Abstract
Eukaryotes have two types of ribosomes containing either 5.8SL or 5.8SS rRNA that are produced by alternative pre-rRNA processing. The exact processing pathway for the minor 5.8SL rRNA species is poorly documented. We have previously shown that the trans-acting factor Rrp5p and the RNA exonuclease Rex4p genetically interact to influence the ratio between the two forms of 5.8S rRNA in the yeast Saccharomyces cerevisiae. Here we report a further analysis of ITS1 processing in various yeast mutants that reveals genetic interactions between, on the one hand, Rrp5p and RNase MRP, the endonuclease required for 5.8SS rRNA synthesis, and, on the other, Rex4p, the RNase III homolog Rnt1p, and the debranching enzyme Dbr1p. Yeast cells carrying a temperature-sensitive mutation in RNase MRP (rrp2-1) exhibit a pre-rRNA processing phenotype very similar to that of the previously studied rrp5-33 mutant: ITS2 processing precedes ITS1 processing, 5.8SL rRNA becomes the major species, and ITS1 is processed at the recently reported novel site A4 located midway between sites A2 and A3. As in the rrp5-Δ3 mutant, all of these phenotypical processing features disappear upon inactivation of the REX4 gene. Moreover, inactivation of the DBR1 gene in rrp2-1, or the RNT1 gene in rrp5-Δ3 mutant cells also negates the effects of the original mutation on pre-rRNA processing. These data link a total of three RNA catabolic enzymes, Rex4p, Rnt1p, and Dbr1p, to ITS1 processing and the relative production of 5.8SS and 5.8SL rRNA. A possible model for the indirect involvement of the three enzymes in yeast pre-rRNA processing is discussed.
Keywords: yeast, pre-rRNA processing, internal transcribed spacer 1, 5.8S rRNA, Rnt1p, debranching enzyme
INTRODUCTION
Biogenesis of eukaryotic ribosomes is a highly coordinated process that largely takes place in the nucleolus, a specialized nuclear structure. It starts with the transcription of two precursor rRNA species, a short one containing the 5S rRNA sequence and a large one encompassing the coding regions for 18S, 5.8S, and 25/28S rRNA, separated by two internal transcribed spacers (ITS1 and ITS2) and flanked by 5′- and 3′-external transcribed spacers (5′- and 3′-ETS; cf. Fig. 1A). The latter precursor transcript undergoes extensive, site-specific 2′-O-methylation and pseudouridylation (Kiss 2001; Decatur and Fournier 2003), after which the spacer regions are removed by an ordered series of endonucleolytic cleavages and exonucleolytic digestion steps that are accompanied by the association of the ribosomal proteins (r-proteins). A large number of studies, mostly carried out in the yeast Saccharomyces cerevisiae, have made it clear that, apart from the r-proteins, a multitude of trans-acting, nonribosomal factors, including various snoRNPs, play a crucial role in the accurate and efficient assembly of functional eukaryotic ribosomes (Kressler et al. 1999; Venema and Tollervey 1999; Warner 2001). Recently it has become clear that these factors are part of two, largely independent, dynamic processing/assembly machineries, one for the small and one for the large subunit (Fatica and Tollervey 2002; Fromont-Racine et al. 2004; Granneman and Baserga 2004; Raué 2004). The inventory of these trans-acting factors is as yet incomplete and their precise role in pre-rRNA processing and assembly is still largely unexplored.
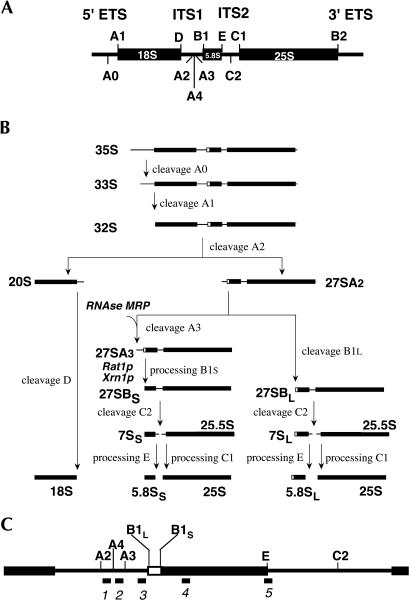
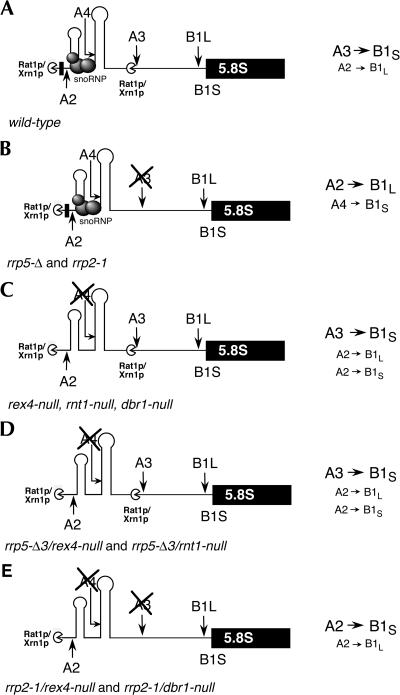
FIGURE 1.
Pre-rRNA processing in Saccharomyces cerevisiae. (A) Structure of the rDNA transcription unit, including the various processing sites. (B) The pre-rRNA processing pathway. The processing steps in ITS1 involving RNase MRP and the 5′ → 3′ exonucleases Xrn1p and Rat1p are indicated. (C) Enlargement of the central portion of the pre-rRNA showing the positions of the different probes used for Northern and primer extension analysis. See Materials and Methods for the sequences of the probes.
As outlined in Figure 1B, the first detectable large pre-rRNA species in yeast is the 35S precursor, which is produced through cleavage of the nascent transcript in the 3′-ETS by Rnt1p, the yeast homolog of bacterial RNAse III (Abou-Elela et al. 1996; Kufel et al. 1999). This precursor, which is part of an 80S–90S pre-ribosomal particle (Dragon et al. 2002; Grandi et al. 2002), undergoes further endonucleolytic processing at sites A0, A1, and A2 that removes the 5′-ETS and produces a 43S and a 66S pre-ribosomal particle containing the small subunit 20S and the large subunit 27SA2 pre-rRNA species, respectively. The 43S pre-ribosome is transported to the cytoplasm, where the 20S pre-rRNA is cleaved at site D to remove the remaining portion of ITS1 (Stevens et al. 1991; Vanrobays et al. 2001).
Conversion of the 27SA2 precursor into the mature 5.8S and 25S rRNA species occurs via two different pathways. Roughly 80% of this precursor is cleaved at site A3 by RNase MRP (Schmitt and Clayton 1993; Chu et al. 1994; Lygerou et al. 1996), and the resulting 27SA3 precursor is processed exonucleolytically into 27SBS pre-rRNA (Henry et al. 1994). The remaining 20% is converted into the 27SBL pre-rRNA, supposedly by an endonuclease that cuts 6 nt upstream from site B1S. We have recently reported the existence of a novel A4 processing site, located midway between sites A2 and A3 (cf. Fig. 1A), that becomes detectable in certain deletion mutants of the trans-acting factor Rrp5p (Eppens et al. 2002). It remains to be established, however, whether processing at this site also occurs in wild-type cells and, if so, at what stage.
The two 27SB precursors undergo the same set of endo- and exonucleolytic processing reactions that produce 5.8SS and 5.8SL as well as 25S rRNA (Mitchell et al. 1996; Allmang et al. 2000; Geerlings et al. 2000; Van Hoof et al. 2000; Faber et al. 2002).
The presence of two forms of 5.8S rRNA with slightly different 5′ ends is a general characteristic of eukaryotic 60S ribosomal subunits, though the ratio between the two forms differs from one organism to another (Henry et al. 1994). However, the physiological relevance of this phenomenon and the manner in which the relative levels of the two 5.8S species are controlled remains obscure. Although, as shown in Figure 1, the common view so far is that the pathways leading to 5.8SS and 5.8SL rRNA, respectively, separate after the cleavage at site A2, mutants that fail to produce 27SA2 pre-rRNA can still generate normal levels of 5.8SL rRNA (Torchet and Hermann-Le Denmat 2000; Vos et al. 2004). On the other hand, a block in A3 processing due to mutations in cis or in trans, while resulting in predominant synthesis of 5.8SL rRNA, does not completely abolish production of the “short” species (Schmitt et al. 1993; Allmang et al. 1996a,b; Eppens et al. 2002). These observations suggest that generation of the two different forms of 5.8S rRNA in their characteristic ratio is likely to be more complicated than apparent from the scheme shown in Figure 1.
In yeast the non-ribosomal protein Rrp5p plays an important role in determining the relative levels of the two 5.8S species, since many of the 12 S1 RNA-binding motifs present in its N-terminal region are crucial for cleavage at site A3. Deletions within this region, therefore, shift production of 5.8S rRNA to mostly the “long” species (Eppens et al. 1999, 2002). The same deletions also led to the detection of processing at site A4 (Eppens et al. 2002). A synthetic lethality screen using one of these rrp5 deletion mutants (rrp5-Δ4) identified the REX4 gene (Eppens et al. 2002), which encodes a protein that is structurally related to a number of nonessential 3′ → 5′ RNA exonucleases including Rex1p, Rex2p, Rex3p, and Pan2p, which are involved in various aspects of snoRNA, rRNA, and mRNA biogenesis and metabolism (Brown and Sachs 1998; Van Hoof et al. 2000). Rex4p does contain the exonuclease signature domain but its enzymatic activity and substrate(s) remain to be identified. Surprisingly, inactivation of Rex4p by itself does not result in a detectable processing phenotype (Van Hoof et al. 2000), but in the rrp5-Δ3 and rrp5-Δ4 mutants it restores cleavage at site A3 and shifts production of 5.8S rRNA back to the short form, indicating that Rex4p is involved, either directly or indirectly, in ITS1 processing.
To obtain more insight into 5.8S rRNA production and the role of Rex4p in ITS1 processing we have analyzed the consequences of inactivating the REX4 gene in cells carrying a temperature-sensitive mutation (rrp2-1) (Shuai and Warner 1991; Chu et al. 1994) in RNase MRP, the enzyme responsible for cleavage at A3. Strikingly, we find that rrp2-1 mutant cells possess a pre-rRNA processing phenotype virtually indistinguishable from that of the previously studied rrp5-Δ3 and -Δ4 mutants, including processing of ITS1 at site A4, that reverts to wild-type upon inactivation of the REX4 gene, except that cleavage at A3 is not restored.
Further experiments showed that the pre-rRNA processing phenotype of the rrp-5-Δ3 and rrp2-1 mutants can similarly be redressed by inactivation of the RNT1 or the DBR1 gene, which encode the eukaryotic homolog of bacterial Rnase III and the lariat debranching enzyme, respectively. These findings suggest that Rex4p, Rnt1p, and Dbr1p are indirectly involved in ITS1 processing via a common factor. We propose that this common factor is an as yet unidentified snoRNA, production of which depends upon all three RNA catabolic enzymes.
RESULTS
Pre-rRNA processing in the rrp2-1 mutant is indistinguishable from that in the rrp5-Δ3 mutant and depends upon Rex4p
We have previously shown that the putative 3′ → 5′ exonuclease Rex4p affects the manner of ITS1 processing in yeast cells carrying a deletion in the N-terminal region of the trans-acting factor Rrp5p that prevents cleavage at site A3, resulting in increased 5.8SL rRNA synthesis at the expense of 5.8SS production (Eppens et al. 2002). To investigate the role of Rex4p in pre-rRNA processing further, we introduced the rex4-null allele from strain YAV41 into strain D308 in which cleavage at site A3 is inhibited by the temperature-sensitive rrp2-1 mutation that inactivates RNase MRP (Shuai and Warner 1991; Chu et al. 1994). It has previously been reported that the rrp2-1 mutation affects pre-rRNA processing in a manner similar to the rrp5-Δ3 mutation: It causes a shift in 5.8S rRNA production from the short to the long species as well as a (partial) reversal of the order in which the internal transcribed spacers are removed, ITS2 being processed prior to ITS1 (Shuai and Warner 1991; Lindahl et al. 1992; Schmitt and Clayton 1993).
The rrp2-1/rex4-null double mutant FVY010A (Table 1) is able to grow at 23°C. Thus, unlike the rrp5-′ mutations, the rrp2-1 mutation is not synthetically lethal with the rex4-null allele. In fact, the growth rate of FVY010A cells at the permissive temperature does not differ significantly from that of the parent D308 strain (data not shown).
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference/Remark |
| YJV103 | Matα/a;ade2/ade2, ade3/ADE3, his3/his3,leu2/leu2, ura3/ura3,trp1/TRP1, SNR10/snr10::LEU2 | Venema and Tollervey 1996 |
| YJV154 | Matα;ade2, leu2, his3, trp1, ura3, Gal::rrp5(URA3) | Eppens et al. 1999 |
| YJV159 | Mata;ade2, ade3, leu2, his3, trp1, ura3 | Segregant of YJV103 |
| YJV204 | Matα;ade2, leu2, his3, trp1, ura3, Gal::rrp5, p(protA-rrp5-Δ3-TRP1) | Eppens et al. 2002 |
| D308 | Matα;ade2, ade3, leu2his3, trp1, ura3, rrp2-1 | Gift of Dr. Tollervey |
| HI227 | Mata;leu2, his3, trp1, lys3, ura3, rnt1::HIS3 | Abou-Elela et al. 1996 |
| Y04999 | Mata;leu2, his3, met15, ura3, dbr1::KANMX4 | Euroscarf |
| YAV41 | Mata;leu2, his3, trp1, ura3, cup1::LEU2, rex4::KAN | Gift of Dr. Van Hoof |
| FVY02C | Matα;leu2, his3, trp1, ura3, rex4::KAN | Eppens et al. 2002 |
| FVY03B | Mata;leu2, his3, trp1, ura3, rex4::KAN | Eppens et al. 2002 |
| FVY07A | Mat ?; leu2, his3, trp1, ura3, Gal::rrp5, rnt1::HIS3, p(protA-rrp5-Δ3-TRP1) | This article |
| FVY010A | Mat ?; leu2, his3, trp1, ura3, rex4::KAN, rrp2-1 | This article |
| FVY20 | Mat ?; leu2, his3, ura3, rrp2-1, dbr1::KANMX4 | This article |
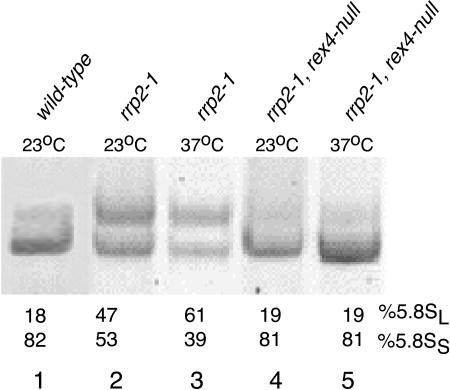
FVY010A and D308 cells were grown in YPD at 23°C and then shifted to 37°C. Total RNA was isolated immediately before and 8 h after the shift and analyzed by PAGE. As can be seen in Figure 2, even at 23°C the rrp2-1 single mutant overproduces the long species to ~50% of the total amount of 5.8S rRNA (Fig. 2, lane 2), while 5.8SL rRNA becomes predominant at 37°C (Fig. 2, lane 3; Lygerou et al. 1994). In the rrp2-1/rex4-null double mutant, however, the 5.8SS: 5.8SL rRNA ratio returns to the value characteristic of wild-type cells irrespective of the growth temperature (Fig. 2, lanes 4,5). Thus, as in the rrp5-Δ3 mutant (Eppens et al. 2002), the preferential use of the long processing pathway in the rrp2-1 strain depends upon the presence of Rex4p.
FIGURE 2.
5.8S rRNA levels in the D308 (rrp2-1; lanes 2,3) and FVY010A (rrp2-1/rex4-null; lanes 4,5) mutant strains. Cultures of the two strains were grown in liquid YPD medium at 23°C to midexponential phase and then shifted to 37°C. Total RNA was isolated from cells harvested immediately before (lanes 2,4) and 8 h after (lanes 3,5) the shift and analyzed on an 8% polyacrylamide gel. The gel was stained with ethidium bromide and the relative levels of 5.8SS and 5.8SL rRNA were determined by scanning.
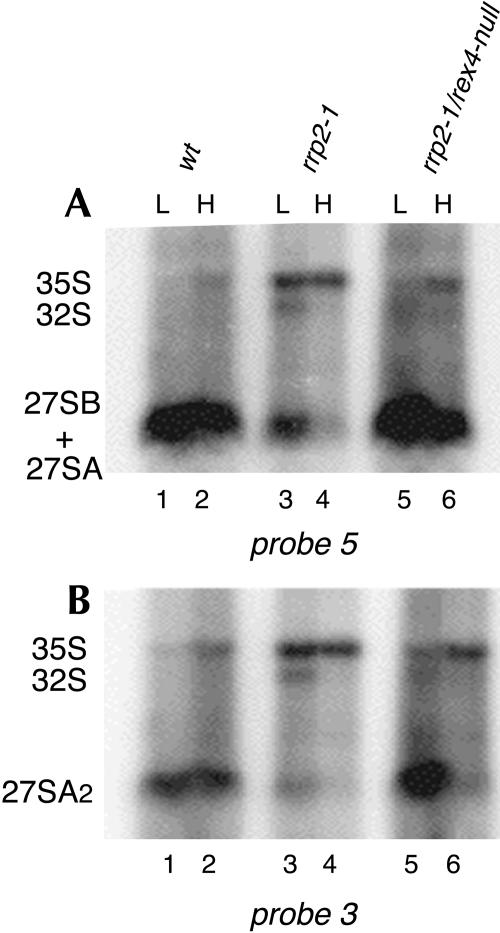
Figures 3 and 4 depict the results of Northern analysis of the same RNA samples using probes that hybridize to various regions between sites A2 and C2 (cf. Fig. 1C). Analysis of the large pre-rRNA species (Fig. 3) shows that in the rrp2-1 cells the level of both the 27SA and 27SB precursors is strongly reduced relative to the wild-type control even at 23°C and these precursors become virtually undetectable at 37°C (Fig. 3, cf. lanes 1,2 and lanes 3,4). In the rrp2-1/rex4-null double mutant at 23°C the levels of these pre-rRNA species return to normal (Fig. 3, lane 5), while at 37°C we see a strongly reduced level of 27SA, a normal level of 27SB, and a slight increase in 35S pre-rRNA (Fig. 3, lane 6). We did not see any significant changes in the levels of 18S and 25S rRNA (data not shown).
FIGURE 3.
Northern analysis of high-molecular-weight pre-rRNA isolated from wild-type YJV159 cells (lanes 1,2), strains D308 (rrp2-1; lanes 3,4), and FVY010A (rrp2-1/rex4-null; lanes 5,6). The strains were grown on YPD medium and shifted from 23°C to 37°C at midexponential phase. Total RNA was isolated from cells harvested immediately before (L) and 8 h after (H) the shift, separated on a 1.2% agarose gel, and subjected to Northern analysis using probes 5 (A) and 3 (B).
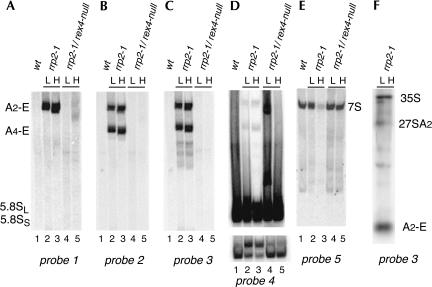
FIGURE 4.
Northern analysis of low-molecular-weight processing intermediates in strains D308 (rrp2-1; lanes 2,3) and FVY010A (rrp2-1/rex4-null; lanes 4,5). Cells were grown on YPD medium and shifted from 23°C to 37°C at midexponential phase. Total RNA was isolated from cells harvested immediately before (L) and 8 h after (H) the shift, separated on an 8% poly-acrylamide gel, and subjected to Northern analysis. (A–E) Northern hybridization with the various probes as indicated. The same gel was hybridized sequentially with probes 4, 3, 1, 2, and 5. Lane 1 contains RNA from exponential growing wild-type cells (YJV159) grown at 23°C. The lower portion of D shows a shorter exposure of the region containing the 5.8S rRNA bands of same blot. (F) Northern hybridization using probe 3 of total RNA isolated from rrp2-1 mutant cells at either 23°C (L) or 37°C (H) and separated on a 1.2% agarose gel to visualize the relative levels of the 27SA2 pre-rRNA and the A2-E fragment.
Further analysis of the samples on an 8% PAA gel (Fig. 4) reveals that the rrp2-1 mutant accumulates a fragment of about 300 nt that hybridizes with probes 1, 2, and 3, located between sites A2 and B1, at both the permissive (L) and nonpermissive (H) temperature (Fig. 4A–C, lanes 2,3), which is absent from the wild-type control (Fig. 4A, lane 1). The same fragment is also detected in the mutant, but not the wild-type cells, by probe 4, located within the mature 5.8S rRNA sequence (Fig. 4D, lanes 2,3). The signal at the same position in Figure 4E, using probe 5, which hybridizes only to precursor species extending beyond site E, corresponds to the normal 7S precursor of 5.8S rRNA (295 nt), as it is also detected in wild-type cells (Fig. 4E, lane 1). We conclude that the fragment in the rrp2-1 cells visualized by probes 1–4 likely extends from site A2 to site E. As shown in Figure 4F, which depicts an agarose gel hybridized with probe 3, the levels of fragment A2-E substantially exceed those of the 27S precursor species. Thus, the almost complete disappearance of the 27SA and 27SB precursors from rrp2-1 mutant cells (Fig. 3) is accompanied by the strong accumulation of a product having the characteristics of a fragment extending from site A2 to the 3′ end of 5.8S rRNA. Together, these data constitute evidence that, like the rrp5-Δ3 mutation (Eppens et al. 2002), the rrp2-1 mutation reverses the normal order of ITS1 and ITS2 processing (Shuai and Warner 1991; Lindahl et al. 1992).
In the rrp2-1/rex4-null double mutant the A2-E fragment cannot be detected at either temperature (Fig. 4A–E), which, together with the reappearance of the 27S species in the double mutant cells (Fig. 3), leads us to conclude that the reverse order of ITS1 and ITS2 processing induced by the rrp2-1 mutation depends upon the presence of Rex4p, as it does in the rrp5-Δ3 mutant (Eppens et al. 2002). Inactivation of the REX4 gene also redresses a further deviation from the wild-type pre-rRNA processing phenotype in the rrp2-1 mutant, namely, the strong reduction in the steady-state level of 7S pre-rRNA of cells grown at 37°C (Fig. 4E, cf. lanes 3 and 5).
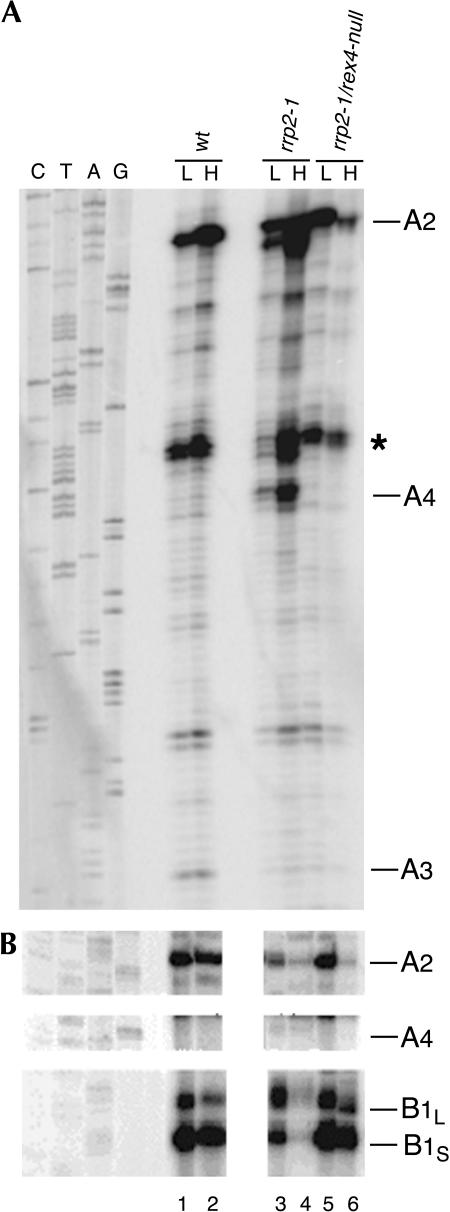
At both 23°C and 37°C probes 2, 3, and 4 detect an additional product in rrp2-1 cells that migrates somewhat faster than the A2-E fragment (Fig. 4B–D, lanes 2,3). This product does not hybridize to either probe 1 (panel A) or probe 5 (panel E), indicating that its 5′ and 3′ ends do not extend beyond sites A4 and E, respectively. The mobility of this fragment indeed corresponds to a product having the size of the A4-E fragment (265 nt). Like the A2-E fragment, the A4-E fragment is absent from rrp2-1/rex4-null mutant cells (Fig. 4A–D, lanes 4,5). These observations extend the resemblance between the processing phenotypes caused by the rrp2-1 and rrp5-Δ mutations even further by including Rex4p-dependent processing at site A4. Previous analysis of pre-rRNA in the original rrp2-1 mutant did detect the A2-E, but not the A4-E fragment (Shuai and Warner 1991; Lindahl et al. 1992; Schmitt and Clayton 1993). The reasons for this discrepancy are not clear.
The conclusions drawn above are fully corroborated by primer extension analysis of total RNA isolated from D308 (rrp2-1) and FVY010A (rrp2-1/rex4-null) cells grown at either 23°C or 37°C (Fig. 5). Using probe 3, we observe a clear stop, corresponding to site A4 with RNA isolated from the rrp2-1 single mutant, in particular at 37°C, while there is no signal at the position of A3 (Fig. 5A, lanes 3,4). In the rrp2-1/rex4-null double mutant the signal corresponding to A4 disappears, although as expected, processing at site A3 is not restored (Fig. 5A, lanes 5,6). These data are unequivocal evidence that in rrp2-1 cells ITS1 is processed at site A4 and that this processing depends upon the presence of Rex4p. Primer extension analysis of the same samples using probe 5 (Fig. 5B) shows that relative to wild-type cells (Fig. 5B, lanes 1,2) the 27SA2 pre-rRNA levels are reduced in rrp2-1 single mutant cells at 23°C (Fig. 5B, lane 3) and even further at 37°C (Fig. 5B, lane 4). Furthermore, the total level of precursors with a 5′ end at B1L or B1S (i.e., 27SB and 7S pre-rRNA) is also strongly reduced at both temperatures and processing at B1L is favored. The rrp2-1/rex4-null double mutant, on the other hand, contains approximately wild-type steady-state levels of these precursors at both temperatures, except for a strong reduction in 27SA2 pre-rRNA at 37°C (Fig. 5B, lanes 5,6). Moreover, double mutant cells grown at either temperature show a wild-type B1S:B1L ratio. Since in the rrp2-1/rex4-null cells there is no processing at either A3 or A4, both the (major) 27SBS and the (minor) 27SBL precursor must be generated from 27SA2 pre-rRNA. We do not detect 27SA4 pre-rRNA in rrp2-1 cells (Fig. 5B, lanes 3,4), suggesting that this precursor is either rapidly processed to 27SB or, more likely, in view of the strong accumulation of the A2-E fragment in these cells (Fig. 4), that A4 processing occurs on the latter intermediate.
FIGURE 5.
Primer extension analysis of pre-rRNA isolated from strains D308 (rrp2-1; lanes 3,4) and FVY010A (rrp2-1/rex4-null; lanes 5,6). Total RNA was isolated from cells immediately before (L) and 8 h after (H) the shift to the nonpermissive temperature and analyzed by primer extension using probe 4 (A) or probe 5 (B). The positions of the stops corresponding to the different processing sites are indicated. Lanes 1 and 2 show the controls using RNA from wild-type YJV159 cells treated in the same manner. The band indicated by the asterisk represents an artificial stop.
In summary, the data presented above further support a role for Rex4p in ITS1 processing and in particular in determining the choice between the processing pathways leading to the two alternative forms of 5.8S rRNA. Moreover, they constitute additional evidence that site A4 is a genuine processing site in yeast ITS1, the use of which depends upon the presence of intact Rex4p. Processing at this site may in fact occur in wild-type cells where, however, it would go unnoticed because of the rapid conversion of most of the 27SA2 precursor in 27SA3 pre-rRNA (see Discussion).
Inactivation of RNT1 restores a wild-type pre-rRNA processing phenotype in rrp5-Δ3 mutant cells
The occurrence of processing at site A4 in rrp2-1 cells, clearly excludes RNase MRP as the responsible nuclease. Interestingly, however, comparison of the region surrounding A4 with that spanning the processing site in the 3′-ETS cleaved by Rnt1p revealed a considerable degree of structural similarity, suggesting another attractive candidate, even though the stem containing the A4 site is not a canonical Rnt1p recognition site (Chanfreau et al. 1998a; Lebars et al. 2001; Lamontagne and Abou Elela 2004) and cleavage appears to occur only on the 5′ side of the stem. However, cleavage of noncanonical substrates by Rnt1p has been reported, though recognition of one such substrate appeared to require a protein chaperone (Giorgi et al. 2001).
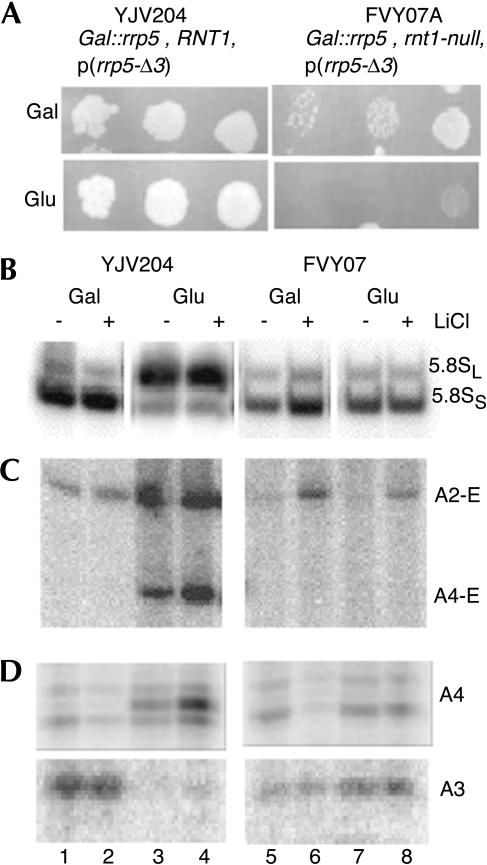
To investigate the possible involvement of Rnt1p in cleavage at A4 we transformed strain FVY07A, which carries an rnt1-null as well as a GAL::rrp5 allele on its genome (Table 1), with a plasmid carrying the rrp5-Δ3 allele behind its own promoter. The resulting transformants can, thus, be rendered dependent upon the mutant Rrp5-Δ3p protein by shifting them from a galactose- to a glucose-based medium. Strain YJV204 [RNT1, GAL::rrp5, p(rrp5-Δ3)] was used as a control. Total RNA was isolated before and 8 h after the shift either directly or after an additional treatment of the culture with 0.2 M LiCl for 1 h to inactivate 5′ → 3′ exonucleases (Dichtl et al. 1997) and thus stabilize processing intermediates, and subjected to Northern analysis.
As shown in Figure 6, combining the rnt1-null and rrp5-Δ3 alleles results in a phenotype strikingly similar to the one exhibited by the rrp5-Δ3/rex4-null mutant (Eppens et al. 2002). First, the rnt1-null mutation is synthetically lethal with the rrp5-Δ3 deletion as FVY07A transformants are unable to grow on glucose (Fig. 6A). Second, the steady-state levels of 5.8SS and 5.8SL rRNA in the rrp5p-Δ3/rnt1-null double mutant cells return to their wild-type value (Fig. 6B, cf. lanes 7,8 and lanes 3,4). Third, RNT1 inactivation causes a strong reduction in the “aberrant” A2-E fragment as well as the disappearance of the A4-E fragment (Fig. 6C, cf. lanes 7,8 and lanes 3,4), indicating restoration of the normal order of ITS1 and ITS2 processing and loss of processing at site A4. Primer extension analysis using probe 3 shown in Figure 6D confirms the loss of a signal at site A4 and restoration of A3 cleavage in the rrp5p-Δ3/rnt1-null double mutant (Fig. 6D, cf. lanes 7,8 and lanes 3,4).
FIGURE 6.
Effect of inactivation of the RNT1 gene on growth and pre-rRNA processing in cells dependent upon the Rrp5-Δ3p mutant protein. Strain YJV204 [Gal::rrp5/RNT1/p(rrp5-Δ3)] and FVY07A [Gal::rrp5/rnt1::KAN/p(rrp5-Δ3)] were grown on galactose-based medium to midexponential phase and plated in 10-fold serial dilutions on either galactose or glucose plates (A). The same strains were grown on galactose-based medium and shifted to glucose-based medium. Samples were taken immediately before and 8 h after the shift, either directly (- LiCl) or after treatment of the culture with 0.2 M LiCl for 1 h (+ LiCl). Total RNA was isolated and subjected to Northern analysis using probe 4 (B) or probe 2 (C) or primer extension analysis using probe 4 (D).
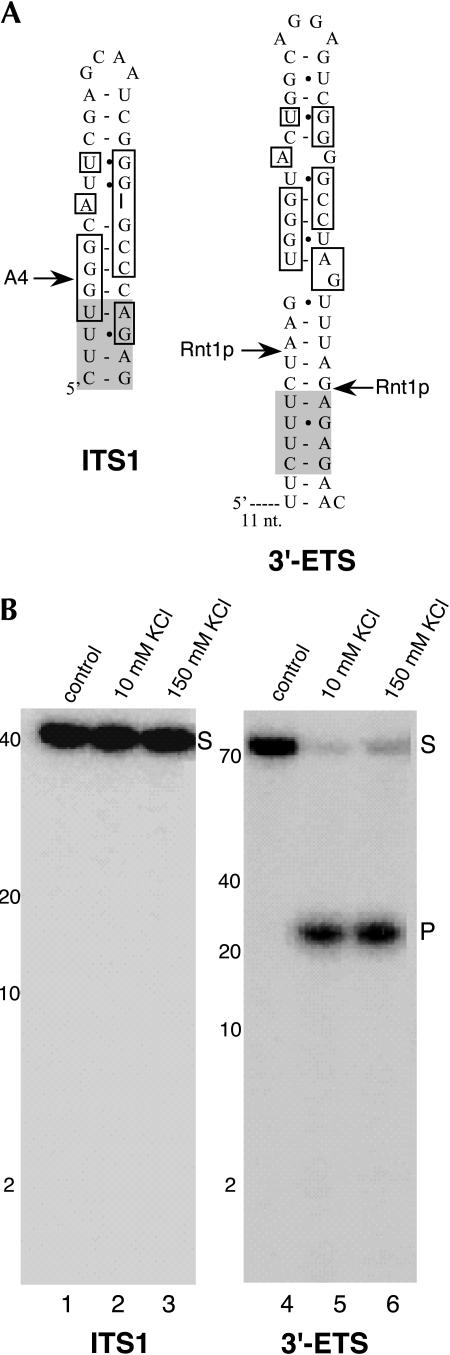
The fact that the signal corresponding to A4 disappears upon inactivation of Rnt1p would support the direct participation of the endonuclease in processing at this site. Therefore, we tested whether Rnt1p is able to cleave a model substrate containing A4 in vitro. To that end we prepared in vitro transcripts (Fig. 7A) representing either the region spanning site A4 or the stem–loop containing the established Rnt1p cleavage site within the 3′-ETS of yeast pre-rRNA (Abou-Elela et al. 1996; Kufel et al. 1999) and, after 5′-end labeling, incubated both transcripts with purified Rnt1p. As shown in Figure 7B, the enzyme efficiently cleaved the 3′-ETS transcript at the known site at both low (10 mM) and high (150 mM) salt concentration (Fig. 7B, lanes 2,3). However, we failed to detect any cleavage of the ITS1 transcript (Fig. 7B, lanes 2,3). Since recruitment of Rnt1p to the noncanonical A4 site may depend upon other factors (Giorgi et al. 2001), we added wild-type cell extract to the in vitro incubation mixture, but without success (data not shown). While these data would suggest that Rnt1p does not directly cleave site A4, they can not be taken as conclusive evidence. For instance, the enzyme might require a factor not present in adequate amounts in the cell extract or this factor might bind to a more distant region of the pre-rRNA lacking from the model substrate.
FIGURE 7.
In vitro analysis of the cleavage of model substrates representing the Rnt1p cleavage site in the 3′-ETS or site A4, respectively. (A) Structural comparison of the model substrates. Conserved nucleotides are boxed. The shaded box highlights the nucleotides in the ITS1 stem that are strongly conserved among various yeast strains (Eppens et al. 2002). (B) Transcripts corresponding to the model substrates were prepared by in vitro transcription, 5′-end labeled, and tested for cleavage by incubating them with 0.8 pmol of recombinant Rnt1p in the presence of two different concentrations of KCl. Cleavage products were fractionated on 20% denaturing polyacrylamide gels. The RNA molecular weight marker is shown on the left. The positions of the substrate (S) and product (P) are indicated.
The pre-rRNA processing phenotype of the rrp2-1 mutant depends upon the lariat debranching enzyme Dbr1p
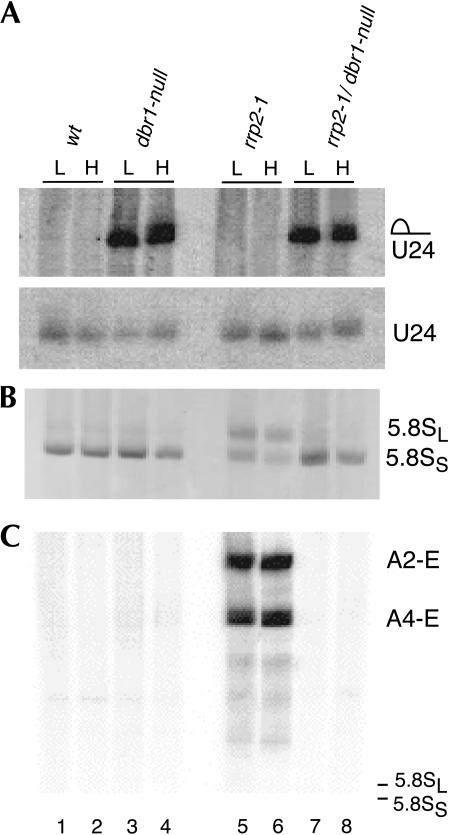
Because both Rnt1p (Chanfreau et al. 1998a,b; Giorgi et al. 2001; Lee et al. 2003) and the Rex4p-related family members Rex1p, Rex2p, and Rex3p (Van Hoof et al. 2000) are involved in snoRNA biogenesis, we speculated that the very similar effect of inactivating either the RNT1 or the REX4 gene on ITS1 processing in rrp5-Δ3 or rrp2-1 cells might arise from a common involvement of the two enzymes in the production of a particular snoRNA. Therefore, we decided to investigate the effect of combining the rrp2-1 mutation with a null allele of the DBR1 gene, which encodes the RNA lariat debranching enzyme required for biogenesis of intron-encoded snoRNAs (Ooi et al. 1998; Petfalski et al. 1998). To that end we crossed strain D308 (rrp2-1) with Y04999 (dbr1::KANMX4) to obtain the double mutant strain FVY20. Northern analysis of total RNA isolated from each of these mutant strains both before and after a shift from 23°C to 37°C is shown in Figure 8. Panel A documents the successful inactivation of Dbr1p as the mutant cells accumulate large amounts of the lariat containing the U24 snoRNA (Ooi et al. 1998). The data shown in panels B and C clearly demonstrate that the pre-rRNA processing phenotype of the rrp2-1 mutant depends, in addition to Rex4p, also upon the presence of Dbr1p. Whereas failure to produce the debranching enzyme has no detectable effect by itself (Fig. 8B,C, cf. lanes 3,4 and lanes 1,2), the lack of Dbr1p in rrp2-1 cells shifts 5.8S rRNA production back to the short form as the predominant species and causes the disappearance of the A2-E and A4-E products (Fig. 8B,C, cf. lanes 7,8 and lanes 5,6), indicating loss of A4 cleavage and restoration of the normal order of ITS1 and ITS2 processing. Together, the data presented in this and our previous (Eppens et al. 2002) article, therefore, link a total of three RNA catabolic enzymes, Rex4p, Rnt1p, and Dbr1p, to ITS1 processing and the relative production of 5.8SS and 5.8SL rRNA.
FIGURE 8.
Effect of inactivation of the DBR1 gene on pre-rRNA processing in rrp2-1 mutant cells. Cultures of YJV159 (wild-type; lanes 1,2), Y04999 (dbr1-null; lanes 3,4), D308 (rrp2-1; lanes 5,6), and FVY20 (rrp2-1/dbr1-null; lanes 7,8) were grown in liquid YPD medium at 23°C to midexponential phase and then shifted to 37°C. Total RNA was isolated from cells harvested immediately before (L) and 8 h after (H) the shift and analyzed on an 8% polyacrylamide gel. (A) Northern analysis using a probe complementary to U24 snoRNA. (B) EtBr staining to visualize 5.8S rRNA. (C) Northern analysis using probe 2. The position of the 5.8S rRNA species as determined by EtBr staining is indicated
DISCUSSION
Over 150 non-ribosomal proteins, as well as several snoRNPs, participate in the biogenesis of ribosomes in eukaryotic cells, not counting the guide snoRNPs involved in modification of the pre-rRNA (Fatica and Tollervey 2002; Fromont-Racine et al. 2004; Raué 2004). Most of these trans-acting factors have been found to cause a well-defined defect in pre-rRNA processing when mutated or inactivated and/or have been detected in pre-ribosomal particles, though not all are essential (Raué 2004). Surprisingly, Rex4p satisfies neither of these two criteria (Van Hoof et al. 2000; Eppens et al. 2002), even though it was identified in a synthetic lethality screen with the bona fide trans-acting factor Rrp5p, and shown to be required for the pre-rRNA processing phenotype caused by mutations in this protein that block cleavage at site A3 (Eppens et al. 2002).
The pre-rRNA processing phenotype caused by the rrp2-1 mutation, which inactivates RNase MRP, the endo-nuclease cutting at site A3 (Lindahl and Zengel 1996; Lygerou et al. 1996), is essentially identical to that of the rrp5-Δ3 and -Δ4 mutants. Not only does this mutation shift production of 5.8S rRNA to the long form and reverse the order of removal of the ITS1 and ITS2 sequences from the 27SA2 pre-rRNA, as reported previously (Shuai and Warner 1991; Lindahl et al. 1992; Henry et al. 1994; Figs. 2–5), but we also observe processing of ITS1 at site A4, first detected in the rrp5-Δ3 and −Δ4 mutants (Eppens et al. 2002). Even more strikingly, as it does in the rrp5-Δ mutants, inactivation of the REX4 gene abolishes all of these effects, except—for obvious reasons—lack of A3 cleavage (Figs. 2–5). These data further strengthen the argument that A4 is a genuine processing site, the use of which depends upon the presence of Rex4p. It seems unlikely, however, that Rex4p is directly responsible for this processing event, since the protein shows significant resemblance to 3′ → 5prime; exonucleases (Van Hoof et al. 2000), whereas, as we have argued before (Eppens et al. 2002), processing at A4 is likely to be endonucleolytic.
A substantial structural similarity between the region containing site A4 and the processing site within the 3′-ETS recognized by the endonuclease Rnt1p (Fig. 7) prompted us to investigate the possible involvement of this endonuclease in ITS1 processing. The finding that the rnt1-null and rrp5-Δ3 mutations are synthetically lethal (Fig. 6A) establishes genetic interaction between the RNT1 and RRP5 genes. Moreover, Northern hybridization as well as primer extension analysis shows that the effect of inactivating Rnt1p on pre-rRNA processing in rrp5-Δ3 cells is identical to that of inactivating Rex4p, including loss of A4 processing and restoration of cleavage at site A3 (Fig. 6). However, recombinant Rnt1p failed to cut a model substrate containing the A4 site in vitro under conditions where it faithfully cleaves an RNA fragment containing its known 3′-ETS cleavage site (Fig. 7B). While this does not completely exclude Rnt1p as the endonuclease responsible for A4 processing, the striking resemblance between the effect of Rex4p and Rnt1p inactivation on ITS1 processing and 5.8S rRNA production in rrp5-Δ3 cells rather suggested a common, indirect involvement of the two enzymes. This idea gains further support from the finding that the pre-rRNA processing phenotype of the rrp2-1 mutant not only depends upon Rex4p (Figs. 3, 4) but also upon the presence of the lariat debranching enzyme Dbr1p (Fig. 8). An obvious common denominator of Rnt1p and Dbr1p is their involvement in snoRNA biogenesis (Chanfreau et al. 1998a,b; Giorgi et al. 2001; Lee et al. 2003). For Rex4p a role in snoRNA biogenesis has yet to be demonstrated, but is plausible in view of the established functions of the other members of the Rex family (Van Hoof et al. 2000). Therefore, we suggest that the involvement of Rex4p, Rnt1p, and Dbr1p in ITS1 processing demonstrated by our data could be due to their nonoverlapping requirement for the formation of a common trans-acting component of the ITS1 processing/assembly machinery, quite possibly an as yet to be identified snoRNP, and propose a model based on this suggestion.
A model
The model, outlined in Figure 9, proposes that the ITS1 processing/assembly machinery containing the postulated snoRNP acts as a barrier to 5′ → 3′ exonuclease digestion of 27SA2 (or A2-E) pre-rRNA by Rat1p and/or Xrn1p, explaining why ITS1 processing shifts to the long pathway upon inhibition of cleavage at site A3 by either the rrp5-Δ3 or rrp2-1 mutation (Fig. 9B). Apparently, processing at sites B1L as well as A4, which according to the model should occur also in wild-type cells, is slow relative to cleavage at C2 and further ITS2 processing, leading to a severe reduction in the levels of the 27SA and 27SB precursors and the accumulation of the A2-E and A4-E fragments (Figs. 4, 5). The former would be converted into 5.8SL rRNA, while exonucleolytic processing of the A4-E fragment by Rat1p and/or Xrn1p could explain the remaining production of 5.8SS rRNA. The latter assumption is supported by the increase in the level of the A4-E fragment observed upon indirect inhibition of these exonucleases by LiCl (Fig. 6C). In wild-type cells highly efficient cleavage of 27SA2 at site A3 obscures processing at site A4.
FIGURE 9.
Model explaining the effect of the various mutations on ITS1 processing in yeast. For reasons of clarity only the postulated snoRNP and not the remainder of the processing/assembly machinery is shown. The position of the snoRNP is arbitrary and does not necessarily indicate its direct interaction with, or position relative to, the spacer. The type size of the processing steps indicated at the right indicates their approximate relative frequency. See text for further details.
Inactivation of Rex4p, Rnt1p, or Dbr1p would, each independently, result in loss of, or at least a substantial lowering of, the level of the snoRNP, either because it reduces production of the mature snoRNA or because the accumulating pre-snoRNA titrates an essential protein component of the snoRNP. The consequent change in the architecture of the processing/assembly machinery would affect ITS1 processing in three distinct ways: loss of processing at A4, removal of the barrier against 5′ → 3′ exonucleolytic digestion of the 27SA2 precursor, and, though only in rrp5-Δ3 cells, reenabling of cleavage at site A3. Apparently, the presence of the Rrp5-Δ3p mutant protein in the ITS1 processing/assembly machinery blocks access of this enzyme to its cleavage site, which is restored by loss of the postulated snoRNP. In otherwise wild-type cells the effects of REX4, RNT1, or DBR1 inactivation would go essentially unnoticed because cleavage at A3 proceeds normally (Fig. 9C). In the rrp5-Δ3 mutants the reenabling of processing at A3 leads to a similar situation (Fig. 9D). In the rrp2-1/rex4-null and rrp2-1/dbr1-null mutants A3 cleavage remains blocked, and thus most of the 27SA2 precursor is rapidly converted into 27SBS pre-rRNA by exonucleolytic digestion, which occurs at a faster rate than processing at B1L (Fig. 9E).
What would be the biological relevance of such a non-essential snoRNP? A possible answer to this important question might lie in our observation that inactivating the REX4 or the DBR1 gene does in fact cause a directly observable phenotype, namely antibiotic hypersensitivity. The growth rate of rex4-null or dbr1-null single mutant cells is significantly retarded by cycloheximide at concentrations that do not affect the wild-type parent strain (A.W. Faber, unpubl.). Such increased antibiotic sensitivity points to a relatively subtle structural change in the (pre-)ribosomes as a result of a disturbance in their assembly (Kressler et al. 1997; Benard et al. 1998; Ho and Johnson 1999; Zabetakis 2000; Moy and Silver 2002). Thus, the postulated snoRNP might have a “hidden” role in keeping ribosome biogenesis on track, which becomes important only under adverse conditions such as exposure of the cells to antibiotics.
We have examined biogenesis of a number of snoRNAs, including U3, U14, snR10, and snR30, in cells carrying an inactivated rex4, rnt1, or drb1 allele, but so far have not been able to identify any alterations common to all three types of mutants.
The model presented above is the result of applying Occam’s razor and we are aware of alternative, even more indirect, explanatory scenarios: e.g., the putative snoRNA might be required for modification of a non-rRNA target that plays a role in pre-rRNA processing or Rex4p, Rnt1p, and Dbr1p might all be required for maturation of a particular mRNA specifying a protein component of the processing/assembly machinery. Hopefully, experiments currently in progress in our laboratory will further clarify these issues.
MATERIALS AND METHODS
Strains and plasmids
Strains used in this study are listed in Table 1. To obtain strain FVY010A (rrp2-1/rex4::KAN) we first crossed YJV154 (Eppens et al. 1999) with YAV41 (rex4::KAN) (Van Hoof et al. 2000), sporulated the resulting diploids, and selected on plates containing uracil and geneticin to obtain FVY02C (Mata, rex4::KAN). The latter was then crossed with strain D308 (Matα, rrp2-1) and rrp2-1/rex4-null double mutants were selected from the spores by checking for resistance against geneticin and inability to grow at 37°C.
To obtain strain FVY07A (GAL::rrp5(URA3), rnt1::HIS3) we crossed YJV154 (Matα, GAL::rrp5(URA3)) with HI227 (Mata, rnt1::HIS3) and selected for spores that were prototrophic for both uracil and histidine as well as unable to grow at 37°C.
Strain FVY20 (rrp2-1, dbr1::KAN) was constructed by crossing strain Y04999 (Mata, dbr1::KANMX4) with strain D308 (Matα, rrp2-1), followed by selection for temperature sensitivity and resistance against geneticin and finally Southern hybridization to ascertain the disruption of the DBR1 gene.
RNA analysis
Isolation of RNA, Northern hybridization, and primer extension analysis were carried out as described previously (Venema et al. 2000). The positions of the different primers used are indicated in Figure 2A. Their sequences were as follows: 1, GATATGAAA ACTCCACAGTG; 2, TTTGGGCATTCGAGCAATCGG; 3, CCAG TTACGAAAATTCTTGTTTTTGAC; 4, CTGCGTTCTTGATCGA TGCG; 5, GAATGTTTGAGAAGGAAATGACGCTC.
In vitro analysis of Rnt1p activity
RNA transcripts used for cleavage were generated and 5′-end labeled with γ-32P-ATP as described before (Lamontagne and Abou Elela 2001; Lamontagne et al. 2003). In vitro cleavage using purified Rnt1p was performed as described by Lamontagne and Abou Elela (2001). Briefly, 0.8 pmol of recombinant Rnt1p were incubated at 30°C with trace amounts of 5′-end labeled RNA in 20 μL of reaction buffer (30 mM Tris-HCl at pH 7.5, 5 mM spermidine, 10 mM MgCl2, 0.1 mM DTT, 0.1 mM EDTA, containing either 10 mM or 150 mM KCl). Reaction products were analyzed by polyacrylamide gel electrophoresis.
Acknowledgments
We thank Drs. Tollervey, Parker, and Van Hoof for providing us with strains and plasmids used in these experiments.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7155904.
REFERENCES
- Abou-Elela, S., Igel, H., and Ares Jr., M. 1996. RNAase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85: 115–124. [DOI] [PubMed] [Google Scholar]
- Allmang, C., Henry, Y., Morrissey, J.P., Wood, H., Petfalski, E., and Tollervey, D. 1996a. Processing of the yeast pre-rRNA at sites A2 and A3 is linked. RNA 2: 60–73. [PMC free article] [PubMed] [Google Scholar]
- Allmang, C., Henry, Y., Wood, H., Morrissey, J.P., Petfalski, E., and Tollervey, D. 1996b. Recognition of cleavage site A2 in the yeast pre-rRNA. RNA 2: 51–62. [PMC free article] [PubMed] [Google Scholar]
- Allmang, C., Mitchell, P., Petfalski, E., and Tollervey, D. 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 28: 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard, L., Carroll, K., Valle, R.C.P., and Wickner, R.B. 1998. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol.Cell. Biol. 18: 2688–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.E. and Sachs, A.B. 1998. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 18: 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Legrain, P., and Jacquier, A. 1998a. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284: 975–988. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., Rotondo, G., Legrain, P., and Jacquier, A. 1998b. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17: 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, S., Archer, R.H., Zengel, J.M., and Lindahl, L. 1994. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl. Acad. Sci. 91: 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur, W.A. and Fournier, M.J. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278: 695–698. [DOI] [PubMed] [Google Scholar]
- Dichtl, B., Stevens, A., and Tollervey, D. 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16: 7184–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon, F., Gallagher, J.E.G., Compagnone-Post, P.A., Mitchell, B.A., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppens, E.A., Rensen, S., Granneman, S., Raué, H.A., and Venema, J. 1999. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA 5: 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppens, E.A., Faber, A.W., Rondaij, M., Jahangir, R.S., van Hemert, S., Vos, J.C., Venema, J., and Raué, H.A. 2002. Deletions in the S1 domain of Rrp5p cause processing at a novel site in ITS1 of yeast pre-rRNA that depends on Rex4p. Nucleic Acids Res. 30: 4222–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber, A.W., van Dijk, M., Raué, H.A., and Vos, J.C. 2002. Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA 8: 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A. and Tollervey, D. 2002. Making ribosomes. Curr. Opin. Cell. Biol. 14: 313–318. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Senger, B., Saveanu, C., and Fasiolo, F. 2004. Ribosome assembly in eukaryotes. Gene 313: 17–42. [DOI] [PubMed] [Google Scholar]
- Geerlings, T., Vos, J.C., and Raué, H.A. 2000. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′ → 3′exonucleases. RNA 6: 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi, C., Fatica, A., Nagel, R., and Bozzoni, I. 2001. Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J. 20: 6856–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., Rybin, V., Baßler, J., Petfalski, E., Strauss, D., Marzioch, M. Schäfer, T., Kuster, B., Tschochner, H., Tollervey, D., et al. 2002. 90S Pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10: 105–115. [DOI] [PubMed] [Google Scholar]
- Granneman, S. and Baserga, S.J. 2004. Ribosome biogenesis: Of knobs and RNA processing. Exp. Cell. Res. 296: 43–50. [DOI] [PubMed] [Google Scholar]
- Henry, Y., Wood, H., Morrissey, J.P., Petfalski, E., Kearsey, S., and Tollervey, D. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13: 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, J.H.-N. and Johnson, A.W. 1999. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 2389–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20: 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler, D., Delacruz, J., Rojo, M., and Linder, P. 1997. Fal1p is an essential dead-box protein involved in 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 7283–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler, D., Doère, M., Rojo, M., and Linder, P. 1999. Synthetic lethality with conditional DBP6 alleles identifies Rsa1p, a nucleo-plasmic protein involved in the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 19: 8633–8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J., Dichtl, B., and Tollervey, D. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamontagne, B. and Abou Elela, S. 2001. Purification and characterization of Saccharomyces cerevisiae Rnt1p nuclease. Methods Enzymol. 342: 159–167. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 279: 2231–2241. [DOI] [PubMed] [Google Scholar]
- Lamontagne, B., Ghazal, G., Lebars, I., Yoshizawa, S., Fourmy, D., and Abou Elela, S.A. 2003. Sequence dependence of substrate recognition and cleavage by yeast RNase III. J. Mol. Biol. 327: 985–1000. [DOI] [PubMed] [Google Scholar]
- Lebars, I., Lamontagne, B., Yoshizawa, S., Abou Elela, S., and Fourmy, D. 2001. Solution structure of conserved AGNN tetraloops: Insights into Rnt1p RNA processing. EMBO J. 20: 7250–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.Y., Lee, A., and Chanfreau, G. 2003. The roles of endonucleolytic cleavage and exonucleolytic digestion in the 5′-end processing of S. cerevisiae box C/D snoRNAs. RNA 9: 1362–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, L. and Zengel, J.M. 1996. RNase MRP and rRNA processing. Mol. Biol. Rep. 22: 69–73. [DOI] [PubMed] [Google Scholar]
- Lindahl, L., Archer, R.H., and Zengel, J.M. 1992. A new rRNA processing mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 20: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou, Z., Mitchell, P., Petfalski, E., Seraphin, B., and Tollervey, D. 1994. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes & Dev. 8: 1423–1433. [DOI] [PubMed] [Google Scholar]
- Lygerou, Z., Allmang, C., Tollervey, D., and Séraphin, B. 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272: 268–270. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., and Tollervey, D. 1996. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes & Dev. 10: 502–513. [DOI] [PubMed] [Google Scholar]
- Moy, T.I. and Silver, P.A. 2002. Requirements for the nuclear export of the small ribosomal subunit. J. Cell Sci. 115: 2985–2995. [DOI] [PubMed] [Google Scholar]
- Ooi, S.L., Samarsky, D.A., Fournier, M.J., and Boeke, J.D. 1998. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: Intron length effects and activity of a precursor snoRNA. RNA 4: 1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petfalski, E., Dandekar, T., Henry, Y., and Tollervey, D. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18: 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raué, H.A. 2004. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevsiae. In The nucleolus (ed. M.O. Olson), pp. 199–222. Kluwer Academic/Plenum Publishers, New York.
- Schmitt, M.E. and Clayton, D.A. 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 7935–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, M.E., Bennett, J.L., Dairaghi, D.J., and Clayton, D.A. 1993. Secondary structure of RNAse MRP RNA as predicted by phylogenetic comparison. FASEB J. 7: 208–213. [DOI] [PubMed] [Google Scholar]
- Shuai, K. and Warner, J.R. 1991. A temperature-sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res. 19: 5059–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, A., Hsu, C.L., Isham, K.R., and Larimer, F.W. 1991. Fragments of the Internal Transcribed Spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′ → 3′ exoribonuclease-1. J. Bacteriol. 173: 7024–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet, C. and Hermann-Le Denmat, S. 2000. Bypassing the rRNA processing endonucleolytic cleavage at site A2 in Saccharomyces cerevisiae. RNA 6: 1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof, A., Lennertz, P., and Parker, R. 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP, and RNase P RNAs in yeast. EMBO J. 19: 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays, E., Gleizes, P.-E., Bousquet-Antonelli, C., Noaillac-Depeyre, J., Caizergues-Ferrer, M., and Gélugne, J.-P. 2001. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essentail non-ribosomal cytoplasmic protein. EMBO J. 20: 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J. and Tollervey, D. 1996. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15: 5701–5714. [PMC free article] [PubMed] [Google Scholar]
- ——— 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 216–311. [DOI] [PubMed] [Google Scholar]
- Venema, J., Vos, H., Faber, A.W., van Venrooij, W.J., and Raué, H.A. 2000. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for the early pre-rRNA processing cleavages and requires box C for its association. RNA 6: 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, H.R., Faber, A.W., De Gier, M.D., Vos, J.C., and Raué, H.A. 2004. Deletion of the three distal S1 motifs of yeast Rrp5p abolishes pre-rRNA processing at site A2 without reducing the production of functional 40S subunits. Eukaryotic Cell (in press). [DOI] [PMC free article] [PubMed]
- Warner, J. 2001. Nascent ribosomes. Cell 107: 133–136. [DOI] [PubMed] [Google Scholar]
- Zabetakis, D. 2000. Inheritance of suppressors of the drug sensitivity of a NSR1 deleted yeast strain. Yeast 16: 147–159. [DOI] [PubMed] [Google Scholar]