp 1992). Full-length chimaeric U2 snRNAs were gel purified and 32P-labeled RNAs were visualized either by autoradiography or with a PhosphorImager (Typhoon, Amersham Biosciences).
U2 snRNP depletion and in vitro reconstitution
HeLa cell nuclear extracts (Dignam et al. 1983) were depleted of U2 snRNPs by affinity selection with bU2 oligonucleotide and streptavidin agarose beads essentially as described previously (Ségault et al. 1995), except that oligonucleotide was used at a concentration of 8 nmol/mL and extract was dialyzed against Dignam’s buffer D (Dignam et al. 1983) without glycerol. Mock-depleted extract was prepared in an identical manner, except that oligonucleotide was omitted. U2 snRNP reconstitution was carried out essentially as described by Ségault et al. (1995). Briefly, 2.4 pmol of the various U2 snRNAs were first reconstituted with 1.9 μg total snRNP proteins in a volume of 12 μL. A 14 μL splicing complementation reaction without pre-mRNA (see below) was then prepared and incubated for 30 min at 0°C. This mixture was used either directly for RNP analysis, or after addition of MINX pre-mRNA (Wolff and Bindereif 1992), for splicing complementation.
In vitro splicing and spliceosomal complex assays
Splicing complementation assays (15 μL) contained 40% nuclear extract, 6.7% (1 μL) U2 snRNP, 2.4 mM MgCl2, 60 mM KCl, 2 mM ATP, 20 mM creatine phosphate, and 50 fmol (1 μL) 32P-labeled pre-mRNA. To analyze splicing of the pre-mRNA, reactions were incubated for 3 h at 30°C. RNA was purified and analyzed on a 15% polyacrylamide-8.3 M urea gel. For E-complex analysis, nuclear extract was first depleted of ATP as described by Das and Reed (1999). Splicing assays (lacking ATP) were then incubated for 10 min at 30°C and 3 μL of 4% sucrose (w/v) containing tracking dyes (loading buffer) were added to a 7 μL aliquot of the splicing reaction. To analyze A-complex formation, splicing was performed as described for E complex, except ATP was added and loading buffer containing 0.2 mg/mL heparin sulphate was subsequently added. Samples were analyzed on a 1.5% low-melting agarose gel (Das and Reed 1999).
Pseudouridine modification assay
The chimaeric Ψm and ΔΨm-U2 RNAs were prepared as described above, except that a [32P]UTP G25-U2 snRNA transcript was used and 3′ end labeling was omitted. After U2 snRNP reconstitution, reaction mixtures were incubated for 3 h at 30°C under splicing conditions. The RNA was purified and then resuspended in 5 μL of 20 mM sodium acetate (pH 5.2) containing nuclease P1 of 200 μg/mL, and then incubated at 37°C for 90 min. Samples were chromatographed on cellulose PEI plates as described (Ségault et al. 1995) and plates were autoradiographed.
Glycerol gradient centrifugation and radioimmunoprecipitation assays
In vitro reconstituted U2 snRNP (140 μL) was prepared with 3′ end-labeled HeLa U2 snRNA or U2 transcript and fractionated on a 10%–30% glycerol gradient (Will et al. 2002). RNA from the odd fractions was purified and analyzed on a 9.6% polyacrylamide-8.3 M urea gel. Immunoprecipitations were performed as described by Will at al. (2002), using 14 μL of in vitro reconstituted U2 snRNPs (see above) per assay and 150 mM NaCl in all buffers. Bound RNA was isolated and analyzed as described above.
Acknowledgments
We thank Utz Fischer and Joan A. Steitz for generously supplying antibodies. We also thank Peter Kempkes and Belinda Hildebrandt for expert technical assistance. We are grateful to Cindy Will for helpful comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Lu 294/12-2) and the Fonds der Chemischen Industrie to R.L.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7186504.
REFERENCES
- Arnez, J.G. and Steitz, T.A. 1994. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33: 7560–7567. [DOI] [PubMed] [Google Scholar]
- Behrens, S.E., Tyc, K., Kastner, B., Reichelt, J., and Lührmann, R. 1993. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol. Cell. Biol. 13: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNA by the spliceosome. In The RNA world (eds. R.F. Gestelandet al.), pp 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Charette, M. and Gray, M.W. 2000. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 49: 341–351. [DOI] [PubMed] [Google Scholar]
- Das, R. and Reed, R. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5: 1504–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, B.K., Xia, L., Palandjian, L., Gozani, O., Chyung, Y., and Reed, R. 1999. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol. Cell. Biol. 19: 6796–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, R., Zhou, Z., and Reed, R. 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell 5: 779–787. [DOI] [PubMed] [Google Scholar]
- Davies, D.R. 1998. Biophysical and conformational properties of modified nucleosides in RNA (Nuclear Magnetic Resonance studies). In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp 85–102. ASM Press, Washington, DC.
- Dignam, J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England, T.E. and Uhlenbeck, O.C. 1978. 3′-terminal labeling of RNA with T4 RNA ligase. Nature 275: 560–561. [DOI] [PubMed] [Google Scholar]
- Fischer, U. and Lührmann, R. 1990. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science 249: 786–790. [DOI] [PubMed] [Google Scholar]
- Gozani, O., Feld, R., and Reed, R. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes & Dev. 10: 233–243. [DOI] [PubMed] [Google Scholar]
- Green, R. and Noller, H.F. 1996. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA 2: 1011–1021. [PMC free article] [PubMed] [Google Scholar]
- Gu, J., Patton, J.R., Shimba, S., and Reddy, R. 1996. Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: Modified nucleotides are clustered in functionally important regions. RNA 2: 909–918. [PMC free article] [PubMed] [Google Scholar]
- Habets, W.J., Hoet, M.H., De Jong, B.A., Van der Kemp, A., and Van Venrooij, W.J. 1989. Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J. Immunol. 143: 2560–2566. [PubMed] [Google Scholar]
- Hamm, J., Darzynkiewicz, E., Tahara, S.M., and Mattaj, I.W. 1990. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell 62: 569–577. [DOI] [PubMed] [Google Scholar]
- Hartmuth, K., Urlaub, H., Vornlocher, H.-P., Will, C.L., Gentzel, M., Wilm, M., and Lührmann, R. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. 99: 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isel, C., Marquet, R., Keith, G., Ehresmann, C., and Ehresmann, B. 1993. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 268: 25269–25272. [PubMed] [Google Scholar]
- Isel, C., Lanchy, J.M., Le Grice, S.F., Ehresmann, C., Ehresmann, B., and Marquet, R. 1996. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 15: 917–924. [PMC free article] [PubMed] [Google Scholar]
- Jacobson, M.R., Rhoadhouse, M., and Pederson, T. 1993. U2 small nuclear RNA 3′ end formation is directed by a critical internal structure distinct from the processing site. Mol. Cell. Biol. 13: 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády, B.E., Darzacq, X., Tucker, K.E., Matera, A.G., Bertrand, E., and Kiss, T. 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 22: 1878–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, T.H., Liu, B., McCully, R.R., and Fournier, M.J. 2003. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11: 425–435. [DOI] [PubMed] [Google Scholar]
- Krämer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65: 367–409. [DOI] [PubMed] [Google Scholar]
- Krämer, A., Gruter, P., Groning, K., and Kastner, B. 1999. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell. Biol. 145: 1355–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge, H. and Richter, J.D. 1995. Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: Implications for translational control of maternal mRNA. EMBO J. 14: 6301–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge, H., Brownlee, G.G., Gershon, P.D., and Richter, J.D. 1998. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 26: 3208–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, M.R. and Steitz, J.A. 1979. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. 76: 5495–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney, P.A., Romfo, C.M., and Nilsen, T.W. 2000. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell 6: 317–328. [DOI] [PubMed] [Google Scholar]
- Meister, G., Buhler, D., Pillai, R., Lottspeich F., and Fischer, U. 2001. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell. Biol. 3: 945–949. [DOI] [PubMed] [Google Scholar]
- Meroueh, M., Grohar, P.J., Qiu, J., SantaLucia Jr., J., Scaringe, S.A., and Chow, C.S. 2000. Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res. 28: 2075–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, J.F. and Uhlenbeck, O.C. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180: 51–62. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. and Sharp, P.A. 1992. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science 256: 992–997. [DOI] [PubMed] [Google Scholar]
- Pan, Z.Q. and Prives, C. 1989. U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing. Genes & Dev. 3: 1887–1898. [DOI] [PubMed] [Google Scholar]
- Reed, R. and Palandijan, L. 1997. Spliceosome assembly. In Eukaryotic mRNA processing (ed. A.R. Krainer), pp 103–129. IRL Press, Oxford, UK.
- Ségault, V., Will, C.L., Sproat, B.S., and Lührmann, R. 1995. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 14: 4010–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wersig, C. and Bindereif, A. 1992. Reconstitution of functional mammalian U4 small nuclear ribonucleoprotein: Sm protein binding is not essential for splicing in vitro. Mol. Cell. Biol. 12: 1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Rümpler, S., Klein Gunnewiek, J., van Venrooij, W.J., and Lührmann, R. 1996. In vitro reconstitution of mammalian U1 snRNPs active in splicing: The U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 24: 4614–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., MacMillan, A.M., Katopodis, N.F., Neubauer, G., Wilm, M., Lührmann, R., and Query, C.C. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20: 4536–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Urlaub, H., Achsel, T., Gentzel, M., Wilm, M., and Lührmann, R. 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homolog and an SF3b DEAD-box protein. EMBO J. 21: 4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, T. and Bindereif, A. 1992. Reconstituted mammalian U4/U6 snRNP complements splicing: A mutational analysis. EMBO J. 11: 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.T., Shu, M.D., and Steitz, J.A. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 17: 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. and Yu, Y.T. 2004. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA 10: 681–690. [DOI] [PMC free article] [PubMed] [Google Scholar]