Abstract
RNA editing in trypanosomes is a post-transcriptional process responsible for correcting the coding sequences of many mitochondrial mRNAs. Uridines are specifically added or deleted from mRNA by an enzymatic cascade in which a pre-edited mRNA is specifically cleaved, uridines are added or removed, and the corrected mRNA is ligated. The process is directed by RNA molecules, termed guide RNAs (gRNA). The ability of this class of small, noncoding RNA to function in RNA editing is essential for these organisms. Typically, gRNAs are transcribed independent of the their cognate mRNA and anneal to form a binary RNA complex . An exception for this process may be cytochrome oxidase subunit II (COII) mRNA since a gene encoding a trans acting gRNA has not been identified. Using an in vitro editing assay we find that the 3′ UTR of COII, indeed, functions as a guide for both the site and number of uridines added to the coding region of the COII mRNA. We further show that the guiding sequence within the COII 3′ UTR can only function in COII editing when contiguous with the editing substrate, indicating that the 3′ UTR of COII lacks sequence or structure information necessary to function as a trans-acting gRNA. While other RNAs have been shown to “guide” RNA processing reactions, our discovery that the COII 3′ UTR directs editing of its cognate mRNA in cis, is a unique function for a 3′ UTR. The findings described here have led us to propose a new model for the evolution of gRNAs in kinetoplastids.
Keywords: Trypanosoma brucei, RNA editing, uridines, insertion editing, COII, 3′UTR
INTRODUCTION
RNA editing, in the mitochondrion of trypanosomes, is a post-transcriptional event in which uridine residues are added to or deleted from mitochondrial mRNAs (Benne et al. 1986). These changes result in the formation of initiation codons, correction of shifts in open reading frames, and in several cases lead to the creation of entire coding sequence with >60% of the mature RNA being formed by inserted uridines (Blum et al. 1990; Alfonzo et al. 1997; Stuart et al. 2002). There is overwhelming evidence that RNA editing in trypanosomes results from an enzymatic cascade performed by multiprotein complexes. Conceptually, this cascade needs to include enzymes that cut the pre-edited mRNA at editing sites, add or delete uridines, and rejoin the edited RNA. High-molecular-weight complexes containing the enzymes proposed to be necessary for RNA editing have now been described (Schnaufer et al. 2003). These activities include an editing site-specific endonuclease that cleaves pre-edited mRNA, a terminal uridyl transferase or an exoribo-nuclease for addition or deletion of uridine residues, respectively, and RNA ligases for rejoining the pre-edited mRNA molecule (Huang et al. 2001; McManus et al. 2001; Schnaufer et al. 2001; Aphasizhev et al. 2003; Ernst et al. 2003; Panigrahi et al. 2003).
The editing reaction garners its specificity from a class of small non-coding RNAs termed guide RNAs (gRNA) that contain sequences that direct the precise site and number of uridines added or deleted. The 5′ portion of a gRNA contains a region of 9–15 nucleotides (nt) that base-pairs with mRNA sequences immediately 3′ to an editing site. Base-pairing between the gRNA and mRNA results in the formation of a short “anchor duplex” (Blum et al. 1990; Alfonzo et al. 1997; Stuart et al. 2002). It has been proposed that proteins necessary for RNA editing are preassembled on gRNAs and pre-edited mRNAs, respectively. Subsequent formation of the anchor duplex between the gRNA/mRNA effectively recruits these separate ribonucleoprotein complexes to form an active editing complex consisting of 15–20 proteins in total (Madison-Antenucci et al. 2002). Alternatively, the formation of the gRNA/pre-mRNA binary complex may facilitate the recruitment and assembly of editing complex proteins (Rusche et al. 1997; Panigrahi et al. 2001). Each gRNA directs the insertion or deletion of multiple uridines at several adjacent editing sites in an mRNA, progressively increasing the extent of gRNA/mRNA complementarity until a perfect duplex is formed. It is likely that the gRNA/mRNA duplex is then melted by the action of an RNA helicase allowing for the binding of another gRNA to direct the editing of upstream portions of the partially edited mRNA (Missel et al. 1997). This process, while seemingly cumbersome, is essential for mitochondrial function in trypanosomes (Schnaufer et al. 2001).
The key to accurate editing of pre-mRNAs lies in the information contained in the gRNA molecules. The gRNAs are encoded by the mitochondrial genome of trypanosomes, an unusual network structure composed of thousands of catenated circular DNAs termed the kinetoplast. The kinetoplast is composed of two classes of interlocked DNA molecules, the maxicircles and minicircles. There are ~50 maxicircles (20–25 kb) per kinetoplast DNA network, each containing genes traditionally associated with mitochondrial genomes including rRNA genes, genes encoding components of the mitochondrial respiratory pathway, and ATP synthase (Feagin et al. 1988a,b). Minicircles (1–2.5 kb) comprise the bulk of the kinetoplast DNA network and encode most of the gRNAs (Blum and Simpson 1990; Pollard and Hajduk 1991). Therefore, the two RNA components of the editing machinery are usually encoded by separate genomes, and gRNAs are trans-acting elements in the editing reaction. An exception has been proposed for the editing of cytochrome oxidase subunit II (COII) mRNA. No trans-acting gRNA for COII editing has been identified; however, a sequence within the 3′ UTR of the pre-edited COII mRNA transcript potentially serves as a cis-acting guide for the limited editing that occurs in this transcript in trypanosomatids (Blum et al. 1990; van der Spek et al. 1991; Kim et al. 1994; Lukes et al. 1994; Blom et al. 1998). Editing of the COII mRNA results in the addition of only four uridines, inserted at three editing sites, and corrects for a frameshift mutation in the COII gene. The proposed guiding function of the COII 3′ UTR presents a unique opportunity to investigate both the origin of guide-directed RNA editing and evaluate structural constraints specific for cis-and trans-guided reactions.
In this paper the role of the 3′ UTR sequence in COII mRNA editing was directly tested using an in vitro editing assay. We show that both the number and sites of uridine insertion are determined by cis-acting 3′ UTR sequences. In addition, we find that the COII 3′ UTR does not function efficiently as a trans-acting gRNA. We propose that the cis-guiding activity found in COII editing may be representative of early events in the establishment of insertional/deletional editing in trypanosomes.
RESULTS
Phylogenetic conservation of COII editing sites and 3′ UTR
Structural and mutational analysis of minicircle-encoded gRNAs has provided some insight into the mechanism of RNA editing (Schmid et al. 1995; Hermann et al. 1997; Cruz-Reyes et al. 2001). Sequences within the 3′ UTR of several kinetoplastid organisms have previously been shown to be consistent with a cis-acting gRNA sequence (Blum et al. 1990; van der Spek et al. 1991; Kim et al. 1994; Lukes et al. 1994; Blom et al. 1998). Using the information gathered by others, we reinvestigated whether covariation of COII editing site and 3′ UTR sequences is consistent with cis-guided COII mRNA editing in kinetoplastids.
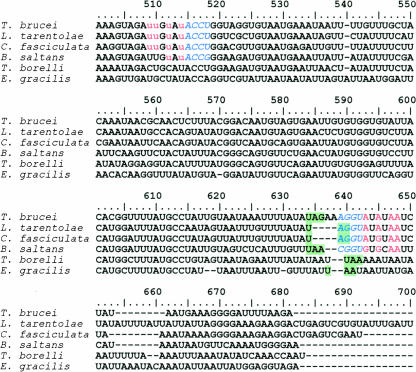
Sequence alignments were performed on pre-edited mRNA sequences from kinetoplastids, which require uri-dine insertional editing for frameshift correction of the COII cryptogene (Fig. 1). Trypanosoma brucei, Leishmania tarentolae, and Crithidia fascilulata all require the insertion of four uridine residues at three editing sites, whereas Bodo saltans requires the insertion of only two uridines, at two editing sites, to correct for a frameshift in the primary transcript. Covariation of sequences within the 3′ UTR and editing sites were consistent with base-pairing interactions between these regions of the COII mRNA. In each case a short, 4-bp duplex was predicted to form between a sequence in the 3′ UTR and a sequence immediately downstream of the first editing site of COII. This is reminiscent of the anchor duplex that forms between trans-acting gRNAs and their cognate mRNAs. In addition, covariation of 3′ UTR and edited mRNA sequences was consistent with the 3′ UTR providing guiding information for insertion of the correct number of uridines at the correct site.
FIGURE 1.
Alignment of edited COII mRNA sequences. Residues shown as red lowercase are inserted post-transcriptionally by editing. Italicized blue residues are involved in the formation of the anchor region between the pre-edited region (nucleotides 515–518) and the putative gRNA (nucleotides 639–642). Stop codons are highlighted green. Uppercase residues shown in red correspond to the putative guiding nucleotides in the 3′ UTR.
Trypanoplasma borelli edits other mitochondrial mRNAs but not COII mRNA (Lukes et al. 1994). Consistent with the role of the 3′UTR in COII editing, no base-pairing was predicted between the 3′ UTR and COII mRNA sequences in T. borelli. Similarly, comparison of COII sequences from non-kinetoplastids that do not edit mitochondrial mRNA, such as Euglena gracilis, revealed both a lack of sequence conservation within 3′ UTR sequences and limited potential base-pairing between the 3′ UTR and protein-coding sequences of the mRNA.
The sequence alignments and phylogenetic comparisons presented here are consistent with the hypothesis that the 3′ UTR of COII functions as a cis-acting guide for RNA editing (Blom et al. 1998). In addition, the limited extent of COII RNA editing suggests that 3′ UTR guiding of RNA editing may represent a recently evolved state in these organisms that is analogous to the molecules involved in the first RNA editing reaction as previously suggested by Blom et al. (1998).
3′ UTR of COII functions as a cis-guiding element
With the exception of COII mRNA editing, it has been proposed that gRNAs are provided to editing machinery in trans (Blum et al. 1990; Pollard et al. 1990). Evidence for the guiding activity of the COII 3′ UTR is based on phylogenetic comparisons and the absence of minicircle or maxi-circle genes encoding a trans-acting gRNA (van der Spek et al. 1991; Kim et al. 1994; Lukes et al. 1994; Blom et al. 1998). Several in vitro studies have shown that when transacting gRNAs are presented in cis they faithfully direct the addition or deletion of uridines at specific sites within the mRNA (Kapushoc and Simpson 1999; Cruz-Reyes et al. 2001). Additionally, the 5′ UTR of some pre-edited mRNAs are known to enhance the activity of nonspecific uridine addition and deletion in adjacent sequences within the mRNA (Connell et al. 1997; Pai et al. 2003). While highlighting the ability of editing events to be influenced by cis regions, these studies have not directly addressed the ability of a 3′ UTR to function as a cis-acting guide. To this end, we developed an in vitro editing assay that uses native COII 3′ UTR sequences to guide uridine insertion at editing sites in the coding sequences of COII mRNA.
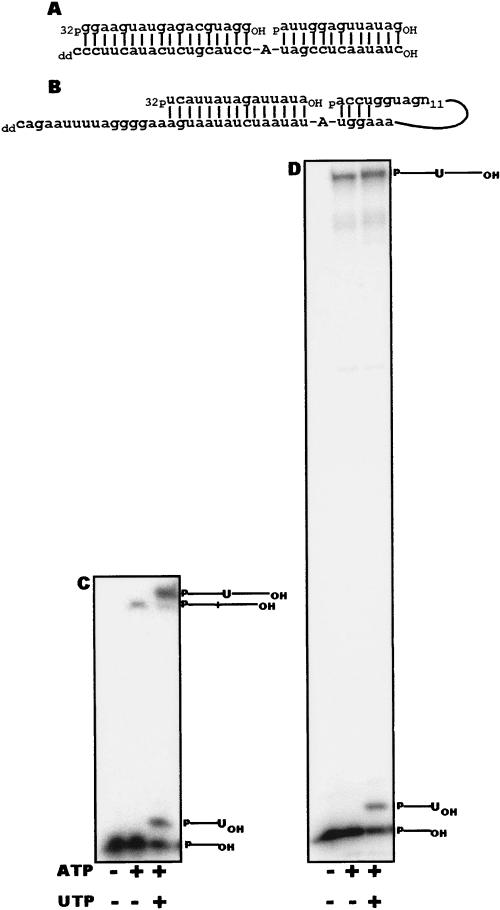
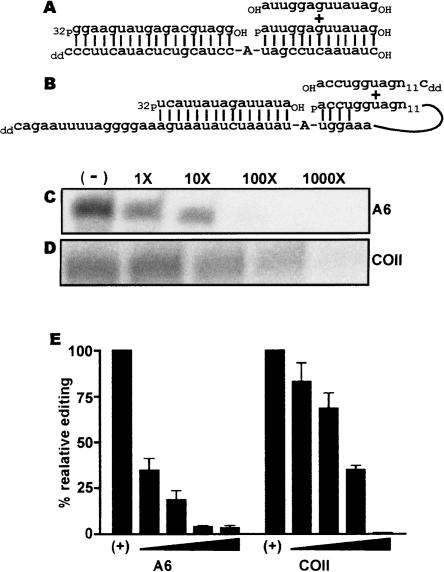
This in vitro editing assay, which eliminates the requirement of endonuclease activity by providing a precleaved editing substrate, is based on a similar assay developed for trans-acting gRNAs (Igo et al. 2000). The substrates used in the precleaved assay for cis- and trans-acting guides differ. For the trans-acting gRNA directed assay, a trimolecular reaction consisting of 5′ and 3′ fragments of the ATPase 6 (A6) mRNA and its cognate gRNA were used. An 18-nt fragment of A6 mRNA, extending to the editing site, was 5′ end-labeled and a second mRNA fragment of 13 nt, containing mRNA sequences 3′ to the editing site, were incubated with a gRNA of 32 nt. The gRNA fragment bridges the two mRNA fragments and is designed to direct the insertion of a single uridine at an A6 editing site (Fig. 2A). A substrate based on COII pre-edited mRNA was designed to investigate the ability of the 3′ UTR to function as a cis-acting gRNA. In this bimolecular reaction, a 5′ radiolabeled COII mRNA fragment was incubated with a 49-nt RNA containing sequences flanking the editing domain of COII pre-edited mRNA and extending into the proposed 3′ UTR guiding sequence of COII (Fig. 2B).
FIGURE 2.
COII 3′ UTR guides an in vitro editing assay. (A) A6 mRNA editing substrate as described previously (Igo et al. 2000). The A6 reaction contains three RNA components; 5′and 3′ cleavage products (upper strand) and a gRNA (lower strand). (B) COII editing substrate with guiding region in cis. Two RNA components are involved in the reaction with the 3′ cleavage product linked covalently to the putative gRNA in the COII 3′UTR. (C) Products from A6 editing (upper panel) and uridine addition to 5′ cleavage product (lower panel); panels are from the same gel. (D) Products from COII editing (upper panel) and uridine addition to the 5′ cleavage product (lower panel); panels are from the same gel. Edited product was observed only in the presence of exogenously added ATP and UTP.
Both trans- and cis-guided editing, with A6 and COII, respectively, showed an ATP/UTP-dependent editing reaction (Fig. 2C,D). In the presence of ATP, but without UTP, a ligation product lacking an added uridine was formed (Fig. 2C,D, upper band). In the presence of ATP and UTP, two products were detected. The smaller is the 5′ mRNA fragment containing a single uridine added at the 3′-terminus (Fig. 2C,D). The larger is the edited product and consists of the ligated 5′ and 3′ mRNA fragments with a single internally added uridine (Fig. 2C,D). These results show the ability of a naturally occurring COII 3′ UTR to act as a cis guide in the specific insertion of uridines into its own mRNA.
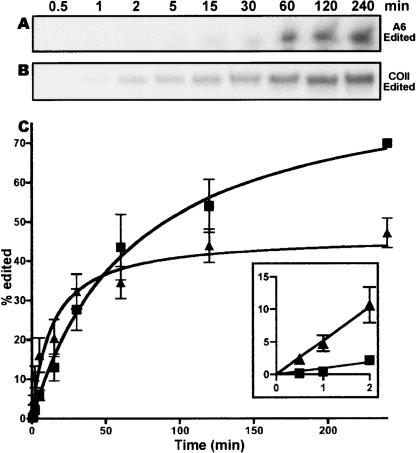
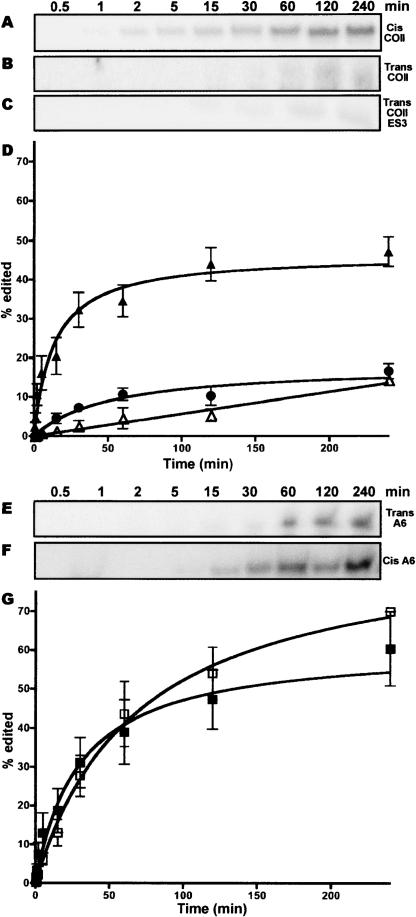
The kinetics of the precleaved editing reactions, with both trans- and cis-acting guides were also evaluated (Fig. 3). Consistent with our fixed time assay, Figure 2, the A6-based substrate shows a higher overall yield of edited product (Fig. 3A,C). However, cis-guided reactions, with the COII mRNA and 3′ UTR, showed more rapid initial kinetics with edited product detected at the 30-sec time point versus 5 min for A6 trans-guided reactions (Fig. 3C, inset). The COII cis-guided reactions reached maximal edited product formation at 50 min, whereas the A6 trans-guided reactions did not reach maximum levels even after 240 min (Fig. 3C). The rapid initial editing kinetics for COII substrates are consistent with an intramolecular reaction, while the slower initial rate of A6 editing might be expected for an intermolecular reaction. These in vitro kinetic differences highlight the ability of the COII 3′ UTR to form active editing complexes more rapidly than the trans A6 gRNA-based reaction.
FIGURE 3.
Kinetic analysis of in vitro editing for both a trans A6 gRNA and cis COII 3′ UTR. (A) Edited product formed for A6 reaction over time 0.5–240 min. (B) Edited product formed for COII based reaction over same time. (C) Percentage of substrate converted to edited product plotted against time on the X-axis for both A6 (▪) and COII (▴) reactions. Percent product formed was calculated by amount of edited product formed over the total amount of radioactivity in each lane as quantified by the Storm 860 PhosphorImager. Initial rate of edited product formation is shown in the inset.
3 ′ UTR of COII has gRNA specificity
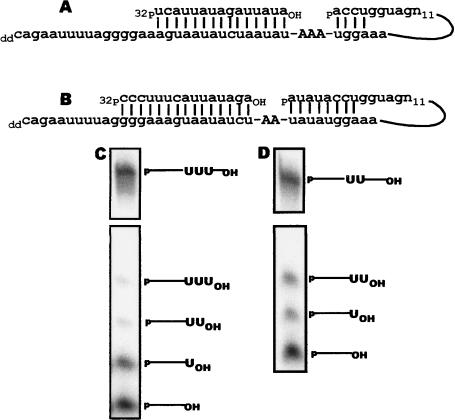
Base-pair interactions between gRNA and cognate mRNA, forming the anchor duplex, provide a molecular basis for editing site recognition. A second critical function of gRNAs is to ensure the correct number of uridines is added or deleted at an editing site. To determine if the 3′ UTR of COII could specify the correct number of uridines added to a site within the COII mRNA, we mutated the number of guiding residues within the 3′ UTR and assayed for both uridine addition to the mRNA 5′ cleavage product (preligated product) and to the fully edited COII mRNA (Fig. 4A,C). In this experiment, the COII 3′ UTR was modified by the addition of two additional adenines (Fig. 4A). We predict this substrate will be edited by the addition of three uridines at editing site 1. Analysis of the 5′cleavage fragment revealed that 27% of the 5′ fragment contained one or two uridines, while 3% contained three uridines (Fig. 4C, lower panel). In the same reactions, 33% of the input RNA was ligated to edited products that contain three uridines, and only 0.5% of the edited RNA contained a single uridine (Fig. 4C, upper panel). In comparison, wild-type COII 3′ UTR, containing a single guiding residue for editing site 1, directed only a single uridine addition to the 5′ cleavage fragment and to edited mRNA (Fig. 2D). Finally, COII 3′ UTR sequences lacking guiding nucleotides failed to mediate the formation of edited product or uridine addition to the 5′ cleavage product (data not shown).
FIGURE 4.
COII 3′ UTR specifies correctly for multiple uridine insertions at multiple sites. (A) Substrate that specifies for three uridine insertions at editing site 1. (B) Substrate used to assay for COII 3′ UTR ability to guide editing at its native third editing site. (C) Editing of COII site 1 with a 3′ UTR guiding the insertion of three uridines. (D) Editing of COII site 3 with a 3′ UTR guiding the insertion of two uridines. The upper and lower panels in C and D are from the same gel and show the fully edited products and the 5′ cleavage products, respectively.
The ability for the 3′ UTR of COII to guide editing at a site other than editing site 1 was investigated using a partially edited substrate in which editing sites 1 and 2 have single uridines added. This substrate requires the addition of two uridines at editing site 3 in COII mRNA (Fig. 4B). In the in vitro precleaved assay, a band corresponding to a specific addition of one uridine to the 5′ cleavage product was detected at 12% of the input substrate (Fig. 4D, lower panel) in addition to a fully edited COII mRNA containing two added uridines (Fig. 4D, upper panel). The amount of COII mRNA edited at the third site was equivalent for that seen with the initial editing site 1 construct, 37% of input (Fig. 2D). Together these results show that the 3′ UTR of COII acts as a gRNA providing editing site selection and specificity and also directing the correct number of uridine residues added to the mRNA.
COII 3′ UTR guides uridine insertion in cis
To determine whether the in vitro editing of COII was facilitated by intramolecular interactions with its own 3′ UTR or through intermolecular reactions with other COII transcripts, we developed a competition assay to distinguish cis- and trans-guided editing. The possibility of a dimer-type editing reaction for COII has been cited as a possible mechanism to allow for trans editing of COII (Kim et al. 1994). In these experiments, increasing amounts of either the A6 or COII 3′ cleavage product were added to a standard precleaved editing assay (Fig. 5).
FIGURE 5.
Trans substrate is not an effective competitor of the COII editing. (A) RNA substrates for A6 editing reaction with the competitor RNA oligonucleotide. (B) RNA substrates for COII editing reaction with the competitor RNA oligonucleotide. (C) Edited product formed for the A6 reactions with increasing amounts of competitor. (D) Edited product formed in COII reactions with increasing amounts of competitor. (E) PhosphorImager analysis of edited product formation in the presence of increasing amounts competitor. The Y-axis is the relative amount of editing in relation to no competitor controls.
The standard A6 precleaved editing reaction requires the assembly of a trimolecular RNA structure containing the 5′ and 3′ mRNA cleavage products and a gRNA spanning editing site 1. To determine whether addition of excess 3′ cleavage product would compete for editing, a 13-nt RNA oligonucleotide identical in sequence to the standard 3′ cleavage product, but with a 5′ hydroxyl group, was added to the A6 precleaved editing assay. The 5′ hydroxylated RNA should effectively compete for gRNA binding with the standard A6 3′ cleavage product but is not a substrate for the ligation; thus, if it assembled into the editing complex an edited product could not be formed. When the hydroxylated cleavage product was added to the reaction at the same concentration of the phosphorylated 3′ cleavage product, a decrease of ~50% was seen in the amount of edited product formation. This degree of inhibition suggests that in the A6 reaction there is no preference for the phosphorylated over the hydroxylated 3′ cleavage product for editing complex assembly or subsequent editing processes (Fig. 5A,C,E). Increasing the competitor concentration to 100-fold that of phosphorylated 3′ cleavage product completely abolished editing.
To determine whether COII editing was dependent on cis-acting elements within the COII 3′ UTR, a competitor RNA oligonucleotide, identical in sequence to the first 22 nt of the COII 3′ cleavage product but lacking the 3′ UTR guiding region of COII, was added to the precleaved COII editing reactions (Fig. 5B,D,E). We used both a competitor with a 5′ hydroxyl and one with a 5′ phosphate (data not shown). The RNA oligonucleotide containing a 5′ phosphate would be a substrate for ligation if recruited into an editing complex. However, if COII editing is guided by cis elements in the 3′ UTR, we expect that competition for the assembly of this trimolecular RNA complex will be much less. Because of the size difference in the competitor RNA and the standard 3′ UTR substrate, cis- and trans-edited products will migrate at 75 or 37 nt, respectively. No significant reduction in edited product formation was seen with either 5′ hydroxylated or phosphorylated 3′ cleavage product until the competitor reached a 100-fold excess to the 3′ cleavage product linked to the 3′ UTR of COII. Even at the highest competitor concentration, no 37-nt product was detected with the 5′ phosphorylated competitor, indicating that inhibition by high competitor concentration was not a consequence of formation of a trimolecular RNA complex (data not shown). The inability of the competitor to efficiently block the editing assay proves that the COII 3′ cleavage product used preferentially is the one linked to the 3′ UTR.
COII 3′ UTR cannot function as a trans gRNA
Based on the results presented here, COII 3′ UTR can function, in cis, to efficiently guide the correct addition of uri-dines, at specific sites, within the COII pre-edited mRNA. Previous studies have shown that linking a trans-acting gRNA to the 3′ end of an mRNA increases editing efficiency (Kapushoc and Simpson 1999; Cruz-Reyes et al. 2001). To determine whether the COII 3′ UTR can function as a trans-acting gRNA, in a trimolecular RNA complex, the precleaved RNA editing assay was modified to use COII mRNA and the COII 3′ UTR as a trans-acting guide (Fig. 6).
FIGURE 6.
COII 3′ UTR does not guide RNA editing in trans. (AC,E,F) Time course of edited product formed in cis COII, trans COII, trans COII-ES3, trans A6, and cis A6, respectively, for an in vitro editing reaction. (D) Percent edited product formed versus time for COII substrate with trans (▵) or cis (▴) acting guide and COII–ES3 substrate (•). (G) Percent edited product formed versus time for A6 substrate with cis- (□) or trans- (▪) acting gRNA.
The trans COII editing reaction contained the 5′ mRNA cleavage product (15 nt), the 3′ cleavage product (22 nt), and the COII 3′ UTR (38 nt). The in vitro reaction was very inefficient with only 14% of the substrate converted after 4 h of incubation. This compares with ~45% conversion of substrate when the COII guide is presented in cis (Fig. 6A,B,D). The amount of edited product formed was roughly equivalent to the amount formed in reactions where the gRNA lacks any guiding nucleotides (data not shown). To determine whether a longer anchor region would allow for the COII 3′ UTR to function in trans, we used an editing substrate similar to the substrate presented in Figure 4B, except with a trans-guiding region. While the longer anchor region did increase the amount of edited product formed, it was still significantly less than that found in the cis reaction (Fig. 6C,D). We also investigated the ability of the A6 gRNA to function in cis by linking this sequence to the 3′ cleavage product of the A6 pre-edited mRNA. The substrate for the A6 reaction was identical to the one shown in Figure 2A, with the exception that the 3′ cleavage product was covalently continuous with the guiding region. The cis A6 reaction produced an edited product at a level comparable to the trans A6 reaction (Fig. 6E–G).
These results indicate that the COII 3′ UTR cannot efficiently function as a trans-acting gRNA. We also find that while the A6 gRNA can function efficiently in cis, this does not significantly increase the overall efficiency of the A6 editing reaction, although the cis A6 reaction does show a increase in the initial rate of conversion when compared to the traditional trans A6 arrangement (Fig. 6D). The inability of the COII 3′ UTR to function in trans is not fully accounted for in the limited amount of base-pairing in the anchor region (Fig. 6C). These results could be possibly due to higher order structural properties important in correct function for a trans gRNA.
DISCUSSION
Mitochondrial mRNA editing is essential to African trypanosomes. This process requires the orchestrated insertion and deletion of hundreds of uridines, at dozens of sites, in primary mRNAs. While trypanosome mRNA editing is a protein-catalyzed enzymatic cascade, RNAs serve as information molecules directing both uridine insertion and deletion. For an RNA sequence to serve as a “guide” in trypanosome RNA editing, it must first direct the recognition of the editing site by the formation of an RNA/RNA duplex and then guide for the correct number of uridine insertions or deletions. In this paper, we show that the 3′ UTR of COII mRNA can function as a guiding sequence for uridine insertion editing in COII mRNA. A phylogenetic comparison (Fig. 1) along with observations previously made of several organisms found a strict covariation of sequences immediately adjacent to the COII editing site and the 3′ UTR (Blum et al. 1990; van der Spek et al. 1991; Kim et al. 1994; Lukes et al. 1994; Blom et al. 1998). Using an in vitro editing assay, we show that the COII 3′ UTR efficiently and specifically guides COII editing reactions (Figs. 2–4). Our results allow us to experimentally demonstrate a unique function for a 3′ UTR, the ability of a 3′ UTR to directly influence the coding potential of its mRNA.
The formation of an active editing complex requires the organization of a binary complex between pre-edited mRNA and a cognate gRNA. The 3′ UTR of COII is recognized as a gRNA in cis but not when it is placed in trans (Fig. 6). We tested whether or not this discrepancy was based solely on the amount of base-pairing in the anchor region by increasing the length of the anchor to 8 bp. This anchor is slightly below the 13-nt anchor found in the A6 substrate tested, and we are currently addressing this difference experimentally. While an increase in the amount of editing product was observed, we could not fully rescue trans COII to the level of a cis reaction. One interpretation of these results is that the 3′ UTR of COII lacks other elements crucial to being recognized as a trans gRNA by the editing machinery. The elements involved could be higher order, secondary or tertiary, interactions contained in the gRNA molecule (Hermann et al. 1997). Exploitation of these differences will allow for a more thorough understanding of the elements crucial to gRNA function.
Untranslated 5′ and 3′ regions in eukaryotic mRNAs play important roles mediating post-transcriptional regulatory events. Localization, stability, translation, and cleavage are influenced by RNA binding proteins that recognize and specifically bind to UTR sequences (Wickens et al. 2002). In trypanosomes, sequences within mRNA 3′ UTRs play critical roles in developmental and cell cycle-specific degradation of mRNAs (Estevez et al. 2001; Avliyakulov et al. 2003; Haile et al. 2003). The localization of regulatory sequences within the UTRs of mRNAs is not surprising, because these sequences are not under the selective pressures imposed by protein coding constraints. While all of these functions affect the protein to be translated from the message, none are known to directly influence the coding capacity of the message.
The ability of the COII 3′ UTR to function as a cis-acting guide for RNA editing is a unique function for a 3′ UTR. However, examples of RNA sequences that “guide” RNA processing reactions are numerous and widespread in plants and animals. Both Group I and Group II intronic sequences contain short stretches of RNA that interact, in cis, with exon/intron splice junctions and intron branch points to facilitate splicing (Bonen and Vogel 2001). These interactions support RNA-catalyzed transesterification reactions resulting in intron excision and exon joining. It has been proposed that Group II introns are the predecessor of modern splicesome-associated RNAs in eukaryotic systems (Cavalier-Smith 1991; Mattick 1994). Highlighting the ability of a cis-guiding RNA to transition to a trans reaction mechanism. As in other internally guided reactions, the COII 3′ UTR defines the specific site and extent of processing within the pre-edited mRNA (Figs. 2–4).
A major question that arises is whether COII cis editing is a unique event in the evolution of gRNAs. Two models for the origin of gRNAs have been proposed. One involves a partial gene duplication event, while the second utilizes the recruitment of random sequences to act as a gRNA (Simpson and Maslov 1999). Both of these models require the transcription of a proto-gRNA prior to the necessity of function. If the proto-gRNA were part of the primary mRNA, then independent transcription would not be a prerequisite feature. Furthermore, our results suggest that a cis-acting sequence can function with much less base-pairing between guide and editing site than found in transacting gRNAs (Fig. 1). It has been proposed that the high percentage of G:U pairing found between an mRNA and cognate gRNA is essential for processive editing to occur (Koslowsky et al. 1991). If gRNAs arose by a gene duplication event of protein-coding sequence, the extensive G:U base-pairing between gRNAs and editing sites could only be accounted for by extensive modification of the duplicated sequence (Simpson and Maslov 1999). The findings reported here lead us to propose that gRNAs originated as cis-acting elements that have been later adapted for a trans mechanism. Once this adaptation occurs and allows for autonomous gRNA molecules, a process of duplication and genetic drift could account for the enormous amount of gRNA diversity seen currently. The major question that arises is what allows a gRNA to act in trans, we are currently seeking possible explanations to these questions. This model is in agreement with previous examples found within the field of RNA processing.
MATERIALS AND METHODS
T. brucei culture and mitochondrial editing complex preparation
Procyclic T. brucei cells were grown at 27°C in SM media supplemented with 10% heat-inactivated fetal bovine serum. Mitochondria were isolated as previously described by hypertonic lysis followed by organelle separation on a 20%–35% Percoll gradient (Harris et al. 1990). Native RNA editing complexes were purified from total mitochondrial lysate on a 10%–30% glycerol gradient as reported previously (Harris et al. 1990).
Precleaved editing assay
RNAs were custom synthesized by Dharmacon Inc. RNAs for A6 reactions were as follows; A6 5′ cleavage product, 5′-GGAAGUA UGAGACGUAGG; A6 3′ cleavage product, 5′-pAUUGGAGUU AUAG; A6 gRNA, 5′-CUAUAACUCCGAUACCUACGUCUCAU ACUUC; and A6 cis gRNA, 5′-pAUUGGAGUUAUAGGGAGGCU AUAACUCCGAUACCUACGUCUCAUACUUCC. RNAs for COII reactions were as follows: COII 5′ cleavage product, 5′-UCAUUA UAGAUUAUA; COII 3′ cleavage product, 5′-pACCUGGUAGG UGUAAUGAAAGGAGG; COII cis UTR, 5′-pACCUGGUAGGUG UAAUGAAAGGAGGGAAAGGUAUAUAAUCUAUAAUGAAAG GGGAUUUUAAGA; and COII 3′ UTR, 5′-GAAAGGUAUAUAA UCUAUAAUGAAAGGGGAUUUUAAGA.
T4 polynucleotide kinase (New England Biolabs) was utilized as described by the manufacturer to label RNAs radioactively. RNAs were purified from unincorporated label by electrophoresis on a 15% denaturing polyacrylamide gel.
In vitro editing reactions were performed essentially as described previously (Igo et al. 2000). That is, 50 fmol of radioactive 5′ cleavage product was mixed with 1 pmol of 3′ cleavage product and 0.5 pmol gRNA in 25 mM HEPES-KOH (pH 7.9), 10 mM Mg(OAc)2, 5 mM CaCl2, 50 mM KCl, 0.5 mM DTT, and 1 mM EDTA for a final reaction volume of 30 μL. Then 1 pmol of 3′ cleavage product:gRNA was added to the editing reactions involving a cis gRNA. RNAs were then heated to 70°C for 2 min followed by cooling at room temperature for 15 min. UTP and ATP were then added to the appropriate reactions at 100 μM and 1 μM, respectively. Then 10 μL of glycerol gradient-purified mitochondrial lysate was added, and the reactions were placed at 27°C for 60 min, unless otherwise noted in the figure. Reactions were terminated by the addition of 70 μL of 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 0.05% NP-40, and 10 μg/mL tRNA as a carrier. Reactions were extracted with phenol chloroform (1:1 [v/v]) followed by a chloroform extraction and subsequently precipitated with 3 volumes of ethanol. Pellets were resuspended in an appropriate volume of water and formamide loading buffer and electrophoresed on 15% polyacrylamide gels containing 8 M urea. Quantitation was performed on a Molecular Dynamics PhosphorImager (model Storm-860).
Acknowledgments
We acknowledge Lynn Sherrer, Torsten Ochsenreiter, Rudo Kieft, Laura Cotlin, and the rest of the Hajduk lab for their helpful discussions and input. This work was supported by the National Institutes of Health grant AI21401 (to S.L.H.).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7170705.
REFERENCES
- Alfonzo, J.D., Thiemann, O., and Simpson, L. 1997. The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 25: 3751–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev, R., Aphasizheva, I., and Simpson, L. 2003. A tale of two TUTases. Proc. Natl. Acad. Sci. 100: 10617–10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avliyakulov, N.K., Hines, J.C., and Ray, D.S. 2003. Sequence elements in both the intergenic space and the 3′ untranslated region of the Crithidia fasciculata KAP3 gene are required for cell cycle regulation of KAP3 mRNA. Eukaryot. Cell 2: 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne, R., Van den Burg, J., Brakenhoff, J.P., Sloof, P., Van Boom, J.H., and Tromp, M.C. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46: 819–826. [DOI] [PubMed] [Google Scholar]
- Blom, D., de Haan, A., van den Berg, M., Sloof, P., Jirku, M., Lukes, J., and Benne, R. 1998. RNA editing in the free-living bodonid Bodo saltans. Nucleic Acids Res. 26: 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, B. and Simpson, L. 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell 62: 391–397. [DOI] [PubMed] [Google Scholar]
- Blum, B., Bakalara, N., and Simpson, L. 1990. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60: 189–198. [DOI] [PubMed] [Google Scholar]
- Bonen, L. and Vogel, J. 2001. The ins and outs of group II introns. Trends Genet. 17: 322–331. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith, T. 1991. Intron phylogeny: A new hypothesis. Trends Genet. 7: 145–148. [PubMed] [Google Scholar]
- Connell, G.J., Byrne, E.M., and Simpson, L. 1997. Guide RNA-independent and guide RNA-dependent uridine insertion into cyto-chrome b mRNA in a mitochondrial lysate from Leishmania tarentolae. Role of RNA secondary structure. J. Biol. Chem. 272: 4212–4218. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Zhelonkina, A., Rusche, L., and Sollner-Webb, B. 2001. Trypanosome RNA editing: Simple guide RNA features enhance U deletion 100-fold. Mol. Cell. Biol. 21: 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, N.L., Panicucci, B., Igo Jr., R.P., Panigrahi, A.K., Salavati, R., and Stuart, K. 2003. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell 11: 1525–1536. [DOI] [PubMed] [Google Scholar]
- Estevez, A.M., Kempf, T., and Clayton, C. 2001. The exosome of Trypanosoma brucei. EMBO J. 20: 3831–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin, J.E., Abraham, J.M., and Stuart, K. 1988a. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 53: 413–422. [DOI] [PubMed] [Google Scholar]
- Feagin, J.E., Shaw, J.M., Simpson, L., and Stuart, K. 1988b. Creation of AUG initiation codons by addition of uridines within cytochrome b transcripts of kinetoplastids. Proc. Natl. Acad. Sci. 85: 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile, S., Estevez, A.M., and Clayton, C. 2003. A role for the exosome in the in vivo degradation of unstable mRNAs. RNA 9: 1491–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, M.E., Moore, D.R., and Hajduk, S.L. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265: 11368–11376. [PubMed] [Google Scholar]
- Hermann, T., Schmid, B., Heumann, H., and Goringer, H.U. 1997. A three-dimensional working model for a guide RNA from Trypanosoma brucei. Nucleic Acids Res. 25: 2311–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.E., Cruz-Reyes, J., Zhelonkina, A.G., O’Hearn, S., Wirtz, E., and Sollner-Webb, B. 2001. Roles for ligases in the RNA editing complex of Trypanosoma brucei: Band IV is needed for U-deletion and RNA repair. EMBO J. 20: 4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo Jr., R.P., Palazzo, S.S., Burgess, M.L., Panigrahi, A.K., and Stuart, K. 2000. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell. Biol. 20: 8447–8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapushoc, S.T. and Simpson, L. 1999. In vitro uridine insertion RNA editing mediated by cis-acting guide RNAs. RNA 5: 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.S., Teixeira, S.M., Kirchhoff, L.V., and Donelson, J.E. 1994. Transcription and editing of cytochrome oxidase II RNAs in Trypanosoma cruzi. J. Biol. Chem. 269: 1206–1211. [PubMed] [Google Scholar]
- Koslowsky, D.J., Bhat, G.J., Read, L.K., and Stuart, K. 1991. Cycles of progressive realignment of gRNA with mRNA in RNA editing. Cell 67: 537–546. [DOI] [PubMed] [Google Scholar]
- Lukes, J., Arts, G.J., van den Burg, J., de Haan, A., Opperdoes, F., Sloof, P., and Benne, R. 1994. Novel pattern of editing regions in mitochondrial transcripts of the cryptobiid Trypanoplasma borreli. EMBO J. 13: 5086–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Grams, J., and Hajduk, S.L. 2002. Editing machines: The complexities of trypanosome RNA editing. Cell 108: 435–438. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. 1994. Introns: Evolution and function. Curr. Opin. Genet. Dev. 4: 823–831. [DOI] [PubMed] [Google Scholar]
- McManus, M.T., Shimamura, M., Grams, J., and Hajduk, S.L. 2001. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA 7: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missel, A., Souza, A.E., Norskau, G., and Goringer, H.U. 1997. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 17: 4895–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai, R.D., Oppegard, L.M., and Connell, G.J. 2003. Sequence and structural requirements for optimal guide RNA-directed insertional editing within Leishmania tarentolae. RNA 9: 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Gygi, S.P., Ernst, N.L., Igo Jr., R.P., Palazzo, S.S., Schnaufer, A., Weston, D.S., Carmean, N., Salavati, R., Aebersold, R., et al. 2001. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21: 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Schnaufer, A., Ernst, N.L., Wang, B., Carmean, N., Salavati, R., and Stuart, K. 2003. Identification of novel components of Trypanosoma brucei editosomes. RNA 9: 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, V.W. and Hajduk, S.L. 1991. Trypanosoma equiperdum minicircles encode three distinct primary transcripts which exhibit guide RNA characteristics. Mol. Cell. Biol. 11: 1668–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, V.W., Rohrer, S.P., Michelotti, E.F., Hancock, K., and Hajduk, S.L. 1990. Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell 63: 783–790. [DOI] [PubMed] [Google Scholar]
- Rusche, L.N., Cruz-Reyes, J., Piller, K.J., and Sollner-Webb, B. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16: 4069–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, B., Riley, G.R., Stuart, K., and Goringer, H.U. 1995. The secondary structure of guide RNA molecules from Trypanosoma brucei. Nucleic Acids Res. 23: 3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer, A., Panigrahi, A.K., Panicucci, B., Igo Jr., R.P., Wirtz, E., Salavati, R., and Stuart, K. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291: 2159–2162. [DOI] [PubMed] [Google Scholar]
- Schnaufer, A., Ernst, N.L., Palazzo, S.S., O’Rear, J., Salavati, R., and Stuart, K. 2003. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell 12: 307–319. [DOI] [PubMed] [Google Scholar]
- Simpson, L. and Maslov, D.A. 1999. Evolution of the U-insertion/deletion RNA editing in mitochondria of kinetoplastid protozoa. Ann. NY Acad. Sci. 870: 190–205. [DOI] [PubMed] [Google Scholar]
- Stuart, K., Panigrahi, A.K., Schnaufer, A., Drozdz, M., Clayton, C., and Salavati, R. 2002. Composition of the editing complex of Trypanosoma brucei. Philos. Trans. R Soc. Lond. B Biol. Sci. 357: 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Spek, H., Arts, G.J., Zwaal, R.R., van den Burg, J., Sloof, P., and Benne, R. 1991. Conserved genes encode guide RNAs in mitochondria of Crithidia fasciculata. EMBO J. 10: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens, M., Bernstein, D.S., Kimble, J., and Parker, R. 2002. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 18: 150–157. [DOI] [PubMed] [Google Scholar]