Abstract
Archaeal pre-tRNAs are characterized by the presence of the bulge–helix–bulge (BHB) structure in the intron stem-and-loop region. A chimeric pre-tRNA was constructed bearing an intron of the archaeal type and the mature domain of the Saccharomyces cerevisiae suppressor SUP4 tRNATyr. This pre-tRNAArchEuka is correctly cleaved in several cell-free extracts and by purified splicing endonucleases. It is also cleaved and ligated in S. cerevisiae cells, providing efficient suppression of nonsense mutations in various genes.
Keywords: BHB, M. jannaschii, RAT1, tap1-1, tRNA splicing
INTRODUCTION
Accuracy in tRNA splicing is essential for the formation of functional tRNAs, and hence for cell viability. In both Eukarya and Archaea correct splicing depends on the specific recognition of tRNA precursors (pre-tRNAs) by tRNA splicing endonucleases, but the mechanisms for determining the cleavage sites appear to differ (Belfort and Weiner 1997; Trotta and Abelson 1999). The eukaryal tRNA splicing endonuclease interacts with the mature tRNA domain, measures the distance from the body of the mature tRNA, and cleaves near a base pair that is formed between nucleotides in the anticodon loop and the intron (the A-I pair) (Mattoccia et al. 1988; Reyes and Abelson 1988). This cleavage generates the 3′ splice site. An independent cleavage event, also at a fixed distance from the mature domain, produces the 5′ splice site (Baldi et al. 1992; Di Nicola Negri et al. 1997; Trotta et al. 1997). In contrast, the archaeal enzyme recognizes a structural element of pre-tRNAs comprised of two bulges of 3 nt each separated by 4 bp, the so-called bulge–helix–bulge (BHB) motif (Daniels et al. 1985; Diener and Moore 1998). This structure functions independently of the part of the molecule that constitutes the mature tRNA.
We recently developed a system in mouse cells and in mice that makes possible the cleavage and ligation of mRNAs having a BHB structure, provided that the archaeal endonuclease from Methanococcus jannaschii is present (Deidda et al. 2003; G. Deidda, N. Rossi, S. Putti, and G.P. Tocchini-Valentini, unpubl.). The BHB-dependent cleavage and ligation work both when the BHB is found in cis and when the BHB is formed, in trans, between appropriately designed RNA molecules and the targeted mRNAs. This method is not limited to mRNA, but, in principle, could be applied to destroy, modify, or restore the formation of regulatory non-coding RNA sequences.
We wish to develop a similar system in Saccharomyces cerevisiae, in view of the fact that RNA interference has not been observed in yeast (Aravind et al. 2000; Wood et al. 2002) and that an elegant genetics is available for that organism. As a first step, we ask whether S. cerevisiae cells are able to splice RNA molecules containing the intrinsically archaeal BHB motif. We use precursors of a suppressor tRNA containing the BHB structure, and we monitor the generation of a functional tRNA by means of its ability to suppress nonsense mutations in appropriate genes in S. cerevisiae cells. We show that yeast cells are able to correctly process, cleave, and ligate tRNA precursors containing the archaeal BHB motif. In addition, we show here that even noncanonical, long pre-tRNA molecules, containing the BHB structure, are cleaved and ligated in yeast, producing functional suppressor tRNA.
RESULTS AND DISCUSSION
SUP4 pre-tRNAArchEuka Tyr is cleaved in vitro
In order to verify the capability of the yeast tRNA splicing endonuclease to recognize and correctly cleave the archaeal bulge–helix–bulge structure in vivo (as already demonstrated in vitro), we used a chimeric suppressor tRNA. We chose the yeast SUP4 tRNATyr, which suppresses nonsense mutations of the ochre type (UAA) by inserting tyrosine (Goodman et al. 1977). SUP4 tRNATyr was modified to bear eukaryotic tRNA structural features as well as the archaeal BHB motif. This hybrid precursor was designated SUP4 pre-tRNAArchEukaTyr (see Fig. 1).
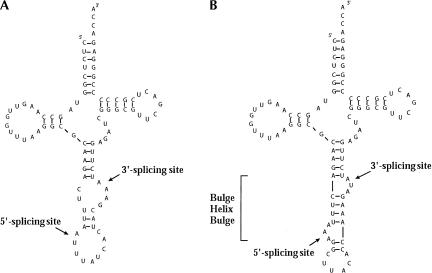
FIGURE 1.
(A) SUP4 pre-tRNATyr. This tRNA precursor carries a mutation in the anticodon that enables it to suppress nonsense mutations of the ochre type (UAA). (B) SUP4 pre-tRNAArchEuka Tyr. An archaeal-type intron of 17 nt forming a BHB structure has been inserted in the anticodon stem-and-loop region of SUP4 pre-tRNATyr.
The SUP4 pre-tRNAArchEuka Tyr consists of two regions derived from yeast SUP4 pre-tRNATyr (from the beginning to nucleotide 39 and from nucleotide 54 to the end) joined by a 17-nt insert that produces a BHB motif typical of archaeal pre-tRNAs. This hybrid pre-tRNA is analogous, in its structure, to other chimeras we used in in vitro experiments, such as the pre-tRNAArchEuka Phe. As we have shown previously, pre-tRNAArchEuka Phe is cleaved by either the eukaryal or the archaeal endonuclease (Fabbri et al. 1998).
If the splicing endonuclease cleaves the SUP4 pre-tRNAArchEuka Tyr and the tRNA ligase ligates the two resulting halves, the product will be indistinguishable from mature SUP4 tRNATyr and will be able to suppress ochre nonsense mutations in appropriate S. cerevisiae strains. Before testing the ability of the SUP4 chimera to suppress nonsense mutations in yeast cells, we examined whether in vitro the SUP4 pre-tRNAArchEuka Tyr chimera behaves like pre-tRNAArchEuka Phe and is cleaved by either the eukaryal or the archaeal endonuclease.
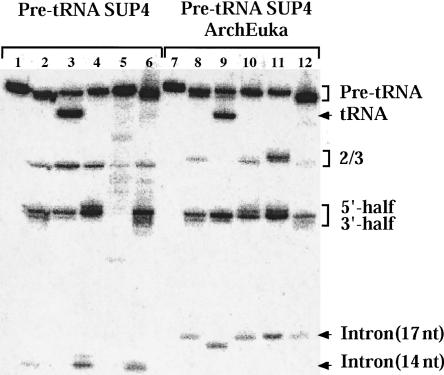
Splicing reactions with natural and chimeric pre-tRNAs were assayed in cell extracts (from yeast or Xenopus laevis) or using purified splicing endonucleases (from X. laevis or M. jannaschii). Figure 2 shows that SUP4 pre-tRNATyr is cleaved correctly by the eukaryal enzymes (lanes 2–4,6) but not by the archaeal enzyme (lane 5), whereas all enzymes precisely cleave the SUP4 pre-tRNAArchEuka Tyr (lanes 8–12). Notice that SUP4 pre-tRNATyr is 93 nt long, while SUP4 pre-tRNAArchEuka Tyr is 96 nt long, because of the presence of the BHB motif. Mature tRNA is present only in lanes 3 and 9, because of the higher ATP concentration (necessary for ligase activity) found in manually prepared nuclei with respect to nuclei prepared en masse. The intron in lane 9 has an additional phosphate group and therefore migrates faster than the intron in lanes 8 and 10–12. This phosphate is added by a kinase, in the presence of ATP, at the 5′-end of the tRNA intron. The intron, in fact, is left with a free OH group after the cleavage by the tRNA-splicing endonuclease, while at the 3′-end of the tRNA 5′-half a 2′,3′-cyclic phosphate remains (Gandini Attardi et al. 1985). In lane 3 the intron is only visible after long exposures and may be degraded in the crude extract. Conceivably, the ArchEuka intron (lane 9) is more stable than its normal counterpart (lane 3) (it is longer by 3 nt and differs in its sequence). The difference in migration of tRNA half-molecules seen when comparing pre-tRNA incubated in extracts (lanes 2,3,6,8,9,12) versus that incubated with pure enzymes (lanes 4,10,11) is due to the heterogeneity at the 3′-end of the precursors, caused by stuttering of the T7 RNA polymerase used to synthesize them. The added nucleotides are then removed by enzymes present in the crude extract, but not in the purified preparation.
FIGURE 2.
In vitro splicing of SUP4 pre-tRNATyr and SUP4 pre-tRNAArchEuka Tyr. 32P-labeled tRNA precursors were incubated in cell-free extracts or with purified splicing endonucleases, electrophoresed in 8 M urea–10% polyacrylamide gels and exposed to PhosphorImager. (Lanes 1–6) SUP4 pre-tRNATyr (93 nt long); (lanes 7–12) SUP4 pre-tRNAArchEuka Tyr (96 nt long). (Lanes 1,7) Nonincubated control; (lanes 2,8) incubation with nuclear extracts of X. laevis oocytes prepared en masse; (lanes 3,9) incubation with nuclear extracts of X. laevis oocytes prepared manually; (lanes 4,10) incubation with purified X. laevis splicing endonuclease; (lanes 5,11) incubation with purified M. jannaschii endonuclease; (lanes 6,12) incubation with S. cerevisiae cell-free extract. “2/3” indicates intermediates in which the 5′-half or 3′-half tRNA is still joined to the intron. The identity of the products was verified by sequencing.
The result shown in Figure 2 confirms that SUP4 pre-tRNATyr, a typical eukaryotic precursor, is not a suitable substrate for the archaeal enzyme, while the chimeric tRNA precursor, featuring an archaeal BHB motif, is recognized and cleaved by both eukaryal and archaeal enzymes, like pre-tRNAArchEuka Phe.
SUP4 pre-tRNAArchEuka Tyr is functional in vivo
The gene encoding SUP4 pre-tRNAArchEuka Tyr was introduced into a yeast strain (PJ17-1A) bearing ochre nonsense mutations in three different loci, met4-1o, lys2-1o, and ade2-1o. These mutations require increasing levels of SUP4 tRNATyr in order to be suppressed by low levels, lys2-1o is suppressed by intermediate levels, and the ade2-1o mutation requires high levels of expressed, that is, met4-1o is suppression of the SUP4 gene (Hawthorne and Leupold 1974). This difference is due to the functionality of the mutated protein in which the tyrosine amino acid is inserted as a replacement for the wild-type amino acid. The use of a multiple mutated strain is important for two reasons: it permits an evaluation of the expression of the SUP4 gene, and it also avoids the complication that revertants of the nonsense mutation would be erroneously confused with true suppression events.
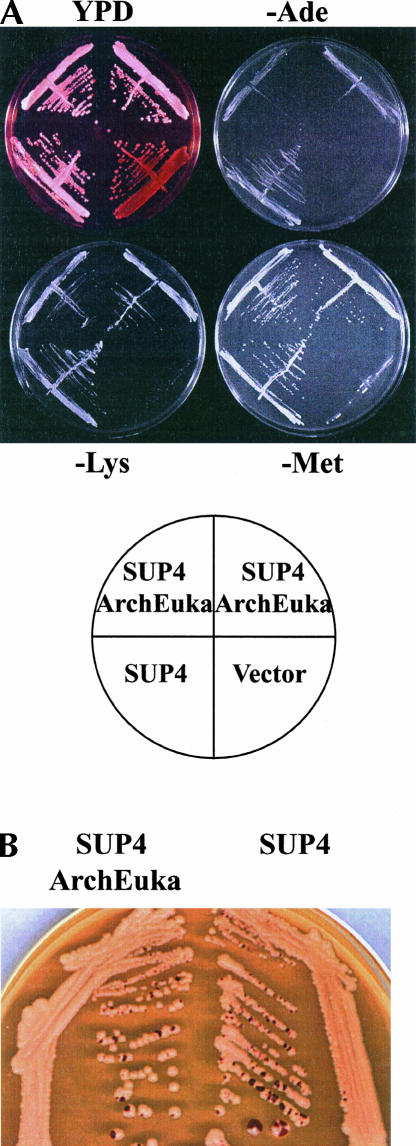
The yeast strain PJ17-1A (met4-1o, lys2-1o, ade2-1o), carrying the centromere-based plasmid YCp50 containing the gene for SUP4 pre-tRNAArchEuka Tyr, was plated onto selective media without methionine, lysine, or adenine. As controls, we used cells with a plasmid encoding SUP4 pre-tRNATyr and with the vector alone. The growth of cells carrying SUP4 pre-tRNAArchEuka Tyr is comparable to that of control cells with SUP4 tRNATyr on -Met and -Lys plates; less growth is obtained on -Ade plates. Cells with the vector alone do not grow on any of the three kinds of selective plates (Fig. 3A).
FIGURE 3.
In vivo suppression in a yeast strain with a plasmid carrying the gene for SUP4 pre-tRNAArchEuka Tyr. (A) The strain PJ17-1A, carrying the YCp50 plasmid with the SUP4 pre-tRNAArchEuka Tyr gene, was streaked onto a rich YPD plate or onto selective plates missing, respectively, methionine, lysine, or adenine, and incubated for 3 d at 30°C. Two different transformants are shown in the same plate, together with transformants containing the SUP4 pre-tRNATyr gene or the YCp50 vector alone, as controls. The scheme of streaking in each plate is shown below. (B) A rich YPD plate is shown to illustrate red sectors in white colonies. A nonsense mutation in the ade2 gene causes the accumulation of a red pigment, while suppression of the mutation restores the white color. Some colonies are white with red sectors, because of the occasional loss of the plasmid.
Another way to determine suppression of the nonsense ade2-1o mutation is to grow the transformed cells on rich media. On these plates, ade2 mutant colonies are red, due to the accumulation of a pigment caused by this mutation; the congenic wild-type ADE2+ strain is white. Depending on the level of suppression, yeast colonies carrying a suppressor tRNA will be white (good suppression level), pink (intermediate level), or red (no suppression at all). Moreover, a well-suppressed colony will be white, but red sectors will appear, especially in large colonies, due to the occasional loss of the plasmid encoding the suppressor tRNA (Di Segni et al. 1993). Colonies with SUP4 pre-tRNAArchEuka Tyr, as well as with SUP4 pre-tRNATyr-encoding plasmids, are white with red sectors (Fig. 3B). This result indicates that the suppression is caused by a tRNA encoded on a plasmid and not by a chromosomal suppressor tRNA gene.
We conclude from this analysis that a hybrid tRNA precursor, bearing features typical of eukaryotic and archaeal pre-tRNAs, produces enough mature tRNA to be functional in suppression of nonsense mutations in yeast. Therefore, the hybrid ArchEuka precursor is a good substrate for the eukaryotic splicing machinery in vivo.
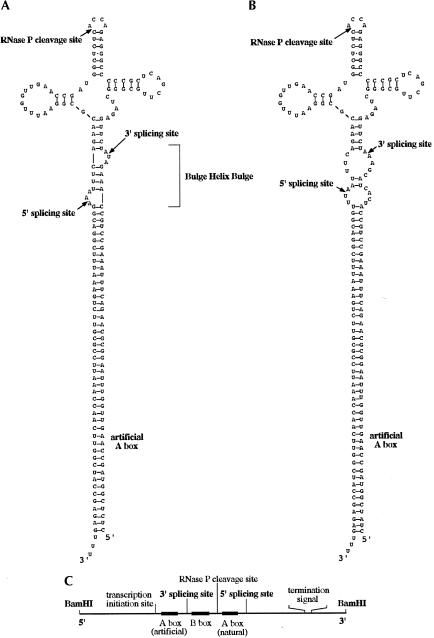
Having shown that BHB-containing pre-tRNAs are processed in S. cerevisiae, we then asked if noncanonical tRNA molecules, carrying the BHB structure, could also be spliced in yeast. To this end, we designed a very long precursor of SUP4 tRNA different from the natural pre-tRNA. In this long precursor, the two regions pairing to form the BHB structure are encoded in two DNA regions that lie far away from each other. The main feature of this RNA, which we named “long pre-tRNAArchEuka,” is the replacement of the intron stem-and-loop of SUP4 pre-tRNATyr by a very long stem that can form a BHB structure. The long stem is open at its end, while the mature 5′-end of the tRNA is joined to an artificially introduced CCA sequence at the 3′-end (see Fig. 4). In order to obtain a mature tRNA from this precursor, a cleavage by RNase P, leaving a mature 5′-end and a 3′-terminal CCA, must occur. We have already shown that a tRNA molecule where the 5′- and 3′-ends are joined together is recognized and correctly cleaved by RNase P (Carrara et al. 1995). In addition, the splicing endonuclease should cut at the correct sites of the BHB motif, followed by ligation of the two halves (Fig. 4A).
FIGURE 4.
(A) Long SUP4 pre-tRNAArchEuka Tyr. After processing by RNase P and splicing of the BHB-containing intron, this long precursor produces a bona fide SUP4 tRNATyr. (B) Long SUP4 pre-tRNATyr. This precursor is like the one in panel A except that the intron does not contain the BHB. (C) Schematic structure of the gene encoding the long tRNA precursor.
In designing such a construct we had to avoid the decreased efficiency of transcription caused by the long distance between the A-box and the B-box of the Pol III transcription promoter (Baker et al. 1987; Fabrizio et al. 1987). Accordingly, we introduced an artificial A-box at the same distance from the B-box as it is in the wild-type SUP4 gene (Fig. 4C). As a control, we designed the same construct without the sequences forming the BHB structure (Fig. 4B).
The genes encoding the long pre-tRNAs were cloned in the low-copy URA3-CEN plasmid YCp50 and transformed into the PJ17-1A strain (met4-1o, lys2-1o, ade2-1o). As mentioned before, the ability of transformants to grow on plates without methionine, lysine, or adenine is an indication of the amount of mature SUP4 tRNATyr and, therefore, of correct splicing.
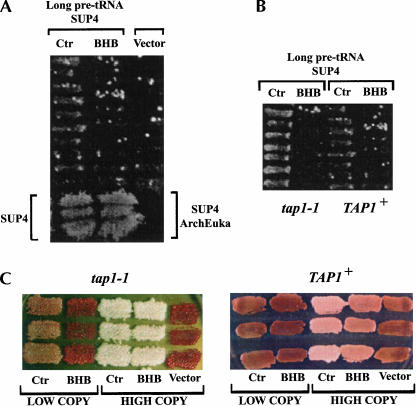
Ura+ transformants were replica-plated on selective media, and growth was checked after 7 d. Some growth was observed on plates lacking methionine, whereas no growth was seen on plates without lysine or adenine. Cells containing the plasmid encoding the control long pre-tRNA, without the BHB structure, grew more than those carrying the plasmid encoding the long pre-tRNA with the BHB, for which growth was hardly visible (Fig. 5A).
FIGURE 5.
(A) Several transformants of the strain PJ17-1A with the low-copy URA3–CEN plasmid YCp50 encoding the long SUP4 pre-tRNAArcheEuka Tyr with the BHB or encoding, as a control (Ctr), the long SUP4 pre-tRNATyr without the BHB were patched on -Ura plates and, after 2 d, replica-plated on -Met plates. The photo was taken after 7 d following the replica. Transformants with the vector alone, as well as transformants with the plasmid encoding SUP4 pre-tRNATyr and SUP4 pre-tRNAArchEuka Tyr are shown. (B) Effect of the tap1-1 mutation. YCp50 plasmids containing the long SUP4 pre-tRNAs (BHB or Ctr) were transformed in the tap1-1 mutant GDS4-16D and compared to transformants in the wild-type TAP1+ (RAT1+) strain PJ17-1A. Growth on -Met plates is shown. (C) Comparison of the suppression level provided by long pre-tRNA (BHB or Ctr) encoded by the high-copy plasmid pYX212 or by the low-copy plasmid YCp50, in the tap1-1 mutant or in the wild-type TAP1+ strain. Growth on rich YPD medium is shown.
The lack of suppression, even for the control construct, could derive, in principle, from the inefficiency in any one of the reactions necessary to get and process the precursor. In order to increase the amount of mature SUP4 tRNA, a yeast mutant strain previously selected, tap1-1 (Aldrich et al. 1993; Di Segni et al. 1993), was used. This mutant is characterized by the ability to increase the expression of a tRNA gene with a defective transcription promoter. Plasmids encoding the long pre-tRNAs were transformed into a tap1-1 mutant strain (GDS4-16D) and, indeed, we obtained increased suppression with the control plasmid, but still very low suppression for the construct bearing the BHB motif (Fig. 5B). The genes encoding the long pre-tRNAs were then cloned into the high-copy 2μ-plasmid pYX212. Using this vector, very good levels of suppression were observed, with both the control plasmid and the construct coding for the BHB structure (Fig. 5C), especially in the tap1-1 mutant. Table 1 summarizes the growth levels on different culture media for all strain/plasmid combinations. The presence of SUP4 tRNATyr was verified by an acidic Northern blot analysis (data not shown). The long pre-tRNA, lacking the BHB structure, gives rise to a higher level of suppression as compared to the BHB-containing long pre-tRNA. This finding is consistent with previous work, where we showed that hybrid ArchEuka pre-tRNAs are less efficiently spliced when they are incubated in cell extracts or are injected into X. laevis nuclei (Fruscoloni et al. 2001).
TABLE 1.
Suppression phenotypes of long pre-tRNAs encoded by high- and low-copy plasmids in different strains
| Growth medium | ||||||||||
| - Ura | - Met | - Lys | - Ade | YPD | ||||||
| Strain/plasmid | ||||||||||
| tap1-1 | TAP1+ | tap1-1 | TAP1+ | tap1-1 | TAP1+ | tap1-1 | TAP1+ | tap1-1 | TAP1+ | |
| pYX212-Ctr | +++ | +++ | +++ | ++ | +++ | ++ | +++ | + | W | P |
| pYX212-BHB | +++ | +++ | +++ | ++ | +++ | + | +++ | +/- | W | P/R |
| pYX212 alone | +++ | +++ | - | - | - | - | - | - | R | R |
| YCp50-Ctr | +++ | +++ | + | +/−− | +/− | - | - | - | P | R/P |
| YCp50-BHB | +++ | +++ | - | - | - | - | - | - | R | R |
High-copy URA3-2μ plasmid pYX212 or low-copy URA3-CEN plasmid YCp50, containing the gene encoding the long pre-tRNA with the BHB structure or without it (Ctr), were transformed in the tap1-1 mutant GDS4-16 or in the wild-type TAP1+(RAT1+) strain PJ17-1A. Transformants were patched onto selective plates lacking uracile (- Ura), methionine (- Met), lysine (- Lys), or adenine (- Ade), or onto rich plates (YPD). Growth was scored after 3 d. (W) white; (P) pink; (R) red. The number of + indicates the relative growth level.
We conclude that long pre-tRNAArchEuka is correctly spliced in yeast cells.
In previous in vitro experiments we could show that the BHB constitutes an autonomous structural element when challenged with the yeast or the X. laevis splicing endonuclease. We were able to discriminate between the eukaryal and the archaeal mode of processing, typical of the enzyme, by increasing the distance of the intron from the mature domain of the pre-tRNA, thereby giving to the eukaryal enzyme the possibility to operate in the mature-domain independent mode. We cannot utilize this method in vivo, since we have to take into account the constraint derived from the need to produce a functional specific tRNA. There is always the requirement to locate the BHB in a fixed position with respect to the mature domain of the tRNA. The experiments reported here, therefore, do not prove that the yeast enzyme can work in vivo in the archaeal way, but they do show that an archaeal structure (the BHB motif) can be correctly spliced in S. cerevisiae cells.
We wish to develop a system in yeast that makes possible the BHB-dependent cleavage and ligation both when the BHB is found in cis and when the BHB is formed, in trans, between appropriately designed targeting RNAs and a targeted mRNA. We will now try to see if yeast cells are able to process a BHB that does not have in its vicinity a correctly positioned mature tRNA domain. If such an attempt fails, as in mouse cells, we will have to express in yeast cells an archaeal splicing endonuclease in order to reconstruct the complete archaeal splicing system dependent on the BHB.
MATERIALS AND METHODS
Strains, plasmids, and growth media
Escherichia coli strain DH5-α was used to propagate plasmid DNA and was grown on LB medium containing ampicillin. S. cerevisiae strains were PJ17-1A (Mata, trp1, ura3-1, ade2-1o, lys2-1o, met4-1o, can1-100o, gal10-1u, his5-2u, leu2-1u) (James and Hall 1990) and GDS4-16D, a derivative of PJ17-1A that carries the mutation tap1-1 in the TAP1/RAT1 gene (Amberg et al. 1992; Aldrich et al. 1993; Di Segni et al. 1993; Kenna et al. 1993). Yeast strains were grown on rich YPD medium or drop-out selective media, prepared according to Ausubel et al. (1998). E. coli was transformed with plasmid DNA using the CaCl2 method or by electroporation; yeast was transformed using the lithium acetate procedure. Bacterial plasmid PUC19 and the yeast shuttle vectors YCp50 (a URA3-CEN plasmid) and pYX212 (a URA3–2μ plasmid; R&D System) were used.
tRNA genes
The SUP4 tRNATyr gene, contained in a 266-bp BamHI fragment inserted in YCp50, was used (Aldrich et al. 1993; Di Segni et al. 1993). The SUP4 tRNAArchEuka Tyr gene was constructed by using eight pairs of annealed oligonucleotides; the long SUP4 tRNATyr gene was constructed with 12 pairs of annealed oligonucleotides. The pairs of oligonucleotides were ligated together and cloned, first into PUC19, between the EcoRI and HindIII sites, and successively into the yeast plasmids. The oligonucleotides were synthesized using the 392 DNA/RNA Synthesizer (Applied Biosystem); they were phosphorylated, annealed, ligated, and digested with restriction enzymes according to standard procedures.
In vitro transcription and processing
tRNA gene sequences were amplified by PCR using two primers bearing, respectively, the T7-promoter sequence at the 5′-end and the sequence complementary to CCA at the 3′-end. Pre-tRNAs were synthesized in vitro by T7 RNA polymerase in the presence of [α-32P]UTP; labeled RNA molecules were purified by phenol-chloroform extraction and ethanol precipitation, followed by electrophoresis on 8 M urea–10% polyacrylamide gels. RNA was eluted from the gel, filtered, and ethanol-precipitated (Di Nicola Negri et al. 1997).
Pre-tRNAs were processed in nuclear extracts of X. laevis stage 6 oocytes prepared en masse (Mattoccia et al. 1979; Gandini Attardi et al. 1990) or manually prepared (Lund and Paine 1990). X. laevis RNA endonuclease was purified and assayed according to Gandini Attardi et al. (1990). M. jannaschii RNA endonuclease was purified according to Li and Abelson (2000) and assayed at 65°C for 30 min in 25 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, and 10% glycerol. Yeast cell extract was prepared and assayed according to Ho et al. (1990). RNA splicing products were purified by phenol-chloroform extraction, ethanol precipitation, and electrophoresis on 8 M urea–10% polyacrylamide gels; the gels were exposed to PhosphorImager and/or to X-ray films at −70°C.
Northern blot analysis was performed under acidic conditions according to Sarkar et al. (1999) and Varshney et al. (1991). A 21-nt-long probe was used for hybridization. The blot was exposed to PhosphorImager after a stringent wash for 1 min at 56°C.
Acknowledgments
We thank M.I. Baldi for continuous advice regarding this work and for assistance with the Northern blot analysis. We also thank P. Fruscoloni and all the members of our laboratory for advice, G. Di Franco for technical assistance, and D. De Simone and R. Matteoni for help with the figures. Our cordial thanks go to Th. Wagner for critically reading the manuscript. We are very grateful to R. Haselkorn for helpful discussions and critical reading of the manuscript. This work was funded by the following projects: “Bio-molecole per la Salute Umana, 5%-L. 95/95” (MURST-CNR), “Genomica Funzionale” (CNR-MIUR), “Progetti Negoziali 2001” (MIUR-FIRB), and “European Networks of Excellence” (EUMORPHIA, MUGEN).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7138805.
REFERENCES
- Aldrich, T.L., Di Segni, G., McConaughy, B.L., Keen, N.J., Whelen, S., and Hall, B.D. 1993. Structure of the yeast TAP1 protein: Dependence of transcription activation on the DNA context of the target gene. Mol. Cell. Biol. 13: 3434–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D.C., Goldstein, A.L., and Cole, C.N. 1992. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 6: 1173–1189. [DOI] [PubMed] [Google Scholar]
- Aravind, L., Watanabe, H., Lipman, D.J., and Koonin, E.V. 2000. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. 97: 11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.M., Seidman, J.G., Smith, J.A., and Struhl, K. 1998. Current protocols in molecular biology, Vol. 2, Sec. 13. John Wiley & Sons, Inc., New York.
- Baker, R.E., Camier, S., Sentenac, A., and Hall, B.D. 1987. Gene size differentially affects the binding of yeast transcription factor tau to two intragenic regions. Proc. Natl. Acad. Sci. 84: 8768–8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi, M.I., Mattoccia, E., Bufardeci, E., Fabbri, S., and Tocchini-Valentini, G.P. 1992. Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science 255: 1404–1408. [DOI] [PubMed] [Google Scholar]
- Belfort, M. and Weiner, A. 1997. Another bridge between kingdoms: tRNA splicing in Archaea and Eukaryotes. Cell 89: 1003–1006. [DOI] [PubMed] [Google Scholar]
- Carrara, G., Calandra, P., Fruscoloni, P., and Tocchini-Valentini, G.P. 1995. Two helices plus a linker: A small model substrate for eukaryotic RNase P. Proc. Natl. Acad. Sci. 92: 2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, C.J., Gupta, R., and Doolittle, W.F. 1985. Transcription and excision of a large intron in the tRNA Trp gene of an archaebacterium, Halobacterium volcanii. J. Biol. Chem. 260: 3132–3134. [PubMed] [Google Scholar]
- Deidda, G., Rossi, N., and Tocchini-Valentini, G.P. 2003. An archaeal endoribonuclease catalyzes cis- and trans-nonspliceosomal splicing in mouse cells. Nat. Biotech. 21: 1499–1504. [DOI] [PubMed] [Google Scholar]
- Di Nicola Negri, E., Fabbri, S., Bufardeci, E., Baldi, M.I., Gandini Attardi, D., Mattoccia, E., and Tocchini-Valentini, G.P. 1997. The eucaryal tRNA splicing endonuclease recognizes a tripartite set of RNA elements. Cell 89: 859–866. [DOI] [PubMed] [Google Scholar]
- Di Segni, G., McConaughy, B.L., Shapiro, R.A., Aldrich, T.L., and Hall, B.D. 1993. TAP1, a yeast gene that activates the expression of a tRNA gene with a defective internal promoter. Mol. Cell. Biol. 13: 3424–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, J.L. and Moore, P.B. 1998. Solution structure of a substrate for the archaeal pre-tRNA splicing endonucleases: The bulge–helix–bulge motif. Mol. Cell 1: 883–894. [PubMed] [Google Scholar]
- Fabbri, S., Fruscoloni, P., Bufardeci, E., Di Nicola Negri, E., Baldi, M.I., Gandini Attardi, D., Mattoccia, E., and Tocchini-Valentini, G.P. 1998. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science 280: 284–286. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., Coppo, A., Fruscoloni, P., Benedetti, P., Di Segni, G., and Tocchini-Valentini, G.P. 1987. Comparative mutational analysis of wild-type and stretched tRNA3Leu gene promoters. Proc. Natl. Acad. Sci. 84: 8763–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruscoloni, P., Baldi, M.I., and Tocchini-Valentini, G.P. 2001. Cleavage of non-tRNA substrates by eukaryal tRNA splicing endonucleases. EMBO Reports 2: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandini Attardi, D., Margarit, I., and Tocchini-Valentini, G.P. 1985. Structural alterations in mutant precursors of the yeast tRNA3Leu gene which behave as defective substrates for a highly purified splicing endoribonuclease. EMBO J. 4: 3289–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandini Attardi, D., Baldi, M.I., Mattoccia, E., and Tocchini-Valentini, G.P. 1990. Transfer RNA splicing endonuclease from Xenopus laevis. Methods Enzymol. 181: 510–517. [DOI] [PubMed] [Google Scholar]
- Goodman, H.M., Olson, M.V., and Hall, B.D. 1977. Nucleotide sequence of a mutant eukaryotic gene: The yeast tyrosine-inserting ochre suppressor SUP4-o. Proc. Natl. Acad. Sci. 74: 5453–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne, D.C. and Leupold, U. 1974. Suppressors in yeast. Curr. Top. Microbiol. Immunol. 64: 1–47. [DOI] [PubMed] [Google Scholar]
- Ho, C.K., Rauhut, R., Vijayraghavan, U., and Abelson, J. 1990. Accumulation of pre-tRNA splicing ‘2/3’ intermediates in a Saccharomyces cerevisiae mutant. EMBO J. 9: 1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P. and Hall, B.D. 1990. ret1-1, a yeast mutant affecting transcription termination by RNA polymerase III. Genetics 125: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna, M., Stevens, A., McCammon, M., and Douglas, M.G. 1993. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol. Cell. Biol. 13: 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. and Abelson, J. 2000. Crystal structure of a dimeric archaeal splicing endonuclease. J. Mol. Biol. 302: 639–648. [DOI] [PubMed] [Google Scholar]
- Lund, E. and Paine, P.L. 1990. Nonaqueous isolation of transcription-ally active nuclei from Xenopus oocytes. Methods Enzymol. 181: 36–43. [DOI] [PubMed] [Google Scholar]
- Mattoccia, E., Baldi, M.I., Carrara, G., Fruscoloni, P., Benedetti, P., and Tocchini-Valentini, G.P. 1979. Separation of RNA transcription and processing activities from X. laevis germinal vesicles. Cell 18: 643–648. [DOI] [PubMed] [Google Scholar]
- Mattoccia, E., Baldi, M.I., Gandini Attardi, D., Ciafrè, S., and Tocchini-Valentini, G.P. 1988. Site selection by the tRNA splicing endonuclease of Xenopus laevis. Cell 55: 731–738. [DOI] [PubMed] [Google Scholar]
- Reyes, V.M. and Abelson, J. 1988. Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55: 719–730. [DOI] [PubMed] [Google Scholar]
- Sarkar, S., Azad, A.K., and Hopper, A.K. 1999. Nuclear tRNA amino-acylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 96: 14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta, C.R. and Abelson, J. 1999. tRNA splicing: An RNA world add-on or an ancient reaction? In The RNA world, 2d ed. (eds. R.F. Gesteland et al.), pp. 561–583. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Trotta, C.R., Miao, F., Arn, E.A., Stevens, S.W., Ho, C.K., Rauhut, R., and Abelson, J.N. 1997. The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89: 849–858. [DOI] [PubMed] [Google Scholar]
- Varshney, U., Lee, C.-P., and RajBhandary, U.L. 1991. Direct analysis of aminoacylation levels of tRNAs in vivo. J. Biol. Chem. 266: 24712–24718. [PubMed] [Google Scholar]
- Wood, V., Gwilliam, R., Rajandream, M.A., Lyne, M., Lyne, R., Stewart, A., Sgouros, J., Peat, N., Hayles, J., Baker, S., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]