Abstract
The human RNA-activated protein kinase PKR is an interferon-induced protein that is part of the innate immune response and inhibits viral replication. The action of PKR involves RNA-dependent autophosphorylation leading to inhibition of translation. PKR has an N-terminal dsRNA-binding domain that can interact non-sequence specifically with long (>33 bp) stretches of dsRNA leading to activation. In addition, certain viral and cellular RNAs containing non-Watson–Crick structures and multiple, shorter dsRNA sections can regulate PKR. In an effort to identify novel binders and possible activators of PKR, we carried out selections on a partially structured dsRNA library using truncated and full-length versions of PKR. A library with 1011 sequences was constructed and aptamers that bound to His6-tagged proteins were isolated. Characterization revealed a novel minimal RNA motif for activation of PKR with the following unified structural characteristics: a hairpin with a nonconserved imperfect 16-bp dsRNA stem flanked by 10–15-nt single-stranded tails, herein termed a “ss-dsRNA motif.” Boundary experiments revealed that the single-stranded tails flanking the dsRNA core provide the critical determinant for activation. The ss-dsRNA motif occurs in a variety of cellular and viral RNAs, suggesting possible novel functions for PKR in nature.
Keywords: dsRNA-binding domain, PKR activation, SELEX
INTRODUCTION
PKR is a 551-residue kinase that is a part of the innate immune response (Fig. 1A). It uses its two N-terminal dsRBMs as a sensor for dsRNA and functions to transduce its presence into the inhibition of translation (Williams 1999). This process is carried out as part of an antiviral response and results in the inability of viruses, which can have dsRNA genomes or replicative intermediates, to replicate. PKR is known to be activated by perfect dsRNAs, requiring a minimum of ~33 bp for activation (Manche et al. 1992). In addition, a number of RNAs containing secondary structural imperfections are known to activate PKR. These include aptamers with bulges, internal loops, hairpin loops, and multistem junctions (Bevilacqua et al. 1998), as well as RNAs with internal loops and pseudoknots (Davis and Watson 1996; Ben-Asouli et al. 2002).
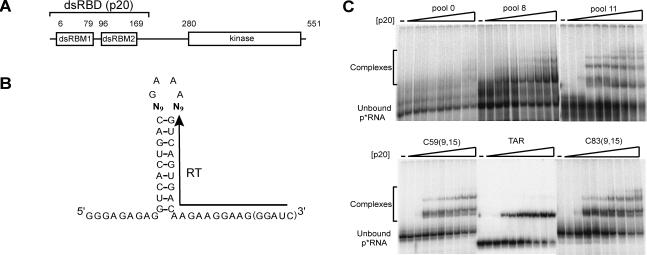
FIGURE 1.
Selection experiment using a partially structured library. (A) Schematic of PKR protein. PKR is 551 residues and has two N-terminal dsRNA-binding motifs (dsRBM1 and dsRBM2) (St Johnston et al. 1992) that comprise its dsRNA-binding domain (dsRBD or p20) and multiple C-terminal kinase subdomains that confer catalytic activity (Hanks et al. 1988). (B) Randomized library constructed to search for RNAs capable of binding to PKR. Eighteen nucleotides were randomized, with nine positions opposite the other nine (bold font). “RT” denotes the binding site for the reverse transcription primer. The five 3′-most nucleotides were only present in the run-off transcripts from the final clones (see Materials and Methods). (C) Gel mobility shifts for p20 binding. Native 10% polyacrylamide gel run at 20°C is shown. Pools 8 and 11 are mixtures of RNAs after eight and 11 rounds of selection, respectively; TAR is a stem–loop RNA substrate for p20 binding containing three bulges (Gunnery et al. 1990; Bevilacqua and Cech 1996). C59(9,15) and C83(9,15) are representative clones after 11 rounds of selection with p20; both have 5′- and 3′-single-stranded tails of 9 and 15 nt, respectively. Concentrations of p20 were 0, 0.01, 0.05, 0.1, 0.2, 0.25, 0.5, 1.0, 2.0, and 5.0 μM. Positions of unbound p*RNA and bound complexes are noted. Binding of RNA to the dsRBD increases with the number of selection rounds, with affinities similar to that of TAR RNA. Similar results were found for in vitro selections for binding to the K296R mutant of full-length PKR (not shown).
One issue that remains unresolved is the range of RNAs, especially shorter ones, capable of activating PKR. The increasing awareness of the importance of short dsRNA molecules in regulating gene expression via RNA interference (RNAi) and inhibition of translation (Ruvkun 2001) in part instigated this re-examination of the RNA activators of PKR. It has been known for some time that RNAs with relatively short (≈14 bp) stems surrounded by other structures can strongly activate PKR (Bevilacqua et al. 1998); however, the critical secondary structural features responsible for activation remain unassigned. Lastly, identification of new classes of activators of PKR might provide a new set of reagents for examining the detailed mechanism of activation and mode of action of this enzyme.
We describe selection experiments on a partially structured dsRNA library. After 11 rounds of selection, aptamers were isolated that were largely RNA hairpins containing internal loops and bulges, with no apparent conservation of sequence or secondary structural defect, as is typical for dsRBM binding sites. These aptamers turned out to be potent activators of the kinase and the length of the single-stranded region flanking the hairpin is shown to provide the critical determinant for activation.
RESULTS
Design of and rationale for a partially structured dsRNA library
Selection experiments were conducted in parallel with two different protein constructs: p20, a 184-residue protein containing the N-terminal dsRBD, and K296R, a full-length PKR protein containing a single, inactivating mutation in the kinase active site (Fig. 1A; Thomis et al. 1992). It was hoped that the RNAs selected to bind to full-length K296R, which would have to be sterically compatible with the presence of the kinase domain, might also be capable of activating wild-type PKR. The reason K296R was chosen over active wild-type PKR is that phosphorylation is known to weaken RNA binding (Jammi and Beal 2001); as such, selection experiments with wild-type PKR might have disfavored isolation of aptamers that were kinase activators.
The library for selection experiments was designed with a 9 × 9 random region flanked by a 9-bp fixed-sequence dsRNA stem and a GAAA sequence (Fig. 1B). Such partially structured libraries turned out to be useful in identifying high-affinity GTP aptamers (Davis and Szostak 2002). The dsRNA stem was flanked on its other end by 9- and 10-nucleotide (nt) fixed-sequence single-stranded tails. The tails comprise a portion of the RT-PCR primer binding sites and were made sufficiently long to facilitate invasion of the stable RNA duplex by the primers. The sequence of the tails was chosen to disfavor any structure formation. As described below, the single-stranded tails, designed out of experimental necessity, turned out to be the critical element for PKR activation.
The sum of the lengths of the dsRNA stem and the random region was forced to be similar to the known minimal length of a p20 binding site, 16–18 bp (Bevilacqua and Cech 1996). A sequence of GAAA was chosen to cap the library since it is a known stable hairpin loop (Woese et al. 1990; Antao et al. 1991). The rationale for designing the library in this fashion was to search for any preferences of PKR for sequence or helical defects within the constraints of a simple dsRNA structure. Also, the library was small enough (418 ≈ 7 × 1010 members) that, in principle, all of the sequence space could be explored. These last two features contrast with an earlier selection for p20 binding to RNA in which a much larger (50 nt) library was used and aptamers with highly unconventional, non-dsRNA features were identified (Bevilacqua et al. 1998).
Selection for RNAs that bind to p20 and K296R
The progress of selection for p20 binding was monitored by a native-gel mobility-shift assay (Fig. 1C). Pool 0 showed essentially no binding, even at the highest (5 μM) concentration of protein. In contrast, pool 8 displayed significant binding for p20, as did pool 11 (Fig. 1C). The RNA from an earlier round, pool 5, showed substantially less binding than pool 8 (data not shown). Gel shifts with K296R for various RNA pools in its selection were qualitatively similar to those with p20 (data not shown).
A direct comparison between p20 binding to aptamers and TAR RNA, a known regulator of PKR with 24 bp and three bulges (Gunnery et al. 1992), was carried out next (Fig. 1C). Although direct quantitative comparison of p20 binding to these three RNAs is complicated by the presence of additional RNA–protein complexes in the aptamer-containing lanes, it can be seen that affinity is similar in all cases, with 0.1 μM p20 required for the onset of observable complex formation. No substantial improvement in binding was observed over the course of the last three rounds of selection, and so selection was halted after 11 rounds. In summary, 11 rounds of selection were sufficient to isolate RNAs from a partially structured dsRNA library capable of binding to p20 (and K296R) with affinities similar to that of a bulged dsRNA regulator of PKR.
Secondary structures of aptamers
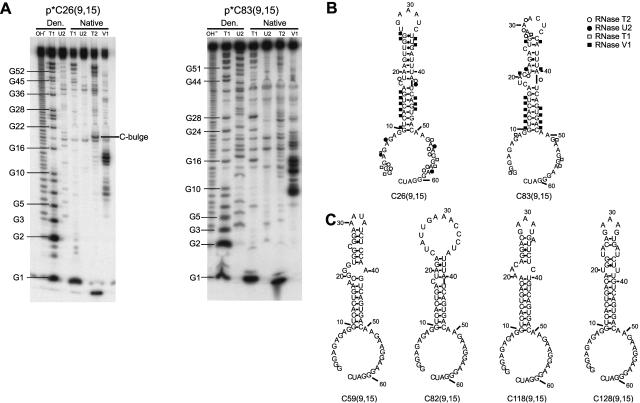
The random regions of ~50 clones from each of the two selections were sequenced, and the secondary structures of their associated transcripts were predicted using mfold v3.1 (Mathews et al. 1999; Zuker et al. 1999). Secondary structures for six representative clones, three from each of the selections, are shown in Figure 2. These particular clones are illustrated since they have obvious stem–loop secondary structures; certain other clones showed folds suggestive of more complex features, including pseudoknots, and were not pursued in this study. The secondary structures of C26(9,15), from selection with K296R, and C83(9,15), from selection with p20, were verified experimentally using structure mapping experiments and were found to be largely in agreement with the mfold predictions (Fig. 2A,B). These two RNAs, which are the focus of the remainder of this study, were chosen for structure mapping. Cleavages by single- and double-strand-specific nucleases (Fig. 2A) in the stem, hairpin loop, bulge, internal loop, and flanking single-stranded regions were consistent with the predicted secondary structures (Fig. 2B). Cleavage of paired A’s near bulges by RNase U2 is likely due to breathing, while occasional cleavage of unpaired nucleotides by RNase V1 near helix ends is likely due to stacking (Lockard and Kumar 1981).
FIGURE 2.
Secondary structures of selected RNAs. (A) Structure mapping of two representative aptamers. Denaturing 12% polyacrylamide gels are shown. p*C26(9,15) is 5′-end labeled clone 26 with 5′- and 3′-tails of 9 and 15 nt, respectively, and is from the selection using K296R; p*C83(9,15) is 5′-end labeled clone 83 with the same tails and is from the selection using p20. Lanes are as follows: OH- is a limited alkaline digest; T1, U2, T2, and V1 are limited digests with ribonucleases specific for single-stranded G’s, A’s, N’s, or double-stranded regions, respectively. “Den.” denotes treatment with nucleases under denaturing conditions, while “Native” denotes treatment with nucleases under nondenaturing conditions. (B) Secondary structural models derived from structure mapping experiments in panel A. Positions of cleavage by single- and double-stranded probes are shown; symbols are provided in the figure. The five 3′-most nucleotides were present in the run-off transcripts from the final clones only. (C) Predicted secondary structures of representative selected RNAs determined using mfold v3.1 (Mathews et al. 1999; Zuker et al. 1999). Clones 59 and 82 are from the selection using p20, and clones 118 and 128 are from the selection using K296R.
Comparison among the secondary structures illustrates a binding motif for the dsRBM containing an imperfect dsRNA stem flanked by single-stranded tails and a hairpin loop (Fig. 2). The double-stranded nature of the base of the stem and the single-stranded nature of the primer binding sites imposed on the starting, partially structured library appear to have been maintained throughout the selection; however, the hairpin loop, which was designed to facilitate formation of a stable GAAA tetraloop, was not always maintained. In addition, no obvious size or sequence conservation of the bulge or internal loop imperfections could be discerned. Thus some of the intended structural bias for the library was maintained during the selections, while other bias was lost. Overall, these experiments supported the dsRBM being able to tolerate a wide variety of secondary structural defects within its binding site, but having little preference for such defects. Moreover, the dsRBM, both alone and in the context of the C-terminal kinase domain, appears capable of tolerating 5′- and 3′-flanking single-stranded nucleotides. We term the identified motif, which consists of an imperfect dsRNA stem–loop flanked by single-stranded tails, an “ss-dsRNA motif.” The lack of discrimination for dsRNA imperfections may have implications for the roles of dsRBMs in vivo (see Discussion). Next, we investigated the ability of various RNAs to activate PKR.
Activation of PKR phosphorylation by perfect dsRNAs
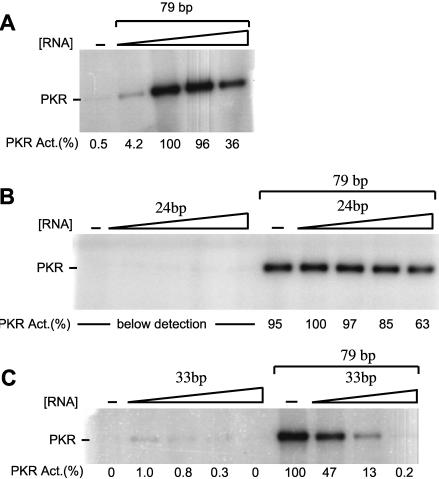
Before testing the ability of the aptamers to activate or inhibit PKR phosphorylation, we tested our PKR phos-phorylation assay on dsRNA modulators without helical imperfections or single-stranded tails. Activation of PKR is known to have a bell-shaped dependence on RNA concentration and to require a minimum of 33 bp (Manche et al. 1992). Double-stranded RNAs of length 24, 33, and 79 bp were prepared (Materials and Methods), and their abilities to regulate PKR were tested. It should be noted that because of the potential importance of single-stranded tails flanking the double-stranded region (see below) we chose to prepare the dsRNAs, including the longest one of 79 bp, in a well-defined fashion. This contrasts with many of the earlier activation assays on PKR which used heterogeneous preparations of RNA for activation such as poly(I):poly(C). Also, activation experiments on TAR were avoided since it is controversial as to whether TAR is an activator or inhibitor of PKR (Gunnery et al. 1990, 1992; Maitra et al. 1994); these conflicting reports may be due to subtle differences in how the TAR RNA was refolded prior to activation, which may cause or prevent RNA dimerization and doubling of RNA length (P.C. Bevilacqua, unpubl.).
As illustrated in Figure 3, 79-bp dsRNA was a potent activator of PKR, as expected. The activation of PKR was highly dependent on added RNA, with only 0.5% reaction in the control lane in the absence of added RNA (Fig. 3A). Moreover, the response of PKR autophosphorylation to 79-bp dsRNA was bell-shaped, showing a maximum near 100 nM dsRNA. These results demonstrate that the preparation of PKR was active and sensitive to the addition of RNA.
FIGURE 3.
Activation and inhibition of PKR by dsRNAs. In vitro activation assays of PKR. Denaturing 10% SDS-polyacrylamide gels are shown. Normalized phosphorylation activities are presented under the gels and were normalized relative to the most reactive lane on the given gel. The position of phosphorylated PKR is noted. (A) In vitro activity assay of PKR by 79-bp dsRNA. Concentrations of 79-bp dsRNA were 0, 0.01, 0.1, 1.0, and 2.5 μM. (B, left set of lanes) In vitro activation of PKR by 24-bp dsRNA. Concentrations of 24-bp dsRNA were 0, 0.01, 0.1, 1.0, and 5.0 μM. Any activation of PKR by 24-bp dsRNA was below the detection limit. (B, right set of lanes) Competition of PKR by 24-bp dsRNA. The concentration of 79-bp dsRNA in all lanes was 0.1 μM, and the concentrations of 24 bp were the same as in the left set of lanes. (C, left set of lanes) In vitro activity assay of PKR by 33-bp dsRNA. Concentrations of 33-bp dsRNA were 0, 0.1, 0.3, 1.0, and 1.5 μM. (C, right set of lanes) Competition of PKR by 33-bp dsRNA. The concentration of 79-bp dsRNA in all lanes was 0.1 μM, and the concentrations of 33 bp were the same as in the left set of lanes, except that the 1.5-μM point was not present.
Next, the effects of 24- and 33-bp dsRNAs were examined. As shown in Figure 3B, 24-bp dsRNA was unable to activate the kinase, consistent with previous reports (Manche et al. 1992). Notably, 24-bp dsRNA was a weak inhibitor of 79-bp dsRNA-dependent activation of PKR (Fig. 3B), which shows that the inability of the 24-bp dsRNA to activate PKR was not due to an inability to bind. Thus, short, perfect dsRNAs without single-stranded tails can inhibit PKR, but they cannot activate it.
As illustrated in Figure 3C, a somewhat longer dsRNA of 33 bp was able to activate the kinase, albeit very weakly (only ≈ 1% the level of 79-bp dsRNA), consistent with previous reports (Manche et al. 1992). The dependence of activation on 33-bp concentration was also bell-shaped (Fig. 3C), showing it can inhibit the kinase at higher concentrations. That 33-bp dsRNA binds to PKR is also supported by its ability to strongly inhibit the 79-bp dsRNA-dependent activation of PKR. As shown in Figure 3C, 0.1 μM 33 bp is able to reduce the autophosphorylation activity of PKR approximately twofold, showing that it is at least 50× (cf. ≈ 50% activities of 0.1 and 5 μM) more potent an inhibitor than 24-bp dsRNA. In summary, the data in Figure 3 show that the length of a perfect dsRNA has a profound effect on its ability to activate and inhibit PKR and that the activation assay is very dependent on RNA. This provided a robust assay to then test the activation ability of the RNA aptamers.
Activation of PKR phosphorylation by RNA aptamers
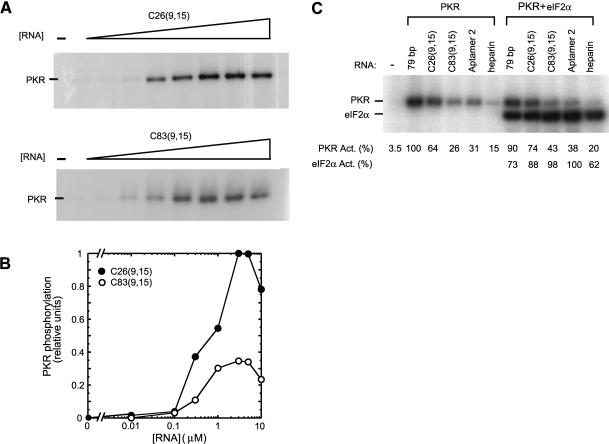
Next, the ability of the aptamers C26(9,15) and C83(9,15) to modulate PKR phosphorylation was tested. Surprisingly, despite having short dsRNA segments of only ≈ 16 bp, both aptamers were potent activators of PKR (Fig. 4A). PKR autophosphorylation showed a bell-shaped response to both aptamers, with approximately twofold greater activity in the presence of C26(9,15) than C83(9,15) (Fig. 4B). As illustrated in Figure 4C, C26(9,15) stimulated autophosphorylation slightly more than 79-bp dsRNA; compare 0.1 μM 79 bp (=100% its maximum, Fig. 3A) with 0.3 μM C26(9,15) (=37% its maximum, Fig. 4A) to give an activity ratio of C26(9,15) / 79bp = (64 / 100) / (37 / 100) = 1.7. Likewise, C83(9,15) stimulated autophosphorylation similarly to 79-bp dsRNA, being only 0.8-fold less active. The concentration of the aptamers required for maximal activation of PKR was somewhat higher, ≈ 3 μM aptamer versus 0.1 μM 79 bp, although reasonable activation by C26(9,15) (≈40% maximal) was seen at concentrations as low as 0.3 μM. Lastly, C26(9,15) and C83(9,15) had similar, or somewhat greater, abilities to activate PKR as optimal concentrations of aptamer 2, the most potent activator from a previous selection on p20 (Fig. 4C; Bevilacqua et al. 1998).
FIGURE 4.
Activation of PKR by aptamers. (A) In vitro activation assay of PKR by selected RNAs. Aptamers C26(9,15) and C83(9,15) were examined for their ability to activate purified PKR kinase. Denaturing 10% SDS-polyacrylamide gels are shown. Concentrations of RNAs were 0, 0.01, 0.1, 0.3, 1.0, 3.0, 5.0, and 10 μM. The position of phosphorylated PKR is noted. Both clones were found to be potent activators of PKR. (B) Quantitation of the gels in panel A. The two experiments were run under identical electrophoresis and exposure conditions, and so were normalized to the same maximum. Note also that C26 and C83 were loaded on the same gel in panel C to facilitate comparison and give similar behavior confirming the reproducibility of the data. Both clones display a classic bell-shaped activation profile. (C) Comparison of PKR activation by C26 and C83, as well as the reference activators: 79-bp dsRNA, aptamer 2 (from a prior selection) (Bevilacqua et al. 1998), and heparin. PKR was incubated with 0.3 μM of C26, C83, and aptamer 2; 0.1 μM of 79-bp dsRNA; and 0.1 unit/μL heparin. The different RNA concentrations allowed similar band intensities to be observed on the gel. For the right panel, reaction mixtures also included 3 μM eIF-2α. Positions of phosphorylated PKR and eIF2α are noted. Normalized phosphorylation activities for PKR and eIF2α are presented under the gel; note that PKR and eIF2α activities were normalized separately.
We then examined the ability of aptamer-activated PKR to phosphorylate purified eIF-2α, the factor which transduces PKR activation into translation inhibition in vivo (Williams 1999). As shown in Figure 4C, C26(9,15)- and C83(9,15)-activated PKR were able to phosphorylate eIF-2α to similar levels as 79-bp dsRNA, aptamer 2, and heparin, another known activator of PKR (Hovanessian and Galabru 1987; George et al. 1996). These results suggest that the ss-dsRNA motif is a plausible regulator of gene expression in vivo.
Molecular features of RNA aptamers required for activation of PKR: Effects of bulge, loop, and tail sequence
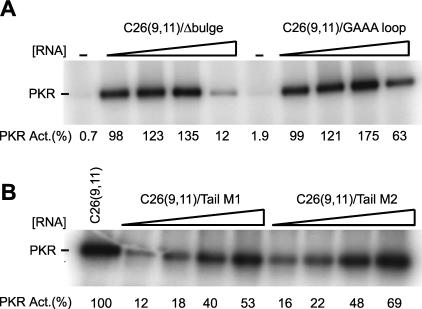
Activation of PKR by aptamers with a dsRNA region of only ≈ 16 bp was surprising given that perfect 24-bp dsRNAs did not activate the kinase. It was therefore of interest to try to uncover the molecular features of the aptamers required for mediating activation. First, we tested the importance of secondary structural defects. These experiments were conducted on C26(9,11); clone 26 was chosen since it is a stronger activator than clone 83 (Fig. 4); an 11-nt 3′-tail was chosen since the last four single-stranded nucleotides are not required for activation (see below), and an 11-nt tail has a slightly lower RNA concentration maximum than a 15-nt tail (0.3–1 μM vs. 3–5 μM) (Figs. 4, 5). Deletion of the C-bulge, the sole secondary structural defect in clone 26, led to similar or somewhat greater PKR phosphorylation activity, as compared with unmodified C26(9,11) (Fig. 5A). Next, the hairpin loop of 5′-UGAAAUC was replaced with 5′-CGAAAG, to give a new loop and closing base pair with a stable and compact structure (Antao et al. 1991). This change led to somewhat greater activity than the unmodified sequence (Fig. 5A). Apparently, the ability of C26(9,11) to activate PKR is not due to a requirement for a secondary structural defect or a certain loop sequence or size. This outcome is consistent with clones 26 and 83, which have unrelated stem defects and loop sequences, both being strong activators of PKR.
FIGURE 5.
Effects of deleting the bulge and varying the hairpin loop and 5′- and 3′-tails on PKR phosphorylation activity. In vitro activation assays of PKR on denaturing 10% SDS-polyacrylamide gels are shown. The position of phosphorylated PKR is noted. (A) Effects of bulge deletion and hairpin loop sequence on PKR activity. C26(9,11)/Δ bulge is a C-bulge-deleted C26(9,11), and C26(9,11)/ GAAA loop is a loop mutant of C26(9,11) in which the UGAAAUC loop was replaced with CGAAAG. Concentrations of RNAs were 0, 0.1, 0.3, 1.0, and 3.0 μM. Normalized phosphorylation activities are presented under the gel, and were normalized relative to the maximum in a C26(9,11) activity assay, which was run under identical electrophoresis and exposure conditions and occurred at 0.3–1.0 μM C26(9,11) (not shown). Deleting the bulge or changing the loop resulted in slightly elevated phosphorylation activities. (B) Effects of 5′-and 3′-flanking sequence on PKR activity. In both cases, the stem and loop were unchanged and the 5′- and 3′-tails were 9 and 11 nt, respectively. For C26(9,11)/Tail M1, most G’s in the 5′- and 3′-tails were changed to C’s, while for C26(9,11)/Tail M2, most G’s were changed to C’s, and all A’s were changed to U’s. (See Materials and Methods for exact sequences.) Concentrations of these RNAs were 0.1, 0.3, 1.0, and 2.0 μM. Normalized phosphorylation activities are presented under the gel and were normalized relative to 0.3 μM C26(9,11) (left-most lane), which is near concentration of C26(9,11) that has maximal activity (0.3–1.0 μM, depending on the experiment). Changing the sequence of the tails had only a minimal effect on phosphory-lation activity.
Next, we tested the importance of the sequence of the single-stranded tails. In the first tail-modified RNA, C26(9,11)/Tail M1, most G’s in the 5′- and 3′-tails of C26(9,11) were replaced with C’s, affording A,C-rich single-stranded tails. In the second modified RNA, C26(9,11)/Tail M2, most G’s and all A’s were changed to C’s and U’s, respectively, affording pyrimidine-rich single-stranded tails (see Materials and Methods for exact sequences). In each case, the sequence of the modified tail was chosen so as to avoid its involvement in RNA secondary structure (i.e., avoid intra-tail, inter-tail, or stem-tail structures), and thereby maintain the single-stranded nature of the tails. Folding of the modified sequences on mfold (Mathews et al. 1999; Zuker et al. 1999) was consistent with only one major secondary structure, similar to that shown in Figure 2B. As shown in Figure 5B, both sequences were efficient activators of PKR, giving a maximal activation within twofold that of C26(9,11). It can be noted that the optimal concentrations of the tail-modified RNAs were somewhat higher (2 μM vs. 0.3–1 μM), suggesting a slight preference for purine-rich tails (Fig. 5). Nevertheless, efficient activation of PKR by the tail-modified RNAs indicates that the ability of clones 26 and 83 to activate PKR is not due to a specific tail sequence.
Molecular features of RNA aptamers required for activation of PKR: Effects of tail length
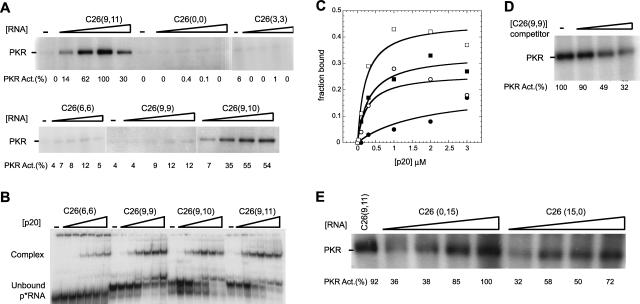
Activation of PKR by the aptamers was not dependent on RNA secondary structural defects, loop sequence, or tail sequence. Next, we examined the dependence of activation on the length of the single-stranded tails; truncated versions of the C26(9,11) sequences were used (see Materials and Methods for exact sequences). As shown in Figure 6, PKR activation has a strong dependence on single-stranded tail length. Essentially no activation was observed for C26(0,0) or C26(3,3). Lengthening the tails by three or six more nucleotides led to weak (12%) activation for C26(6,6) and C26(9,9). However, lengthening the 3′-tail by only two more nucleotides, in C26(9,10) and C26(9,11), restored efficient activation, with maximal relative levels of 55 and 100%. These data show that the single-stranded tails are critical elements in mediating the activation of PKR by short dsRNAs.
FIGURE 6.
Effects of varying the length of the 5′- and 3′-tails on PKR phosphorylation activity. The lengths of the 5′- and 3′-tails flanking the stem–loop of C26 are provided in the figure. (A) Effects of tail length on PKR activity. In vitro activation assays of PKR on denaturing 10% SDS-polyacrylamide gels are shown. Concentrations of RNAs for the first five samples were 0, 0.1, 0.3, 1.0, and 3.0 μM; in a few instances, the 3.0-μM point was replaced with 5 μM RNA, which does not affect the conclusions. Concentrations of the sixth sample, C26(9,10), were 0.1, 0.3, 1.0, and 2.0 μM; the 0-μM point was run on another portion of this gel (not shown) and had 0% activity. The position of phosphorylated PKR is noted. Normalized phosphorylation activities are presented under the gels. The figure is a composite of five different gels. For each gel, a separate activation assay was performed using C26(9,11) (only one of which is shown) and used to normalize the activity of the sample on that particular gel. Flanking sequence is seen to have a marked effect on PKR activity. (B) Effects of tail length on binding to p20. Native 15% polyacrylamide gel run at 13°C is shown. Concentrations of p20 were 0, 0.03, 0.1, 0.3, 1.0, 2.0, and 3.0 μM. Positions of unbound p*RNA and bound complexes are noted. (C) Representative plots of fraction RNA bound versus p20 concentration for data in panel B. Symbols are as follows: C26(6,6) (•), C26(9,9) (○), C26(9,10) (▪), and C26(9,11) (□). Values of Kd and fmax were determined by nonlinear least squares fitting (Kaleidagraph, Synergy Software) to a simple hyperbolic equation (see Materials and Methods), except in the case of C26(6,6) where the value of fmax was fixed at 0.2 in order to have the fit converge. A microshift was detected just above the unbound p*RNA. This microshift has been seen before (Bevilacqua and Cech 1996), although its nature is not entirely clear. The microshift above the main unbound p*RNA was scored as free RNA, which biases the Kd toward higher values. Because the nature of the microshift is unclear, the following Kd values must be treated as semiquantitative. Values of fmax and Kd from the fits were as follows: C26(6,6) 0.2 (fixed in fit) and 2 μM; C26(9,9) 0.25 and 0.3 μM; C26(9,10) 0.3 and 0.3 μM; C26(9,11) 0.5 and 0.2 μM. Binding is only weakly dependent on the length of flanking sequence. (D) Inhibition of PKR activity by C26(9,9). In vitro activation assays of PKR on a denaturing 10% SDS-polyacrylamide gel are shown. C26(9,11) at 0.3 μM was challenged with increasing concentrations (0.3, 1.0, and 2.0 μM) of C26(9,9) RNA. Normalized phosphorylation activities are presented under the gels. A weak activator of PKR can serve as an inhibitor. (E) Effects of having only one tail on PKR activity. In vitro activation assays of PKR on a denaturing 10% SDS-polyacrylamide gel are shown. C26(0,15) has a 15-nt 3′-tail but no 5′-tail, while C26(15,0) has a 15-nt 5′-tail but no 3′-tail; see Materials and Methods for exact sequences. Concentrations of tail-deleted RNAs were 0.1, 0.3, 1.0, and 2.0 μM, and the concentration of C26(9,11) was 0.3 μM. Normalized phosphorylation activities are presented under the gel. Flanking sequence on either the 5′- or 3′-end is sufficient to activate PKR.
In an effort to uncover the basis for the stimulatory effect of the single-stranded tails, we carried out native-gel mobility-shift assays. As shown in Figure 6B, C26(6,6) and C26(9,9), which were poor activators of PKR, were able to bind efficiently to p20; this is especially the case for C26(9,9), whose Kd and fmax are similar to those for C26(9,11) (Fig. 6C). Moreover, C26(9,9) is an efficient inhibitor of PKR activation by C26(9,11) (Fig. 6D), directly showing that its inability to strongly activate PKR is not due to an inability to bind. Apparently the single-stranded tails do not confer activation merely by promoting more efficient binding.
Lastly, we tested whether both tails were required for mediating activation. Either a 15-nt 5′ or 3′ tail was retained, while the other tail was removed. As shown in Figure 6E, both transcripts were efficient activators, indicating that both tails do not have to be present simultaneously in order to activate PKR.
DISCUSSION
Selection experiments were carried out on a partially structured dsRNA library using truncated and full-length versions of PKR (Fig. 1). We chose a partially structured library in hopes of identifying sequences or structures closely related to simple dsRNAs. It can be noted that Davis and Szostak (2002) used partially structured RNA libraries to identify high-affinity GTP aptamers, and that these aptam-ers were very active (Carothers et al. 2004). Use of a partially structured library contrasts with an earlier selection for dsRBD binders and PKR activators in which an unstructured N50 library was used and aptamers with unusual and complex structures were identified (Bevilacqua et al. 1998). The interest in simpler dsRNA or dsRNA-like molecules arose in part from the recent identification of important biological roles for short dsRNAs (Ruvkun 2001), as well as an interest in uncovering any underlying sequence specificity for the dsRBD. As described below, the activating RNAs shared characteristics with the input sequences, as expected.
The outcome of the selection did not lead to any obvious sequence specificity (Fig. 2). Indeed, bulge, hairpin loop, and internal loop defects with a wide range of sizes and sequences were found. It appears that the dsRBD of PKR is tolerant of a wide range of defects, as previously suggested (Tian et al. 2004). (As a caveat, a minor, slower mobility band is present in the C59 and C83 gel shift lanes, but largely absent from the C26 lane, suggesting large bulges may have a subtle affect on protein stoichiometry.) The molecular basis for this promiscuity may lie in the geometry of a PKR–dsRNA complex. The crystal structure of a related complex between dsRNA and a dsRBM from a Xenopus RNA-binding protein indicated that the protein binds to one face of a largely undistorted dsRNA helix, with no wrapping around it (Ryter and Schultz 1998). In addition, experiments from our lab showed that the dsRBD binds to RNAs containing bulge defects by straightening of the helix (Zheng and Bevilacqua 2000). Thus, one possibility is that the dsRBD binds to a face of the aptamer without imperfections, extruding any defects away from the binding interface and thereby straightening the helix. This scenario would help explain the ability of the dsRBD to tolerate such a wide range of defects in its binding site.
The most unanticipated outcome of this study was the identification of a novel class of RNA activators of PKR. Experiments on PKR activation by perfect dsRNAs (Fig. 3) confirmed the studies of Manche and coworkers (1992): A minimum of 33 bp of dsRNA is needed to weakly activate PKR; long dsRNAs, such as 79 bp, strongly activate PKR; and short dsRNAs, such as 24 bp, only inhibit the kinase. Thus, it came as a surprise to find that hairpin aptamers with only ≈16-bp stems were potent activators of the kinase, activating PKR to similar, or somewhat greater, extents than 79-bp dsRNA (Figs. 4, 6). Characterization of a series of variants showed that this effect arises from the length of the flanking single-stranded tails, and not from the presence of secondary structural defects, a specific hairpin loop size or sequence, or the sequence of the tails (Fig. 5). A minimum of 11 nt in the tails was found to lead to activation, and the tails could be present on either the 5′- or 3′-ends, or both (Fig. 6). Discovery of the tails as the activating element was serendipitous since they were originally included in the selections for cloning purposes alone; nevertheless, it is clear from these studies that the tails provide the critical feature for activation. We propose the term “ss-dsRNA” for this PKR-activating motif.
That the tails are attached to a ≈16-bp stem, which is the minimal length of dsRNA known to support PKR binding (Bevilacqua and Cech 1996), supports a preliminary mechanistic model wherein the stem binds to the dsRBD and “presents” the tails to the kinase domain, which leads to activation. It is ironic to note that ssRNA, albeit in the context of a dsRNA stem, should activate a kinase long known as the dsRNA-activated kinase. Curiously, the tails do not enhance binding appreciably, as supported by native gel experiments on hairpins (Fig. 6B–D) and duplexes (Bevilacqua and Cech 1996), pointing to some other role for the tails in activation. In the future, it should prove interesting to determine the mechanistic role of the single-stranded tails in activation. Are they the counterpart of long stretches of dsRNA, or do they serve a new role? Do they play a role in mediating the conformational change proposed to activate the kinase (Wu and Kaufman 1997; Nanduri et al. 2000; Vattem et al. 2001)?
The finding that PKR can be activated by short dsRNA-containing molecules raises the question as to possible biological relevance of the ss-dsRNA motif. MicroRNAs (miRNAs) are ≈22-nt RNAs that bind to the 3′-untranslated regions of mRNAs and control gene expression by inhibiting translation or inducing RNA interference (RNAi) (Ruvkun 2001). Two recent papers examined the effect of small-interfering RNAs (siRNAs) on the interferon system (Bridge et al. 2003; Sledz et al. 2003). Both studies reported nonspecific changes in expression of interferon-stimulated genes due to delivery of siRNAs (Moss and Taylor 2003), which have 2- to 3-nt 3′ overhangs. One confusing point is that Sledz et al. (2003) saw activation of PKR by a synthetic 21-bp siRNA. This observation disagrees with our findings and those of others (Manche et al. 1992) that 24 bp of perfect dsRNA (Fig. 3) and short ssRNA overhangs do not activate the kinase (Fig. 6A). Perhaps the intermolecular nature of their siRNAs system leads to alternative RNA folds not populated by the hairpins in our study. Resolution of these discrepancies will require additional biochemical experiments.
There are several other possible biological functions for the ss-dsRNA motif. First, the motif may resemble the dsRNA replicative intermediates of RNA viruses. Such intermediates have double-stranded regions of limited size and are flanked by nonstructured regions (Richards et al. 1984). The fact that the ss-dsRNA motif requires a stem of only 16 bp suggests that such intermediates may activate PKR. Second, the duplex between a miRNA and mRNA contains bulges and loops, as well as flanking single-stranded regions (Ruvkun 2001). This raises the possibility that PKR, which both inhibits translation and was shown herein to be activated by motifs highly similar to these sites, is involved, at least some of the time, in transducing this miRNA–mRNA signature. The lack of sequence conservation in the ss-dsRNA motif would allow PKR to be activated by a wide variety of such sequences, as required by the innate immune response. In addition, Dicer, a protein that contains 1 dsRBM (Bernstein et al. 2001; Hutvagner et al. 2001), processes imperfect stem–loop precursor miRNAs into mature miRNAs, raising the possibility that it recognizes such imperfect substrates in a similar fashion to PKR. The observation that Dicer processes substrates with nonconserved bulge and loop defects supports this notion (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001; Tian et al. 2004). It should prove interesting to see the extent to which cellular and viral RNAs use, or avoid using, the ss-dsRNA motif in order to activate, or avoid activating, PKR.
MATERIALS AND METHODS
Protein preparation
Selection experiments were performed with either p20 or K296R. The p20 construct is an ≈20-kDa N-terminal truncation of PKR that has an N-terminal (His)6 tag, a thrombin cleavage site, and residues 1–184 of PKR, which contain the dsRBD. The (His)6 tag has been shown to not interfere with binding to RNA (Bevilacqua and Cech 1996). p20 was cloned into the T7 expression vector pET14b (Novagen), transformed into Escherichia coli BL21(DE3)pLysS cells, and purified as previously described (Bevilacqua and Cech 1996). K296R was cloned into pET-28a (Nova-gen) and otherwise treated identically as p20. Wild-type PKR, used for phosphorylation assays, was cloned into pET-28a as well, although it was treated with a subsequent dephosphorylation step (described below). The concentration of proteins was determined spectrophotometrically (Gill and von Hippel 1989).
RNA tail mutants: Shorthand notation and sequences
Most RNA stem–loops were flanked by different lengths of single-stranded tails. To simplify presentation of the data, a shorthand notation was adopted. As an example, “C26(6,6)” refers to selected RNA clone 26, which contains 6 nt upstream and 6 nt downstream of the stem–loop. Sequences of tails for C26 variants were as follows: C26(3,3) 5′-tail: GAG, 3′-tail: AAG; C26(6,6) 5′-tail: GGAGAG, 3′-tail: AAGAAG; C26(9,9) 5′-tail: GGGAGAGAG, 3′-tail: AAGAAGGAA; C26(9,10) 3′-tail: AAGAAGGAAG; C26(9,11) 3′-tail: AAGAAGGAAGG; C26(9,15) and C26(0,15) 3′-tail: AAG AAGGAAGGGAUC; C26(15,0) 5′-tail: GGAGGAGGGAGAGAG. Two clones were prepared to test for any sequence dependence of the tails on PKR activation. In tail mutant 1, referred to as “C26(9,11)/Tail M1”, most (see below for exceptions) G’s in the tails were changed to C’s, while in tail mutant 2, referred to as “C26(9,11)/Tail M2”, most G’s and all A’s in the tails were changed to C’s and U’s, respectively. Sequences were as follows: C26(9,11)/Tail M1 5′-tail: GGGACACAG, 3′-tail: AACAAC CAACC; C26(9,11)/Tail M2 5′-tail: GGCUCUCUG, 3′-tail: AUC UUCCUUCC; for both C26(9,11)/Tail M1 and C26(9,11)/Tail M2, the first few G’s were kept to promote efficient T7 transcription (Milligan and Uhlenbeck 1989), while the bases immediately flanking the base of the stem were left unchanged since dangling ends contribute to helix stability and conformation (Turner and Bevilacqua 1993).
Preparation, purification, and radiolabeling of RNA
DNA oligonucleotides were from IDT Inc., and were gel purified prior to use. A 79-bp dsRNA was prepared by annealing the non–self-complementary RNA products of two separate T7 transcription reactions, which were done using T7 kits from Ambion, Inc. The dsDNA fragment for transcription of the top strand (TS) was prepared by PCR amplification of a pUC19 template using the following set of primers:
TSP79-T7: 5′-GCTAATACGACTCACTATAGGGTTTTCCCAGT CACGACG, and BSP79: 5′-GGTCGACTCTAGAGGATC.
Likewise, the dsDNA fragment for transcription of the bottom strand (BS) was prepared by PCR from pUC19 using the following set of primers:
TSP79: 5′-GGGTTTTCCCAGTCACGACG, and BSP79-T7: 5′-GCTAATACGACTCACTATAGGTCGACTCTAGA GGATC.
The two 79mer RNAs had the following sequences:
TS79R: 5′-GGGUUUUCCCAGUCACGACGUUGUAAAACGAC GGCCAGUGAAUUCGAGCUCGGUACCCGGGGAUCCUCU AGAGUCGACC, and
BS79R: 5′-GGUCGACUCUAGAGGAUCCCCGGGUACCGAGC UCGAAUUCACUGGCCGUCGUUUUACAACGUCGUGACU GGGAAAACCC.
All RNAs were gel purified by denaturing PAGE (Bevilacqua and Cech 1996), and concentrations were determined spectrophoto-metrically. Equal concentrations of purified TS79R and BS79R were annealed in TEN100 (10 mM Tris [pH 7.5], 1 mM EDTA, and 100 mM NaCl) for 1 min at 95°C followed by cooling on the bench for 10 min. Annealed duplexes were stored at -20°C and used at room temperature immediately after thawing. Although these particular sequences were designed to favor the duplex, it is possible that there are small concentrations of species other than perfect dsRNA in the reaction such as single-stranded RNAs or structured RNAs with imperfections, which can never be avoided altogether in a noncovalent nucleic acid complex.
Double-stranded 33-bp RNAs were prepared by T7 transcription from a hemi-duplex (Milligan and Uhlenbeck 1989), while double-stranded 24-bp RNAs were prepared chemically (Bevilacqua and Cech 1996). Twenty-four- and 33-bp dsRNA were annealed as described for 79 bp. The sequences of 24-bp and 33-bp dsRNA were as follows (BS is the perfect complement to the TS): TS24R: 5′-GGGUUCCCUGGUUUCGGUCUCUCG, and TS33R: 5′-GGAGGGUUCGCCUGGUGUUCGGUCUGCUUGUUC.
These sequences were designed with none or one A’s in the top strand to avoid self-structure and favor the duplex. Formation of the duplex was directly observed for the 24-bp dsRNA (Bevilacqua and Cech 1996).
In vitro selected RNAs were initially prepared by T7 transcription of a BamHI-linearized plasmid and gel purified (Bevilacqua et al. 1998). To 5′-end label, RNAs were treated with calf intestinal phosphatase to remove the 5′-triphosphate, reacted with poly-nucleotide kinase and [γ-32P] ATP, repurified by gel electrophoresis, excised from the gel, eluted overnight in TEN250 (10 mM Tris [pH 7.5], 1 mM EDTA, and 250 mM NaCl) at 4°C, ethanol precipitated, and resuspended in TE (10 mM Tris [pH 7.5], 1 mM EDTA).
For certain experiments, C26(9,15) and C83(9,15) RNAs were cleaved by RNase H (Boehringer Mannheim) in the presence of chimeric 2′-O-methyl RNA oligonucleotides (from Integrated DNA Technologies and PAGE purified) to remove the 5′-tail, 3′-tail, or both. The chimeras for cleavage of the 5′- and 3′-ends of the RNA were (5′-m[CAGUGA]d[CCTC] m[UCUCCC]) and (5′-m[GAUCCCUUCCUUC]d[TTGT]), respectively, where “d” signifies a 2′-deoxy sugar, and “m” a 2′-O-methyl sugar. RNase H cleavage reactions were done as published (Lapham and Crothers 1996; Lapham et al. 1997; Chadalavada et al. 2000). Briefly, the chimera was annealed to body-labeled RNA by heating for 3 min to 95°C and cooling to room temperature. The chimera was present at 10× the RNA concentration to ensure complete hybridization of the RNA. RNase H was added to 5 units per 100 μL reaction, and incubated for 3 h at 30°C in 20 mM HEPES-KOH (pH 9.0), 50 mM KCl, and 10 mM MgCl2. The cleaved RNA products were purified by 6% denaturing PAGE. To minimize dimerization, RNA samples were renatured by heating for 1 min to 95°C, followed by cooling on the bench top for all the experiments below.
Selection procedure
Selection procedures followed the general approaches described elsewhere (Tuerk and Gold 1990; Bartel and Szostak 1993; Bevilacqua et al. 1998).
Preparation of the randomized DNA template
The randomized DNA template was prepared by PCR using primers TS1 and BS1 to amplify DNA template, Template1. TS1: 5′-GGGGGAATTCTAATACGACTCACTATAGGGAGAGAGGTCA CTGA; BS1: 5′-CTTCCTTCTTGTCACTGA; Template1: 5′-GGG AGAGAGGTCACTGAC-N(9)-GAAA-N(9)-GTCAGTGACAAGA AGGAAG, where “N” is an equimolar mixture of the four phos-phoramidites. The resultant dsDNA fragment had an upstream EcoRI cloning site, a T7 promoter, and an 18-nt random region. A downstream BamHI cloning site was introduced after the last round of selection by overlap extension with an extended bottom strand primer, BS2. BS2: 5′-CGCGGATCCCTTCCTTCTTGT CACTGA. The BamHI site was omitted during the selection process to keep the single-stranded tails from being any longer than necessary. The PCR reaction was carried out in a volume of 1.0 mL and performed for six cycles of three-temperature amplification (1 min at 95°C, 1 min at 54°C, and 1 min at 72°C). The PCR reaction was extracted with 1 vol of chloroform:isoamyl alcohol (24:1), ethanol precipitated, washed in 70% ethanol, and stored in TE.
Preparation of the randomized RNA library
T7 transcription was in the presence of [α-32P] ATP in a total volume of 1 mL (Bevilacqua et al. 1998). After reaction for 4 h at 37°C, 60 μL of 0.5 M EDTA was added to chelate the Mg2+. The solution was ethanol precipitated, dissolved in 100 μL of TE, and desalted over a Sephadex G-25 spin column. The eluate was digested with 50 units of DNase I (Pharmacia) for 20 min at 37°C. The resulting solution was phenol extracted, ethanol precipitated, and redissolved in 100 μL of HE (10 mM Hepes [pH 7.0], 0.1 mM EDTA). RNA concentration was determined spectrophotometri-cally. This pool of RNAs is referred to as pool 0 RNA.
Selection of the p20- or K296R-binding RNAs
Selection experiments were carried out at 22°C similar to those previously described (Bevilacqua et al. 1998). The RNA pool was first cleared with nickel(II) nitrilotriacetic acid agarose resin. Then herring sperm DNA and protein were added at concentrations of 0.1 mg/mL and 1 μM, respectively. Binding was allowed to proceed in solution for 5 min followed by addition of 100 μL of a 50% slurry of nickel(II) nitrilotriacetic acid agarose resin in 1× selection buffer (SxnB) (150 mM NaCl, 25 mM Hepes [pH 7.5], and 5 mM 2-mercaptoethanol). This mixture was manually shaken for 10 min at 22°C, followed by centrifugation for 10 s at 14,000 rpm. The supernatant was removed, and the resin was washed 10× with 500–600 μL of 1× SxnB for each wash. The RNA was eluted with two or three 100 μL portions of 1× elution buffer (150 mM NaCl, 25 mM Hepes [pH 7.5], 5 mM 2-mercaptoethanol, and 250 mM imidazole). The eluate fractions were pooled and phenol extracted. Glycogen carrier (20 μg from Boehringer Mannheim) was added, and the RNA was ethanol precipitated and washed with 70% ethanol. The pellet was dissolved in 50 μL of TE, and passed over a Sephadex G-25 spin column. The RNA was ethanol precipitated, washed with 70% ethanol, dried, and dissolved in 10 μL of TE. Fractions were collected at all steps and quantitated via liquid scintillation counting.
RT-PCR to convert the selected RNA into dsDNA template
Selected RNA (4 μL) was annealed with BS1 and cDNAs were obtained by reverse transcription for 30 min at 60°C using AMV reverse transcriptase (Life Sciences). To this solution, 50 μL of a master mix containing buffer and TS1 was added. This mixture was heated for 1 min at 95°C prior to addition of Taq polymerase. PCR was performed for 20 cycles of three-temperature amplification: 1 min at 95°C, 1 min at 54°C, and 1 min at 72°C.
After 11 rounds of selection, the RNA was converted into cDNA using BS2 and amplified by PCR. PCR fragments were digested with EcoRI and BamHI and cloned into pUC19. Sequences of isolated clones were determined by dideoxy sequencing, using gels with 20% formamide and 7 M urea. Approximately 100 clones were sequenced, half from selection with p20 and half from selection with K296R.
Binding assays
Native-gel mobility-shift assays were carried out to determine the binding affinity between p20 and certain 5′-32P-labeled RNA. Experiments were similar to previously described (Bevilacqua and Cech 1996; Zheng and Bevilacqua 2000), using excess protein and trace amounts (nM) of radiolabeled RNA, p*RNA. Gel shifts were in the presence of 0.1 mg/mL tRNA competitor. Fits were to
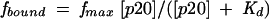 |
where fmax is the maximal fraction of RNA bound, and Kd is the dissociation constant. Experiments were in triplicate and trends were reproducible.
Structure mapping
5′-End labeled RNA was renatured by heating for 1 min at 95°C in TE and incubating for 10 min at 22°C. RNAs were digested with the appropriate nuclease under either native (1 h at 22°C in 1× SxnB) or denaturing conditions (15 min at 50°C in 8.3 M urea). Nuclease concentrations were chosen to give limited RNA hydrolysis: 1 U/μL RNase T1, 1 U/μL RNase U2, 0.02 U/μL RNase T2, and 0.07 U/μL RNase V1. Sequencing lanes for A and G were prepared by limited hydrolysis with RNases U2 and T1, respectively, under denaturing conditions. All-nucleotide lanes were prepared by treatment with alkali. Samples were fractionated on a denaturing polyacrylamide gel.
PKR activation assays
The ability of various RNAs to activate or inhibit PKR kinase was determined by PKR autophosphorylation and eIF-2 phosphoryla-tion assays. PKR was first dephosphorylated by treating with λ-PPase (NEB), as described (Matsui et al. 2001). Subsequently, λ-PPase was inactivated by treatment with freshly prepared sodium orthovanadate (Matsui et al. 2001). Next, 10 μCi [γ-32P] ATP, 0.6 μM dephosphorylated PKR, and RNA were incubated in 20 mM Hepes (pH 7.5), 4 mM MgCl2, 100 mM KCl, and 1 mM ATP for 30 min at 30°C. In certain reactions, 2 μM of purified eIF-2α was added and incubated another 5 min (Krishnamoorthy et al. 2001). Reactions were stopped by adding SDS loading buffer and analyzed on a 10% SDS-polyacrylamide gel. Heparin, a known activator of PKR (Hovanessian and Galabru 1987; George et al. 1996), was used for certain experiments. Heparin was from Sigma and used at a concentration of 0.1 U/μL without any further purification. Data were quantified on a PhosphorImager (Molecular Dynamics). Normalization was to the highest value of counts for a given reaction set. Background was subtracted from all bands before normalization.
Acknowledgments
We thank Elena Sineva and Professors Craig Cameron and Tom Dever for the gift of purified eIF-2α, and Craig Cameron for helpful discussions. This work was supported by a grant from the National Institutes of Health (GM58709).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7150804.
REFERENCES
- Antao, V.P., Lai, S.Y., and Tinoco Jr., I. 1991. A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 19: 5901–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. and Szostak, J.W. 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418. [DOI] [PubMed] [Google Scholar]
- Ben-Asouli, Y., Banai, Y., Pel-Or, Y., Shir, A., and Kaempfer, R. 2002. Human interferon-γ mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell 108: 221–232. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C. and Cech, T.R. 1996. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35: 9983–9994. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C., George, C.X., Samuel, C.E., and Cech, T.R. 1998. Binding of the protein kinase PKR to RNAs with secondary structure defects: Role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry 37: 6303–6316. [DOI] [PubMed] [Google Scholar]
- Bridge, A.J., Pebernard, S., Ducraux, A., Nicoulaz, A.L., and Iggo, R. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34: 263–264. [DOI] [PubMed] [Google Scholar]
- Carothers, J.M., Oestreich, S.C., Davis, J.H., and Szostak, J.W. 2004. Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 126: 5130–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadalavada, D.M., Knudsen, S.M., Nakano, S., and Bevilacqua, P.C. 2000. A role for upstream RNA structure in facilitating the catalytic fold of the genomic hepatitis delta virus ribozyme. J. Mol. Biol. 301: 349–367. [DOI] [PubMed] [Google Scholar]
- Davis, J.H. and Szostak, J.W. 2002. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl. Acad. Sci. 99: 11616–11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S. and Watson, J.C. 1996. In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3′ untranslated regions of human α-tropomyosin. Proc. Natl. Acad. Sci. 93: 508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, C.X., Thomis, D.C., McCormack, S.J., Svahn, C.M., and Samuel, C.E. 1996. Characterization of the heparin-mediated activation of PKR, the interferon-inducible RNA-dependent protein kinase. Virology 221: 180–188. [DOI] [PubMed] [Google Scholar]
- Gill, S.C. and von Hippel, P.H. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182: 319–326. [DOI] [PubMed] [Google Scholar]
- Gunnery, S., Rice, A.P., Robertson, H.D., and Mathews, M.B. 1990. Tat-responsive region RNA of human immunodeficiency virus 1 can prevent activation of the double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. 87: 8687–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnery, S., Green, S.R., and Mathews, M.B. 1992. Tat-responsive region RNA of human immunodeficiency virus type 1 stimulates protein synthesis in vivo and in vitro: Relationship between structure and function. Proc. Natl. Acad. Sci. 89: 11557–11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks, S.K., Quinn, A.M., and Hunter, T. 1988. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52. [DOI] [PubMed] [Google Scholar]
- Hovanessian, A.G. and Galabru, J. 1987. The double-stranded RNA-dependent protein kinase is also activated by heparin. Eur. J. Biochem. 167: 467–473. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Jammi, N.V. and Beal, P.A. 2001. Phosphorylation of the RNA-dependent protein kinase regulates its RNA- binding activity. Nucleic Acids Res. 29: 3020–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy, T., Pavitt, G.D., Zhang, F., Dever, T.E., and Hin-nebusch, A.G. 2001. Tight binding of the phosphorylated α subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 21: 5018–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Lapham, J. and Crothers, D.M. 1996. RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA 2: 289–296. [PMC free article] [PubMed] [Google Scholar]
- Lapham, J., Yu, Y.T., Shu, M.D., Steitz, J.A., and Crothers, D.M. 1997. The position of site-directed cleavage of RNA using RNase H and 2′-O- methyl oligonucleotides is dependent on the enzyme source. RNA 3: 950–951. [PMC free article] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Cae-norhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Lockard, R.E. and Kumar, A. 1981. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 9: 5125–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra, R.K., McMillan, N.A.J., Desai, S., McSwiggen, J., Hovanessian, A.G., Sen, G., Williams, B.R.G., and Silverman, R.H. 1994. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology 204: 823–827. [DOI] [PubMed] [Google Scholar]
- Manche, L., Green, S.R., Schmedt, C., and Mathews, M.B. 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12: 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Matsui, T., Tanihara, K., and Date, T. 2001. Expression of unphos-phorylated form of human double-stranded RNA-activated protein kinase in Escherichia coli. Biochem. Biophys. Res. Commun. 284: 798–807. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F. and Uhlenbeck, O.C. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180: 51–62. [DOI] [PubMed] [Google Scholar]
- Moss, E.G. and Taylor, J.M. 2003. Small-interfering RNAs in the radar of the interferon system. Nat Cell Biol. 5: 771–772. [DOI] [PubMed] [Google Scholar]
- Nanduri, S., Rahman, F., Williams, B.R., and Qin, J. 2000. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 19: 5567–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, O.C., Martin, S.C., Jense, H.G., and Ehrenfeld, E. 1984. Structure of poliovirus replicative intermediate RNA. Electron microscope analysis of RNA cross-linked in vivo with psoralen derivative. J. Mol. Biol. 173: 325–340. [DOI] [PubMed] [Google Scholar]
- Ruvkun, G. 2001. Molecular biology. Glimpses of a tiny RNA world. Science 294: 797–799. [DOI] [PubMed] [Google Scholar]
- Ryter, J.M. and Schultz, S.C. 1998. Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17: 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz, C.A., Holko, M., de Veer, M.J., Silverman, R.H., and Williams, B.R. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5: 834–839. [DOI] [PubMed] [Google Scholar]
- St Johnston, D., Brown, N.H., Gall, J.G., and Jantsch, M. 1992. A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. 89: 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomis, D.C., Doohan, J.P., and Samuel, C.E. 1992. Mechanism of interferon action: cDNA structure, expression, and regulation of the interferon-induced, RNA-dependent P1/eIF-2a protein kinase from human cells. Virology 188: 33–46. [DOI] [PubMed] [Google Scholar]
- Tian, B., Bevilacqua, P.C., Diegelman-Parente, A., and Mathews, M.B. 2004. The double stranded RNA binding motif: Interference and much more. Nat. Rev. Mol. Cell Biol. (in press). [DOI] [PubMed]
- Tuerk, C. and Gold, L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- Turner, D.H. and Bevilacqua, P.C. 1993. Thermodynamic considerations for evolution by RNA. In The RNA world (eds. R.F. Geste-land and J.F. Atkins), pp. 447–464. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Vattem, K.M., Staschke, K.A., Zhu, S., and Wek, R.C. 2001. Inhibitory sequences in the N-terminus of the double-stranded-RNA- dependent protein kinase, PKR, are important for regulating phosphory-lation of eukaryotic initiation factor 2α (eIF2α). Eur. J. Biochem. 268: 1143–1153. [DOI] [PubMed] [Google Scholar]
- Williams, B.R. 1999. PKR; a sentinel kinase for cellular stress. Oncogene 18: 6112–6120. [DOI] [PubMed] [Google Scholar]
- Woese, C.R., Winker, S., and Gutell, R.R. 1990. Architecture of ribo-somal RNA: Constraints on the sequence of “tetra-loops”. Proc. Natl. Acad. Sci. 87: 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. and Kaufman, R.J. 1997. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem. 272: 1291–1296. [DOI] [PubMed] [Google Scholar]
- Zheng, X. and Bevilacqua, P.C. 2000. Straightening of bulged RNA by the double-stranded RNA-binding domain from the protein kinase PKR. Proc. Natl. Acad. Sci. 97: 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M., Mathews, D.H., and Turner, D.H. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. In RNA biochemistry and biotechnology (eds. J. Barcisze-wski and B.F.C. Clark), pp. 11–43. NATO ASI Series, Kluwer Academic, Dordrecht, The Netherlands.