Abstract
The signal recognition particle (SRP) is a cytosolic ribonucleoprotein complex that guides secretory proteins to biological membranes in all organisms. The SRP RNA is at the center of the structure and function of the SRP. The comparison of the growing number of SRP RNA sequences provides a rich source for gaining valuable insight into the composition, assembly, and phylogeny of the SRP. In order to assist in the continuation of these studies, we propose an SRP RNA nomenclature applicable to the three divisions of life.
Keywords: signal recognition particle, SRP, nomenclature, comparative sequence analysis, secondary structure
INTRODUCTION
Signal recognition particle (SRP) is a ribonucleoprotein complex that binds to ribosomes during the synthesis and translocation of secretory proteins. SRP directs the polypeptide toward the proper cellular compartment by recognizing first the nascent signal sequence, and then the SRP receptor in the membrane. Translation continues when SRP dissociates from the ribosome/nascent chain complex (RNC), thus allowing the delivery of the secretory protein into the translocation channel (for review, see Lütcke 1995; Keenan et al. 2001; Wild et al. 2002; Zwieb and Eichler 2002; Nagai et al. 2003; Zwieb 2003).
Mammalian SRP consists of the SRP RNA (typically about 300 nucleotides long) and of six proteins named SRP9, SRP14, SRP19, SRP54, SRP68, and SRP72 according to their estimated molecular weights in kilodaltons (Walter and Blobel 1983). SRP9 and SRP14 form the SRP9/14 heterodimer. SRP68 and SRP72 exist bound to each other as SRP68/72. All SRP proteins bind or are in close proximity to the SRP RNA.
The assembled SRP is composed of two structurally and functionally separable domains. The small domain is also named “Alu”, as it contains SRP RNA helices 2–4 and a portion of helix 5, which are homologous to the human repetitive Alu element (Ullu et al. 1982). Together with SRP9/14, these terminal regions of the SRP RNA modulate the elongation rate of the secretory protein. The large domain, also known as the S (for SRP “specific”) domain, is composed of proteins SRP19, SRP54, SRP68/72, SRP RNA helices 6–8, and a moderately conserved portion of helix 5 (Siegel and Walter 1988). Cross-linking experiments demonstrate the close proximity between the nascent polypeptide and SRP54 of the ribosome-bound SRP, indicating that the large domain captures the signal peptide (Lütcke et al. 1992; Zopf et al. 1993).
Due to the spatial separation of the small and large domain by the variable portion of SRP RNA helix 5 (Gundelfinger et al. 1983; Larsen and Zwieb 1991), the mammalian SRP has an overall elongated dumbbell-like shape. The length of the SRP (230–240 Å) as measured by electron microscopy (Andrews et al. 1987) is appropriate for bridging the distance between the ribosome interface and the polypeptide exit site (Zwieb 1989). In the recent visualization of the ribosome–SRP complex by cryoelectron microscopy (Halic et al. 2004), the large domain of SRP is indeed positioned near the nascent peptide exit site, whereas the translation–modulation function of the small SRP domain is elegantly explained by its ability to reach into the elongation factor binding site.
Most bacteria contain an SRP of significantly reduced complexity composed of a small SRP RNA (4.5S RNA) and a homolog of protein SRP54, also called Ffh or P48 (Bernstein et al. 1993). The bacterial SRP RNA forms a simple hairpin that is phylogenetically related to helix 8. NMR has been used to investigate the structure of the conserved portion of Escherichia coli SRP RNA helix 8 (Schmitz et al. 1996), and X-ray crystallography has solved the structure of the core of the E. coli SRP composed of helix 8 and SRP54 (Batey et al. 2000). Although most bacterial SRPs lack the small domain and a functional connection between translation control and signal peptide recognition, certain gram-positive bacteria encode relatively large SRP RNAs. For example, the secondary structure of the 271-nucleotide SRP RNA (scRNA) of Bacillus subtilis is remarkably similar to that of the eukaryotic or archaeal SRP RNAs (Larsen and Zwieb 1991). The histone-like HBsu 10-kDa protein may provide SRP9/14-like functions in these larger bacterial SRPs (Nakamura et al. 1999).
Archaeal SRP RNAs closely resemble their mammalian homologs (Larsen and Zwieb 1991), but only two SRP proteins, namely SRP19 and SRP54, are believed to be part of the archaeal SRP (Bhuiyan et al. 2000). The crystal structure of the large-domain SRP RNA of Methanococcus jannaschii in complex with protein SRP19 (Hainzl et al. 2002) reveals tertiary RNA–RNA contacts equivalent to those observed between the loops of helix 6 and helix 8 of the human SRP (Kuglstatter et al. 2002; Oubridge et al. 2002).
Understanding the molecular biology of SRP RNA is critically dependent on our knowledge of its structure. The availability of high-resolution data obtained by X-ray crystallography and NMR is expected to be limited to the SRP domains of a few organisms. In contrast, comparative sequence analysis is an all-inclusive approach for gaining insights into the structure and function of macromolecules. The previously presented SRP RNA secondary structures are based on the comparison of a small number of bacterial, archaeal, and eukaryotic sequences (Larsen and Zwieb 1991). Since then, the discovery of many more SRP RNA sequences permitted a gradual refinement of the initial models (e.g., see Rosenblad et al. 2003). Nevertheless, three distinct SRP RNA nomenclatures still exist, separated according to their association with the bacterial, archaeal, or eukaryal division. Furthermore, due to their considerable diversity, determining the secondary structures of the protozoan and fungal SRP RNAs has been proven to be difficult.
Recently, the methods for identifying SRP RNA sequences in the genomes of protozoa and fungi have been improved significantly (Rosenblad et al. 2004), and compatible secondary structures have been determined experimentally (van Nues and Brown 2004). On the basis of these advances, we propose a unified SRP RNA nomenclature applicable to every organism.
RESULTS AND DISCUSSION
SRP RNA sequences
The NCBI databases (Pruitt et al. 2003) were searched with BLAST (Altschul et al. 1990) using representative known SRP RNA sequences as queries. SRP RNAs with less pronounced homology were identified as described in Regalia et al. (2002) by combining pattern searches and covariance models using RNABOB (accessible at www.genetics.wustl.edu/eddy/software/#rnabob) and COVE (Eddy and Durbin 1994). The divergent protozoan SRP RNA sequences were from the unicellular green alga Chlamydomonas reinhardtii, the colorless euglenoid Entosiphon sulcatum, the parasites Eimeria tenella and Theileria annulata, and three Plasmodium species. Fungal SRP RNA sequences were from Candida albicans, Candida dubliniensis, Neurospora crassa, Aspergillus nidulans, Yarrowia lipolytica, Schizosaccharomyces pombe, several Saccharomyces species, as well as the osmo-tolerant yeast Zygosaccharomyces rouxii. The primary sources of most of these sequences are acknowledged in Rosenblad et al. (2004) and van Nues and Brown (2004).
Molecules termed sRNA-85 and sRNA-76 coisolate with the SRPs of Leptomonas collosoma and Trypanosoma brucei, respectively (Beja et al. 1993; Ben-Shlomo et al. 1999). Cross-linking and a possible complementarity between sRNA-85 and the 5′ end of the L. collosoma SRP RNA indicate that sRNA may function in place of SRP9 or SRP14. However, the relationship between sRNA and SRP RNA has not been firmly established yet, and thus, the sRNA sequences of the Trypanosomatids are excluded from our analysis.
Comparative sequence analysis
Previously established procedures (Larsen and Zwieb 1991) were used for the initial alignment of the SRP RNA sequences. Close relatives were aligned on the basis of primary structure similarity. This step was followed by aligning each group of sequences collectively against other groups. Sets of conserved nucleotides were then identified and used to align the more variable regions. Finally, regions with little or no primary structure were inspected visually to identify common secondary structural elements. The search for secondary structure features was assisted by MFOLD (Zuker 2003) accessible at http://www.bioinfo.rpi.edu/applications/mfold. Manual adjustments to the SRP RNA alignment were made using the program BioEdit available at http://jwbrown.mbio.ncsu.edu/RNaseP/info/programs/BIOEDIT/bioedit.html.
Base pairs were confirmed or disproved by observing covariances and compensating base changes (CBCs). (A CBC supports the existence of a Watson-Crick or G-U pair, because, during evolution, random single mutations would not have been corrected for by a second change unless required for survival of the organism.) The number of CBCs and mismatches were counted, and a base pair was considered supported if there was at least twice as much positive as negative evidence. The supported base pairs were indicated in the alignment by uppercase letters. Invariant residues provided no information with regard to the existence or absence of a base pair. The alignment and base-pairing rules are implemented in rnadbtools (accessible at http://www.bioinf.au.dk/rnadbtool/), a suite of computer programs that examines base-pair consistency, proposes helix extensions, and suggests alternative supported base pairs (Gorodkin et al. 2001). An abridged version of the alignment containing 96 nonredundant full-length SRP RNA sequences can be obtained from the SRP database online at http://psyche.uthct.edu/dbs/SRPDB/SRPDB.html.
SRP RNA nomenclature
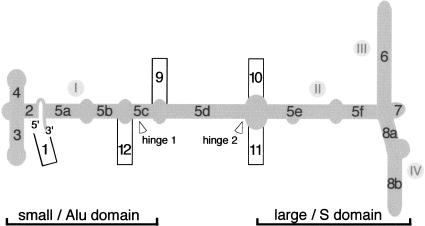
For compatibility with the existing literature, we maintained the original helix numbering system consisting of helices 1–8 (Larsen and Zwieb 1991). The four recently discovered helical insertions in helix 5 were numbered from 9 to 12 (Fig. 1). Segments within a helix were assigned letters in alphabetical order, similar to the conventions used for group 1 introns (Burke et al. 1987). We believe that the terms “Alu” and “S” domain, while originally derived from the relation of mammalian SRP RNA to the Alu repetitive sequences and the SRP RNA-specific insertion, remain useful.
FIGURE 1.
The proposed SRP RNA nomenclature. The mammalian SRP RNA secondary structure is sketched in gray. The 5′ and 3′-ends are indicated. Helices are numbered from 1 to 12 with helical sections labeled with letters a–f. The approximate boundaries of the small (Alu) and the large (S) domain are indicated at the bottom. The use of the terms “domains I” to “domain IV” (indicated by gray dots with roman numbers) is discouraged, as these regions designate RNA secondary structure elements that do not conform to the accepted functional, biochemical, or phylogenetic definition for a domain. In agreement with the conventional numbering of RNA starting from the 5′-end, hinge 1 is encountered first and located near the small domain, whereas hinge 2 is closer to the large domain. (This numbering is different from that used by Halic et al. 2004.)
Representative examples of the updated SRP RNA secondary structures, extracted from the alignment, are shown in Figure 2 in the familiar format. The characteristic properties of each helix and the connecting bulges and loops are discussed below.
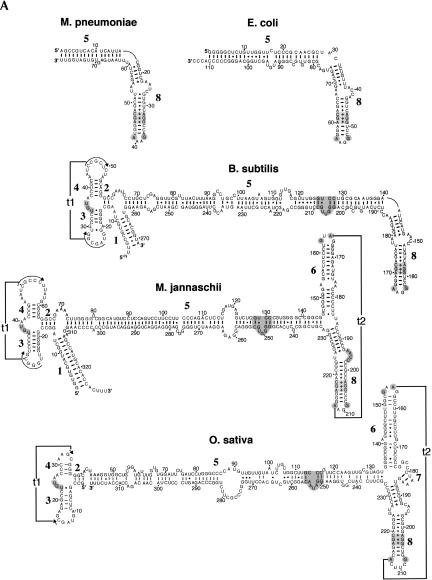
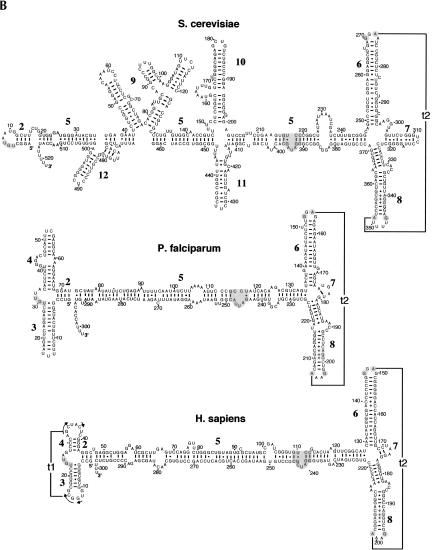
FIGURE 2.
Representative SRP RNA secondary structures. Examples are shown for the bacteria Mycoplasma pneumoniae, Escherichia coli, and Bacillus subtilis, the archaeon Methanococcus jannaschii, the plant Oryza sativa (A), as well as the yeast Saccharomyces cerevisiae, the protozoon Plasmodium falciparum, and Homo sapiens (B). The 5′- and 3′-ends are labeled as such. Helices are numbered from 1 to 12. Residues are numbered in increments of 10. Base pairs supported by comparative sequence analysis (Larsen and Zwieb 1991; Rosenblad et al. 2004) or experiment (Batey et al. 2000; Nissen et al. 2001) are drawn as lines (Watson-Crick), filled circles (G-U pairs), or plus signs (non-Watson-Crick base pairs). Two tertiary interactions between the loops of helices 3 and 4 (t1), and helices 6 and 8 (t2) are indicated. The UGU motif in the small domain, the 5e motif, the conserved G and A of the tetraloop of helix 6, and the three noncanonical base pairs as well as G and A in helix 8 are highlighted in gray (for details, see Fig. 3).
SRP RNA features
Helix 1 pairs the terminal regions in the SRP RNAs of the archaeal and the large bacterial SRP RNAs of Bacillus, Listeria, and Staphylococcus (Fig. 2A). Helix 1 contains between three (Pyrodictium occultum) and 13 (M. jannaschii) base pairs and displays considerable variability in the number of base pairs, even among closely related species. One possible function of helix 1 may be to prevent the unfolding of the SRP RNA under the extreme environmental conditions in which some of these organisms thrive.
Helix 2 consists of three or four base pairs in the archaeal, the majority of eukaryotic, and the larger bacterial SRP RNAs. In the yeasts, the length of helix 2 varies from three (Y. lipolytica, S. pombe) to six base pairs in Saccharomyces kluyveri. In the archaeal and most of the eukaryotic SRP RNAs, base pairing on the 5′ strand of helix 2 is continuous with helix 3, although in some sequences one residue is inserted between the two helices.
Helix 3 is composed of six or seven base pairs in the majority of the SRP RNAs that possess a small domain. In Plasmodium, helix 3 is extended by several base pairs (Fig. 2B). The loop of helix 3 contains six residues in most SRP RNA and is involved in a tertiary interaction (t1) with helix 4. Helix 3 is absent in the fungal SRP RNA sequences known to date.
The proximal base pair of helix 3 is frequently a G–U. This uridine contributes to a motif (marked gray in Fig. 2) with the consensus sequence UGUNR (see also Fig. 3). Typically, the motif connects helices 3 and 4, but it is preserved also in the fungal SRP RNAs which lack helices 3 and 4. In Saccharomyces and related species, a G–U is likely to form the closing base pair of helix 2, but due to its conserved nature, this pairing is neither supported nor disproved (Fig. 2B). The three-dimensional structure of the motif is characterized by a U-turn that is important for the binding of SRP9/14 (Weichenrieder et al. 2000). This sequence also forms the major footprint of the SRP14 homodimer in the S. cerevisiae SRP (Strub et al. 1999). Its conserved nature and broad distribution among the SRP sequences suggests that this sequence is required for the functionality of all of the larger SRP RNAs, even in organisms that lack homologs of SRP9/14.
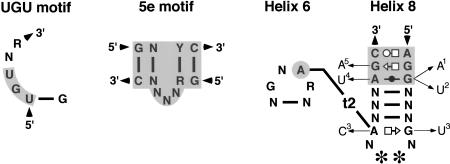
FIGURE 3.
Conserved SRP RNA features. Shown are the UGU motif (left), the 5e motif (middle), and the conserved portions of helices 6 and 8 (right). Invariant or highly conserved bases are indicated by the letters A, C, G, and U; (N) any base; (R and Y) purine and pyrimidine, respectively. Nucleotides present in the SRP RNAs of plants and yeasts are indicated by stars. Noncanonical pairings in helix 8 are shown using the geometric nomenclature of Leontis and Westhof (2001). The tertiary interaction (t2) between the helix 6-tetraloop and helix 8 is indicated. Arrows pointing to subscripted letters represent rare nucleotide changes in (1) Buchnera aphidicola, Deinococcus radiodurans, Neurospora crassa, and Vibrio cholerae; (2) Yarrowia lipolytica; (3) Staphylococcus aureus, Staphylococcus epidermidis, and Lactococcus lactis; (4) Schizosaccharomyces pombe, Aspergillus nidulans; and (5) Tetrahymena rostrata and Tetrahymena thermophila.
Helix 4 is typically composed of four base pairs and a variable number of loop nucleotides. This helix is absent in the fungal RNAs and shortened in Entosiphon and Trypanosoma. An internal loop appears be to present in the more extended helix 4 of the Plasmodium species (Fig. 2B), resulting in two sections, 4a and 4b. (These helical sections are indicated in the alignment provided at the SRPDB.)
Helices 4 and 2 are continuous on their 3′ strands, due to their coaxial arrangement, which is supported by the even distribution of CBCs along the base pairs of the two helices, as well as the crystal structure of the small domain of the human SRP (Weichenrieder et al. 2000). One residue appears to be inserted between helices 2 and 4 in the SRP RNAs of the intracellular protozoan parasite Eimeria tenella and the colorless euglenoid E. sulcatum, but it is currently unclear whether this insertion disrupts helix stacking.
Helix 5 is separated into helical sections 5a–5f by several internal symmetric or asymmetric loops (Fig. 1). As discussed below, additional helical structural elements are inserted into helix 5 in the SRP RNAs of Saccharomyces and other yeasts (Fig. 2B). Sections 5c and 5d, located between the two hinges, connect the small with the large SRP domain. This portion of helix 5 is likely to play a role in the separation of the two functional SRP domains by a distance that is appropriate for the interactions between the SRP and the ribosome (Halic et al. 2004).
Nucleotides corresponding to positions 101–128 and 222–251 of human SRP RNA (Fig. 2B) are constituents of the large SRP domain. About 10 base pairs adjoining helices 6 and 8 display moderate levels of conservation among the eukaryotic SRP RNAs. The presence of the 5e motif (Regalia et al. 2002; Fig. 3) may reflect the need to preserve the RNA-binding sites for SRP68/72. An asymmetric loop of unknown function (corresponding to nucleotides 225–236 in S. cerevisiae, Fig. 2B) is present in the SRP RNAs of Saccharomyces and Z. rouxii.
Helix 6 occurs exclusively in the SRP RNAs of eukaryotes and archaea. Its extensively base-paired character is supported by numerous CBCs. A larger than average number of base pairs is observed in helix 6 of most Saccharomyces species and in Z. rouxii. The 5′-portion of helix 6 of the SRP RNAs of the trypanosomatid L. collosoma and the colorless euglenoid E. sulcatum contain functionally unexplained insertions of CCUCC and CU, respectively.
The apical tetraloop of helix 6 has the consensus sequence GNAR (N is any nucleotide, R is a purine). Although a few exceptions occur in Methanococcus voltae (UUAA), the two Thermoplasma species (AAAG), and Chlamydomonas (UGAU), a loop of four residues and the adenosine at the third position of the tetraloop, are strictly conserved. The crystal structures of the large domain of M. jannaschii SRP RNA with SRP19 (Hainzl et al. 2002; Oubridge et al. 2002) show that this adenosine is involved in a tertiary interaction (t2) with helix 8 (Fig. 3). T2 is required for high-affinity binding of protein SRP19 (Yin et al. 2004).
Helix 7 is exclusive to the eukaryotic SRP RNAs and is usually very short. While CBCs support helix 7, it is absent in the crystal structures (Kuglstatter et al. 2002; Oubridge et al. 2002), indicating that helix 7 might form only temporarily. A significantly larger helix is preceded by a short single strand in Saccharomyces and Z. rouxii (Fig. 2B). Although the phylogenetic relationship of these features is unclear, the single strand may correspond to mammalian helix 7, while the longer helix may represent an insertion peculiar to Saccharomyces and its relatives.
Helix 8, divided into sections 8a and 8b separated by an asymmetric loop, is a prominent aspect of every SRP RNA. Within section 8b, an invariant noncanonical A-C and two highly conserved unusual G-G and G-A pairs (Fig. 3) form a flattened minor groove to accommodate protein SRP54 (Batey et al. 2000). Minor deviations from this pattern occur in the bacteria Buchnera, Deinococcus radiodurans, and Vibrio cholerae, the ciliate Tetrahymena, and the yeasts Y. lipolytica, N. crassa, A. nidulans, and S. pombe. The terminal loop of helix 8 is typically a purine-rich tetraloop, but replaced with a hexanucleotide loop in the SRP RNAs of plants, Saccharomyces, and Z. rouxii. Irrespective of the number of residues within the loop, the first residue is almost always a guanosine, whereas the last residue is a highly conserved adenosine. The conservation of these two residues (G198 and A201 in human SRP RNA, Fig. 2B) is explained by their near coplanar arrangement with A149 of helix 6 (Kuglstatter et al. 2002).
Helices 9–12
In analogy to what has been observed in the ribosomal RNAs (Gerbi 1996) the expansion between helices 6 and 8 (discussed above), as well as four helical insertions into helix 5 (Fig. 1) are responsible for the larger size of the SRP RNAs of Saccharomyces species, Z. rouxii, and other yeasts (Fig. 2B). Helices 9–12 are the most variable portions of the yeast SRP RNAs, both at the primary and secondary structure level. By combining phylogenetic and biochemical analyses, two models have been put forward that differ in the structure and the positions of these insertions (Rosenblad et al. 2004; van Nues and Brown 2004). The structures presented here not only take into account the available expanding phylogenetic information, but are also consistent with previous nuclease sensitivity data (van Nues and Brown 2004) and more recent site-directed mutagenesis studies (R.W. van Nues and J.D. Brown, unpubl.).
Helix 9 is inserted between 5c and 5d near hinge 1 (Fig. 1) and separated into four sections (labeled 9a–9d in the alignment). Section 9a contains the internal RNA polymerase III box B motif crucial for transcription of the S. cerevisiae SRP RNA (Dieci et al. 2002).
Helices 10 and 11. The extensively base-paired helix 10 is present in Z. rouxii, the two SRP RNAs of Y. lipolytica, and the Saccharomyces species, but is truncated in S. kluyveri. The two sections are separated by a variable length loop, which, in some cases (e.g., S. castellii and Z. rouxii), has the potential to base pair. Opposite from helix 10, helix 11 extends from helix 5 in all Saccharomyces species with the exception of S. kluyveri, and may also be present in Y. lipolytica. The junction of helices 10 and 11 with helix 5 may correspond to hinge 2 (Fig. 1) and contains unpaired nucleotides that may contribute to the flexibility required for simultaneously contacting the translation elongation-factor binding and nascent chain exit sites of the ribosome (Halic et al. 2004).
Helix 12, comprised of two sections, is of variable size, located near hinge 1 (Fig. 1), and a constituent of all Saccharomyces SRP RNAs (except for S. castellii, which lacks one section) and Z. rouxii.
Tertiary interactions
The tertiary interaction between the loops of helix 3 and helix 4 in the SRP RNAs of archaea and some bacteria (labeled t1 in Fig. 2) involves up to six well-supported base pairs. Direct evidence for specific pairings has been provided by the crystal structure of the human Alu domain in complex with SRP9/14 (Weichenrieder et al. 2000). The SRP RNAs of fungi and yeasts lack this feature, because the loops are absent or significantly reduced. The helix 3/4 tertiary interaction is missing also in most of the lower eukaryotes. Remnants of t1 may exist in the plant SRP RNAs, but the phylogenetic evidence is weak. We speculate that in the Plasmodium SRP RNAs, the coexisting enlarged helices 3 and 4 (Fig. 2B) may arrange in parallel fashion without an extensive pairing of the loops in order to preserve a tightly folded small SRP domain. Micrococcal nuclease data (van Nues and Brown 2004) support a compact organization of the yeast Alu domain. The lack of more detailed information precludes attempts to suggest tertiary interactions that might determine the three-dimensional arrangement of the yeast Alu domain including insertion elements 9 and 12. However, the footprint of SRP14 on an abbreviated S. cerevisiae RNA indicates weaker protections within helix 9, in addition to the strong protection covering the conserved UGUAA motif (Strub et al. 1999).
A hydrogen bonded A-A pair forms between the loops of helices 6 and 8 of all archaeal and eukaryotic SRP RNAs (tertiary interaction t2 in Figs. 2 and 3). This pairing has been shown to exist in the crystal structures of the large domain of SRP RNA with SRP19 (Oubridge et al. 2002) and the ternary complex with SRP54 (Kuglstatter et al. 2002). Model building (data not shown) suggests that the near coplanar triple configuration of two adenosines and a guanosine residue forms even in the plant and Saccharomyces SRP RNAs, despite that fact that these RNAs contain a hexanucleotide loop that is composed of four pyrimidines flanked by G and A (Fig. 2B).
Outlook
Newly discovered SRP RNA sequences and recent progress in the development of tools capable of finding SRP RNAs in divergent genomes allow the prediction of reliable secondary structures for all SRP RNAs. The previous nomenclature has been expanded to provide a unified basis for the identification and alignment of all SRP RNAs, including the protozoan and yeast sequences. This advance is expected to help in the design of automated procedures for the extraction of SRP RNAs from genome sequences, the refinement of the secondary structures, as well as the discovery of interactions with SRP proteins and other cellular components. Ultimately, this effort will lead to a better understanding of SRP-mediated protein targeting in all organisms.
Acknowledgments
We thank Jan Gorodkin (The Royal Veterinary and Agricultural University, Frederiksberg, Denmark) for help with rnatbtools. This work is supported by NIH grant GM-49034 to C.Z., a Medical Research Council (UK) Senior Non-clinical Research Fellowship to J.D.B., and EU grant QLK3-CT-200-00082 to T.S.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7203605.
REFERENCES
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andrews, D.W., Walter, P., and Ottensmeyer, F.P. 1987. Evidence for an extended 7SL RNA structure in the signal recognition particle. EMBO J. 6: 3471–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey, R.T., Rambo, R.P., Lucast, L., Rha, B., and Doudna, J.A. 2000. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287: 1232–1239. [DOI] [PubMed] [Google Scholar]
- Beja, O., Ullu, E., and Michaeli, S. 1993. Identification of a tRNA-like molecule that copurifies with the 7SL RNA of Trypanosoma brucei. Mol. Biochem. Parasitol. 57: 223–229. [DOI] [PubMed] [Google Scholar]
- Ben-Shlomo, H., Levitan, A., Shay, N.E., Goncharov, I., and Michaeli, S. 1999. RNA editing associated with the generation of two distinct conformations of the trypanosomatid Leptomonas collosoma 7SL RNA. J. Biol. Chem. 274: 25642–25650. [DOI] [PubMed] [Google Scholar]
- Bernstein, H.D., Zopf, D., Freymann, D.M., and Walter, P. 1993. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc. Natl. Acad. Sci. 90: 5229–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan, S., Gowda, K., Hotokezaka, H., and Zwieb, C. 2000. Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res. 28: 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J.M., Belfort, M., Cech, T.R., Davies, R.W., Schweyen, R.J., Shub, D.A., Szostak, J.W., and Tabak, H.F. 1987. Structural conventions for group I introns. Nucleic Acids Res. 15: 7217–7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci, G., Giuliodori, S., Catellani, M., Percudani, R., and Ottonello, S. 2002. Intragenic promoter adaptation and facilitated RNA polymerase III recycling in the transcription of SCR1, the 7SL RNA gene of Saccharomyces cerevisiae. J. Biol. Chem. 277: 6903–6914. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. and Durbin, R. 1994. RNA sequence analysis using covariance models. Nucleic Acids Res. 22: 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi, S.A. 1996. Expansion segment: Regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In Structure, evolution, processing, and function in protein synthesis (eds. R.A. Zimmermann and A.E. Dahlberg), pp. 71–87. CRC Press, New York.
- Gorodkin, J., Zwieb, C., and Knudsen, B. 2001. Semi-automated update and cleanup of structural RNA alignment databases. Bioinformatics 17: 642–645. [DOI] [PubMed] [Google Scholar]
- Gundelfinger, E.D., Krause, E., Melli, M., and Dobberstein, B. 1983. The organization of the 7SL RNA in the signal recognition particle. Nucleic Acids Res. 11: 7363–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl, T., Huang, S., and Sauer-Eriksson, A.E. 2002. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature 417: 767–771. [DOI] [PubMed] [Google Scholar]
- Halic, M., Becker, T., Pool, M.R., Spahn C.M., Grassucci, R.A., Frank, J., and Beckmann, R. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814. [DOI] [PubMed] [Google Scholar]
- Keenan, R.J., Freymann, D.M., Stroud, R.M., and Walter, P. 2001. The signal recognition particle. Annu. Rev. Biochem. 70: 755–775. [DOI] [PubMed] [Google Scholar]
- Kuglstatter, A., Oubridge, C., and Nagai, K. 2002. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat. Struct. Biol. 9: 740–744. [DOI] [PubMed] [Google Scholar]
- Larsen, N. and Zwieb, C. 1991. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 19: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis, N.B. and Westhof, E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke, H. 1995. Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur. J. Biochem. 228: 531–550. [DOI] [PubMed] [Google Scholar]
- Lütcke, H., High, S., Römisch, K., Ashford, A.J., and Dobberstein, B. 1992. The methionine-rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J. 11: 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, K., Oubridge, C., Kuglstatter, A., Menichelli, E., Isel, C., and Jovine, L. 2003. Structure, function and evolution of the signal recognition particle. EMBO J. 22: 3479–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K., Yahagi, S., Yamazaki, T., and Yamane, K. 1999. Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J. Biol. Chem. 274: 13569–13576. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B., and Steitz, T.A. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 98: 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge, C., Kuglstatter, A., Jovine, L., and Nagai, K. 2002. Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol. Cell 9: 1251–1261. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D., Tatusova, T., and Maglott, D.R. 2003. NCBI reference sequence project: Update and current status. Nucleic Acids Res. 31: 34–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalia, M., Rosenblad, M.A., and Samuelsson, T. 2002. Prediction of signal recognition particle RNA genes. Nucleic Acids Res. 30: 3368–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad, M.A., Gorodkin, J., Knudsen, B., Zwieb, C., and Samuelsson, T. 2003. SRPDB: Signal recognition particle database. Nucleic Acids Res. 31: 363–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad, M.A., Zwieb, C., and Samuelsson, T. 2004. Identification and comparative analysis of components from the signal recognition particle in protozoa and fungi. BMC Genomics 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, U., Freymann, D.M., James, T.L., Keenan, R.J., Vinayak, R., and Walter, P. 1996. NMR studies of the most conserved RNA domain of the mammalian signal recognition particle (SRP). RNA 2: 1213–1227. [PMC free article] [PubMed] [Google Scholar]
- Siegel, V. and Walter, P. 1988. Each of the activities of signal recognition particle (SRP) is contained with a distinct domain: Analysis of biochemical mutants of SRP. Cell 52: 39–49. [DOI] [PubMed] [Google Scholar]
- Strub, K., Fornallaz, M., and Bui, N. 1999. The Alu domain homolog of the yeast signal recognition particle consists of an Srp14p homodimer and a yeast-specific RNA structure. RNA 5: 1333–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu, E., Murphy, S., and Melli, M. 1982. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell 29: 195–202. [DOI] [PubMed] [Google Scholar]
- van Nues, R.W. and Brown, J.D. 2004. Saccharomyces SRP RNA secondary structures: A conserved S-domain and extended Alu-domain. RNA 10: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P. and Blobel, G. 1983. Disassembly and reconstitution of the signal recognition particle. Cell 34: 525–533. [DOI] [PubMed] [Google Scholar]
- Weichenrieder, O., Wild, K., Strub, K., and Cusack, S. 2000. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408: 167–173. [DOI] [PubMed] [Google Scholar]
- Wild, K., Weichenrieder, O., Strub, K., Sinning, I., and Cusack, S. 2002. Towards the structure of the mammalian signal recognition particle. Curr. Opin. Struct. Biol. 12: 72–81. [DOI] [PubMed] [Google Scholar]
- Yin, J., Huang, Q., Pakhomova, O., Hinck, A., and Zwieb, C. 2004. The conserved adenosine in helix 6 of signal recognition particle RNA initiates SRP assembly. Archaea 1: 257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf, D., Bernstein, H.D., and Walter, P. 1993. GTPase domain of the 54-kD subunit of the mammalian signal recognition particle is required for protein translocation but not for signal sequence binding. J. Cell Biol. 120: 1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb, C. 1989. Structure and function of signal recognition particle RNA. Prog. Nucleic Acid Res. Mol. Biol. 37: 207–234. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Signal recognition particle-mediated protein targeting. Recent Res. Develop. Mol. Biol. 1: 205–224. [Google Scholar]
- Zwieb, C. and Eichler, J. 2002. Getting on target: The archaeal signal recognition particle. Archaea 1: 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]