Abstract
Small inhibitory RNAs (siRNAs) are produced from longer RNA duplexes by the RNAse III family member Dicer. The siRNAs function as sequence-specific guides for RNA cleavage or translational inhibition. The precise mechanism by which siRNAs direct the RNA-induced silencing complex (RISC) to find the complementary target mRNA remains a mystery. Some biochemical evidence connects RNAi with translation making attractive the hypothesis that RISC is coupled with the translational apparatus for scanning mRNAs. Such coupling would facilitate rapid alignment of the siRNA antisense with the complementary target sequence. To test this hypothesis we took advantage of a well-characterized translational switch afforded by the ferritin IRE-IRP to analyze RNAi mediated cleavage of a target mRNA in the presence and absence of translation. Our results demonstrate that neither active translation nor unidirectional scanning is required for siRNA mediated target degradation. Our findings demonstrate that nontranslated mRNAs are highly susceptible to RNAi, and blocking scanning from both the 5′ and 3′ ends of an mRNA does not impede RNAi. Interestingly, RNAi is about threefold more active in the absence of translation.
Keywords: RNA interference (RNAi), small interfering RNA (siRNA), translation, iron responsive element (IRE)
INTRODUCTION
RNA interference (RNAi) is a phenomenon in which double-stranded RNA triggers the silencing of target gene expression by inducing sequence-specific target mRNA degradation (Fire et al. 1998). An important intermediate in the RNAi pathway are small interfering RNAs (siRNAs) which are duplexes of 21–23 nucleotides containing two unpaired nucleotides at the 3′-end of each strand (Hamilton and Baulcombe 1999; Zamore et al. 2000). Strong inhibition of target gene expression using synthetic or expressed siRNAs has been demonstrated in mammalian cells by many groups (Elbashir et al. 2001; McManus and Sharp 2002; Scherer and Rossi 2003).
A critical step in RNAi function is target site recognition, in which the antisense strand derived from the siRNA serves as a guide for RISC to find the complementary target sequence (Hammond et al. 2000; Martinez et al. 2002). The precise mechanism by which this process occurs remains to be determined. There are at least two general models: random access and directed scanning. Very small amounts of siRNA can efficiently induce targeted mRNA degradation (Dykxhoorn et al. 2003), therefore it is highly likely that RISC functions catalytically.
In considering mechanisms by which RISC targets mRNAs, a unidirectional or bidirectional scanning process could facilitate more rapid alignment of the siRNA anti-sense with the complementary target sequence. If a scanning model is correct, it is likely that RISC function would be coupled with some cellular process that effectively scans mRNAs, such as translation (Merrick 1992; Sachs et al. 1997). Some biochemical evidence already exists which connects RNAi with translation in that RISC components have been found to co-purify with translation factors such as ribosomal proteins L5, L11, as well as 5S rRNA (Hammond et al. 2001; Caudy et al. 2002; Ishizuka et al. 2002; Caudy et al. 2003; Pham et al. 2004). Furthermore, siRNAs were found associated with polyribosomes in the protozoan Trypanosome brucei (Djikeng et al. 2003). We therefore asked whether or not the mammalian siRNA pathway of RNAi is linked with translation. As a model system for addressing this question, we have taken advantage of the factors involved in the regulation of ferritin translation (Theil 1990; Hentze and Kuhn 1996). When the intracellular iron concentration is low, the iron regulatory protein 1 (IRP-1) binds to an iron-responsive element (IRE) located in close proximity to the cap structure of the ferritin mRNAs. This IRP1-IRE complex represses translation by blocking the recruitment of the small ribosomal subunit to these mRNAs (Muckenthaler et al. 1998). To take advantage of this relatively simple system, we have created a reporter gene construct under the regulation of IRE/IRP-1 system. This IRE-IRP-1 system can be utilized as a molecular switch to regulate translation, thereby allowing the testing of siRNA function in the absence of active translation. An additional feature of this system is that it allows the manipulation of a specific reporter gene construct without impacting on global translation. We demonstrate here that neither active translation nor unidirectional scanning is required for siRNA-mediated target degradation. The implications of our findings are that nontranslated mRNAs are susceptible to RNAi, suggesting that RISC identification of target RNAs is not actively linked with mRNA translation.
RESULTS
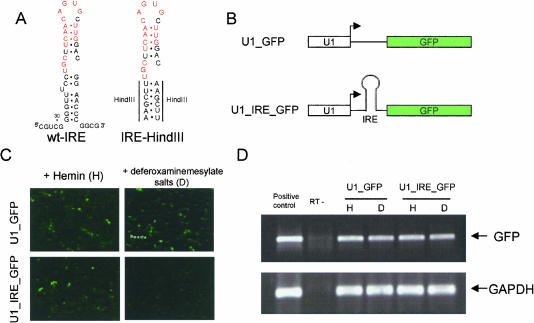
In an effort to regulate translation of a reporter gene construct, we designed a vector using the Pol II U1 promoter to drive expression of an EGFP gene in which the iron response element (IRE) was inserted upstream of the EGFP coding sequence. This construct is designated as pU1-IRE-GFP. A similar construct, pU1-GFP, lacking the IRE was constructed as a control (Fig. 1A,B). Notably, the IRE is inserted 28 base pairs (bp) downstream of the cap site. Close proximity to the cap structure has been shown to be important for the IRE/IRP-1 system to function as a translational regulator (Koloteva et al. 1997). Translational regulation of the pU1-IRE-GFP was achieved by the addition of Hemin (as a source of iron) or deferoxaminemesylate salt (as an iron chelator) following transfection of the reporter plasmid into HEK293 cells. EGFP expression from pU1-IRE-GFP was greatly reduced when the iron concentration was reduced with deferoxaminemesylate salt (Figs. 1C and 2B,C). RT-PCR analyses of the IRE-GFP mRNA revealed that the mRNA level remains unchanged from conditions in which iron is present (Fig. 1D), demonstrating the repression of EGFP expression is solely due to a block in translation. EGFP expression from the pU1-GFP control vector was not affected by the presence or absence of iron (Fig. 1C,D). Consistent with previous reports, our data confirmed that the IRE-IRP1 system functions as a translational switch (Hentze et al. 1987).
FIGURE 1.
Iron response element (IRE) and IRP regulate translation in HEK293 cells. (A) sequence and computer-modeled secondary structure of the human ferritin H-chain IRE and synthetic oligonucleotides used in this study. The sequence recognized by the iron regulatory protein (IRP), is highlighted in red. (B) Designs of the two constructs used in this study. (C) Three-hour post-transfection of the constructs in HEK293 cells, Hemin (50 μM final concentration), or deferoxaminemesylate salts (100 μM final concentration) were added to the medium and uptake was allowed to proceed for 21 h. EGFP expression 24 h after transfection is depicted. (D) EGFP mRNA levels analyzed by RT-PCR. GAPDH, served as internal control, and the RT-PCR products are also depicted. U1-GFP plasmid DNA and total RNA from nontransfected HEK293 cells were used as standards and positive controls for EGFP and GAPDH, respectively. To control for DNA contamination, minus RT PCR reactions were carried out for each RNA preparation.
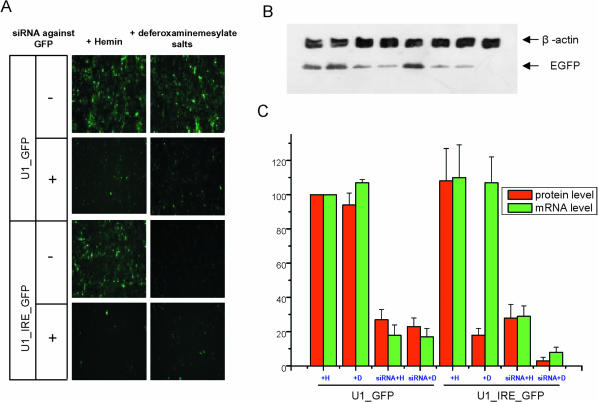
FIGURE 2.
siRNA mediated RNAi is fully functional when translation of the target mRNA is active or blocked. (A) EGFP expression from U1_GFP and U1_IRE_GFP under various conditions. Hemin (iron source) labeled as H or deferoxaminemesylate salts (iron chelator) labeled as D and siRNA targeting the EGFP coding region were added to the culture medium as indicated. (B) Cells were lysed 24 h after treatment, and levels of EGFP and an internal control β-actin were obtained by sequential immunoblotting using anti-eGFP and anti-β-actin antibodies. Lanes (from left to right): U1_GFG plus Hemin; U1_GFP plus deferoxaminemesylate salts; U1_GFP treated with siRNA; U1_GFP treated with both siRNA and deferoxaminemesylate salts; U1_IRE_GFP plus Hemin; U1_IRE_GFP plus deferoxaminemesylate salts; U1_IRE_GFP treated with siRNA; U1_IRE_GFP treated with both siRNA and deferoxaminemesylate salts. (C) Summary of EGFP expression and mRNA levels for each sample. The mean levels of EGFP expression were measured by Dot density assays of the immunoblots (B) after normalization with the internal control β-actin. The EGFP mRNA levels were determined using real time RT-PCR after normalization with GAPDH mRNA levels. EGFP expression and mRNA levels from the U1-GFP plus Hemin in the absence of siRNA were assigned a value of 100 percent. Quantitations of EGFP values and mRNA relative to the U1-GFP values are plotted. The results are derived from averaging values from triplicate experiments. Values for EGFP expression for all of the samples are (from left to right): 100 ± 0%, 93.9 ± 6.6%, 27.3 ± 6.1%, 22.8 ± 5.1%, 108.1 ± 18.8%, 17.7 ± 4.4%, 27.5 ± 8.3%, 3.1 ± 2.1%. Values for mRNA level of all samples are (from left to right): 100 ± 0%, 107.4 ± 1.7%, 18.5 ± 6.4%, 16.6 ± 5.3%, 110.0 ± 19.2%, 107.3 ± 15.3%, 28.9 ± 6.2%, 8.1 ± 2.6%.
To address the question of whether or not RNAi function is coupled with translation, a synthetic siRNA targeting the EGFP coding region was tested for RNAi in the pU1-IRE-GFP and pU1-GFP cells in the presence and absence of iron. In co-transfections of the siRNA with the expression vectors this siRNA effectively reduced EGFP expression from both constructs under both high and low iron concentrations (Fig. 2). These results demonstrate that siRNA mediated RNAi is fully functional when translation of the target mRNA is active or severely blocked by greater than 80% (Fig. 2B). Interestingly, quantitative real-time RT-PCR analyses of the EGFP mRNA revealed that the siRNA induced about a fourfold reduction of the actively translated IRE-GFP mRNA whereas an approximate 13-fold reduction of this mRNA was achieved when translation was blocked. These results suggest that in the absence of translation the mRNA may be more accessible to RNAi. This increase in siRNA efficacy during translational inhibition was consistently observed in three independent experiments (Fig. 2). Moreover, the mRNA levels analyzed in these studies were normalized with GAPDH mRNA levels, which were unchanged; demonstrating specificity in the siRNA-mediated targeting of EGFP.
There have been several reports suggesting that high concentrations of siRNA can result in off-target effects or activate genes in the interferon pathway (Sledz et al. 2003; Scacheri et al. 2004). To validate the specificity of our results, we tested another synthetic siRNA targeting a different sequence in EGFP. For these studies we utilized a synthetic siRNA that we have independently demonstrated to be a potent trigger for RNAi-mediated destruction of the EGFP mRNA (D. Kim, M. Behlke, S. Rose, M. Chang, S. Choi, and J.J. Rossi, unpubl.). With this siRNA we utilized a final concentration of only 2 nM and repeated the transfection experiments described above. The results obtained with this more potent siRNA trigger were similar to those obtained with the first siRNA although the second siRNA worked at a much lower concentration (data not presented).
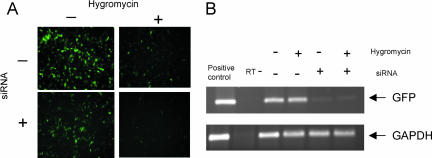
To further substantiate the conclusion that siRNA triggered RNAi is translation independent, we blocked translation via a different approach, using the translational elongation inhibitor Hygromycin B (Gonzalez et al. 1978). As we observed using the IRE-IRP1 approach, a significant reduction of target mRNA was achieved by siRNA treatment in Hygromycin B treated cells (Fig. 3).
FIGURE 3.
Translational elongation mediated by Hygromycin does not protect transcripts from RNAi mediated degradation. (A) Three hours after U1_GFP was transfected into HEK293 cells, Hygromycin (10 nM final concentration) was added to the medium with or without siRNA against EGFP. EGFP expression was monitored 21 h later. (B) EGFP mRNA levels were analyzed by RT-PCR as described in the legend to Figure 2.
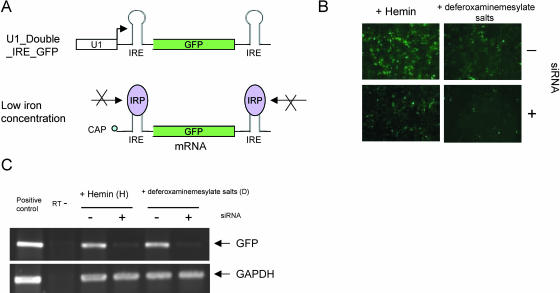
Although translation is not required for siRNA, it is possible that RISC may scan messages 3′ to 5′. To test this possibility we designed a construct harboring the IRE in both the 5′-UTR and 3′-UTRs of the EGFP mRNA. Under conditions of low iron the IRP-1 should bind to both elements that in turn would block a unidirectional scan from either end (Fig. 4A). Utilizing this double IRE containing construct we observed efficient RNAi under both conditions of high and low iron (Fig. 4B,C). These data suggest that unidirectional scanning of mRNAs, initiating either at the 5′ or 3′ ends of the transcripts, is not the mechanism by which RISC functions.
FIGURE 4.
RNAs containing IRE duplications at the 5′ and 3′ end are still susceptible to siRNA triggered RNAi. (A) Schematic representation of the Double IRE construct and how it would be protected when the iron concentration is low. (B) Three hours after the U1_Double_IRE_GFP was transfected into HEK293 cells, Hemin (iron source labeled as H) or deferoxaminemesylate salts (iron chelator labeled as D) were added to the medium with or without siRNA against EGFP. EGFP expression was monitored 24 h post-transfection. (C) EGFP mRNA levels are analyzed by RT-PCR as described in the legend to Figure 2.
DISCUSSION
There are several cellular mechanisms in which an RNA guide sequence is used to identify the target RNA sequence. These include translation (Sachs et al. 1997), pre-mRNA splicing (Nilsen 1994), RNA editing (Smith et al. 1997), telomere synthesis (Cech 2004) and snoRNA mediated pre-rRNA site-specific modifications (Lafontaine and Tollervey 1998). For each of these processes, a complex of proteins associates with the guide RNA that provides the specificity for the process. In the case of RNAi, the antisense strand selected from the siRNA duplex guides RISC to the target sequence. Based upon the reported potencies of RNAi, identification of the target is an efficient process, which presumably would make it a diffusion independent process. Since message-specific degradation by RNAi has been purported to be a cytoplasmic process (Zeng and Cullen 2002), we devised a set of experiments to ask whether or not RISC is associated with the translational apparatus, thereby providing a scaffold for scanning mRNAs from the 5′ to 3′ end. Several relevant but conflicting sets of data have been published in the past few years. Zamore and coworkers previously showed that RNAi activity is not attenuated when translation is blocked by several antibiotics in Drosophila lysates, indicating translation is uncoupled from RNAi activity in vitro in Drosophila (Zamore et al. 2000). In contrast, there are observations that untranslated mRNAs are recalcitrant to RNAi in Drosophila oocytes in vivo (Kennerdell et al. 2002). The insensitivity to RNAi of maternal, untranslated mRNAs in oocytes has been attributed to protective mechanisms that sequester these mRNAs from both translation, and hence RNAi (Kennerdell et al. 2002). Other studies utilizing RNAi to target viruses with RNA genomes are also contradictory. Hu and colleagues provided evidence that HIV-1 RNA genomes encapsulated in viral particles during the early stages of viral entry and reverse transcription are relatively insensitive to RNAi (Hu et al. 2002) whereas Jacque and coworkers observed that genomic HIV RNA within a nucleoprotein complex is still susceptible to RNAi (Jacque et al. 2002). In the present study we took advantage of a well-characterized translational switch afforded by the ferritin IRE-IRP1 and compared RNAi results from this system with antibiotic mediated inhibition of translation. Our results demonstrate that an untranslated target is as susceptible, if not more susceptible to RNAi than a translated target, indicating that in live cells active translation is not required for siRNA function. Interestingly, inhibition of translation initiation rendered by the IRE-IRP1 complex would leave fewer ribosomes on the target mRNA. In contrast, Hygromycin mediated arrest of translational elongation would presumably leave more ribosomes along the target mRNA (Todorov et al. 1977). RNAi function was not impaired in either case, demonstrating the recruitment of RISC to the target mRNA does not require active translation.
In addition to investigating the link between translation and RNAi, we took advantage of the ferritin IRE-IRP-1 system to test whether or not unidirectional 5′ to 3′ or 3′ to 5′ scanning was required for RNAi. Collectively the results we have obtained suggest that RNAi can take place in the absence of active translation apparatus and does not involve a unidirectional scanning process. Our results are not in total contradiction with the biochemical evidence that links RISC to components of the translational apparatus, but they clearly demonstrate that active translation is not required for RNAi. The actual mechanism by which RISC efficiently finds the target sequence is still unknown. An attractive possibility is that RNAi may be linked to another RNA degradation pathway, the recently described RNA processing bodies, or P bodies (Sheth and Parker 2003). These could serve as sites for co-localizing mRNAs with the degradation components. The problem of how mRNAs get to P bodies is currently unknown. RNAi functions to reduce target RNA levels by impacting on the steady state levels of the target mRNA. Thus, RNAs may be cycled through P-bodies following a round of translation or cycled to P-bodies when translation is blocked, providing a possible explanation for our results. The observation that siRNA efficacy is increased during translational inhibition implies that RNAi and translation are not totally independent processes. One rational hypothesis is that one or more cellular factors are shared by the translation apparatus and RISC, thereby establishing a competitive relationship between those two pathways. Finally, the observation that micro RNAs which normally direct translational inhibition can be converted to siRNAs that direct cleavage by simply changing the base-pairing properties with the target (Doench et al. 2003) suggests that these two pathways share a common mechanism for target identification. It would seem that in the case of micro RNAs, the block to translation may also occur prior to ribosome scanning of the mRNA. Further understanding of the intracellular localization of RISC complexes and their relation to the translational machinery is of paramount importance.
MATERIALS AND METHODS
DNA constructs and siRNAs
Both strands of IRE were chemically synthesized (sense strand: CCCCAAGCTTTGCTTCAACAGTGCTTGGACAAGCTTGGGG; antisense strand: CCCCAAGCTTGTCCAAGCACTGTTGAAGCA AAGCTTGGGG), annealed, purified, and restriction digested by HindIII. The products were inserted into a HindIII site 28 bp downstream of transcription start site of the human U1 snRNA promoter, which was cloned upstream of an EGFP-coding sequence in a pBluescript vector. A similar approach was used to generate the double IRE construct by insertion of another IRE-like sequence within a unique XhoI site downstream of the EGFP stop codon. The siRNAs were synthesized by the City of Hope DNA/ RNA synthesis facility. The sequence of the sense strand of siRNA-GFP is 5′-GCUGACCCUGAAGUUCAUCdTdT; the antisense strand sequence of siRNA-GFP is 5′-GAUGAACUUCAGGGUCA GCdTdT.
Hemin (Cat No. 51280) and Deferoxaminemesylate (Cat No. D9533) were purchased from SIGMA-ALDRICH, Inc.
Cell culture and transfections
Adherent HEK-293 cells were grown in 10% FBS in DMEM, supplemented with glutamine in the presence of antibiotics. All transfection assays were done using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. HEK-293 cells at ninety percent confluency were transfected in six-well plates with 0.5 μg reporter gene construct and 50 nM siRNA (final concentration) unless specified otherwise. Carrier DNA was used to bring the total amount of nucleic acid transfected to each well to 4 μg.
RNA extraction and real-time RT-PCR
Total RNA was isolated using RNA STAT-60 (TEL-TEST B, Inc.) following the product protocol. The DNA-free kit (Ambion Cat # 1906) was used to purify total RNA from contaminating DNA. Oligo-dT was used to produce the cDNAs using a commercial Reverse Transcriptase kit (Invitrogen) following manufacturer’s protocol. cDNA from each sample was used as template for the subsequent PCR and real-time PCR analyses. Real-time PCR reactions were performed in a Bio-Rad Light Cycler using Cyber Green to monitor amplification. The PCR primers are: 5′-ACG TAAACGGCCACAAGTTC (sense) and 5′-AAGTCGTGCTGCTT CATGTG (antisense). GAPDH was used as internal control, and was amplified using the following primers: 5′-CATTGACCTCA ACTACATG (sense) and 5′-TCTCCATGGTGGTGAAGAC (anti-sense).
Serial dilutions of the U1-GFP plasmid were used to produce the standard curve and to determine the copy number of GFP transcripts in the samples. Serial dilutions of total RNA from nontransfected HEK293 cells were used to produce the standard curve for GAPDH. In all of the assays, linear correlation coefficients (r) for the standard curve were greater than 0.97.
SDS-PAGE and Western blotting
Cell extract samples were denatured in loading buffer (final concentration: 50 mM Tris, 5% SDS, 10% glycerol, 2% 2-mercapto-ethanol, 0.01% Bromophenol blue dye) for 5 min at 95°C and separated in 10% SDS-PAGE gels. The denatured proteins were then electro-transferred onto a PVDF membrane which was blocked with 5% fat-free milk powder in PBS and 0.5% Tween 20 for 1h. An anti-EGFP antibody (diluted 1:4000, Clonetech), and an anti-β-actin antibody (diluted 1:2500, Sigma) were used sequentially. Following three washes of 5 min in PBS, a secondary antibody (HRP-anti-mouseIgG, Sigma, dilution 1:10000) was incubated with the blots for 1 h at room temperature, followed by three 5-min washes in PBS. Antibody-bound proteins were visualized using the ECL Western blotting analysis system (Amersham, RPN2109).
Acknowledgments
This work was supported by NIH grants to JJR: AI29329, AI42552, and HL074704. The authors wish to thank Dongho Kim for providing the EGFP siRNAs and members of the Rossi Lab for critical discussions of this work.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7158605.
REFERENCES
- Caudy, A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy, A.A., Ketting, R.F., Hammond, S.M., Denli, A.M., Bathoorn, A.M., Tops, B.B., Silva, J.M., Myers, M.M., Hannon, G.J., and Plasterk, R.H. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414. [DOI] [PubMed] [Google Scholar]
- Cech, T.R. 2004. Beginning to understand the end of the chromosome. Cell 116: 273–279. [DOI] [PubMed] [Google Scholar]
- Djikeng, A., Shi, H., Tschudi, C., Shen, S., and Ullu, E. 2003. An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA 9: 802–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn, D.M., Novina, C.D., and Sharp, P.A. 2003. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4: 457–467. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Gonzalez, A., Jimenez, A., Vazquez, D., Davies, J.E., and Schindler, D. 1978. Studies on the mode of action of Hygromycin B, an inhibitor of translocation in eukaryotes. Biochim. Biophys. Acta. 521: 459–469. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A, Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hentze, M.W. and Kuhn, L.C. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. 93: 8175–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M.W., Caughman, S.W., Rouault, T.A., Barriocanal, J.G., Dancis, A., Harford, J.B., and Klausner, R.D. 1987. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science 238: 1570–1573. [DOI] [PubMed] [Google Scholar]
- Hu, W.Y., Myers, C.P., Kilzer, J.M., Pfaff, S.L., and Bushman, F.D. 2002. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 12: 1301–1311. [DOI] [PubMed] [Google Scholar]
- Ishizuka, A., Siomi, M.C., and Siomi, H. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 16: 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacque, J.M., Triques, K., and Stevenson, M. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418: 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell, J.R., Yamaguchi, S., and Carthew, R.W. 2002. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes & Dev. 16: 1884–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koloteva, N., Muller, P.P., and McCarthy, J.E. 1997. The position dependence of translational regulation via RNA-RNA and RNA-protein interactions in the 5′-untranslated region of eukaryotic mRNA is a function of the thermodynamic competence of 40 S ribosomes in translational initiation. J. Biol. Chem. 272: 16531–16539. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D.L. and Tollervey, D. 1998. Birth of the snoRNPs: The evolution of the modification-guide snoRNAs. Trends Biochem. Sci. 23: 383–388. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3: 737–747. [DOI] [PubMed] [Google Scholar]
- Merrick, W.C. 1992. Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 56: 291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler, M., Gray, N.K., and Hentze, M.W. 1998. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol. Cell 2: 383–388. [DOI] [PubMed] [Google Scholar]
- Nilsen, T.W. 1994. RNA-RNA interactions in the spliceosome: Unraveling the ties that bind. Cell 78: 1–4. [DOI] [PubMed] [Google Scholar]
- Pham, J.W., Pellino, J.L., Lee, Y.S., Carthew, R.W., and Sontheimer, E.J. 2004. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94. [DOI] [PubMed] [Google Scholar]
- Sachs, A.B., Sarnow, P., and Hentze, M.W. 1997. Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell 89: 831–838. [DOI] [PubMed] [Google Scholar]
- Scacheri, P.C., Rozenblatt-Rosen, O., Caplen, N.J., Wolfsberg, T.G., Umayam, L., Lee, J.C., Hughes, C.M., Shanmugam, K.S., Bhattacharjee, A., Meyerson, M., et al. 2004. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. 101: 1892–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, L.J. and Rossi, J.J. 2003. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 21: 1457–1465. [DOI] [PubMed] [Google Scholar]
- Sheth, U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz, C.A., Holko, M., de Veer, M.J., Silverman, R.H., and Williams, B.R. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5: 834–839. [DOI] [PubMed] [Google Scholar]
- Smith, H.C., Gott, J.M., Hanson, M.R. 1997. A guide to RNA editing. RNA 3: 1105–1123. [PMC free article] [PubMed] [Google Scholar]
- Theil, E.C. 1990. Regulation of ferritin and transferrin receptor mRNAs. J. Biol. Chem. 265: 4771–4774. [PubMed] [Google Scholar]
- Todorov, I.N., Smal’ko, P., and Galkin, A.P. 1977. Polysomal apparatus as a reflection of function interactions between translation and transcription processes during protein synthesis inhibited by cycloheximide. Biokhimiia 42: 2149–2159. [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]
- Zeng, Y. and Cullen, B.R. 2002. RNA interference in human cells is restricted to the cytoplasm. RNA 8: 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]