Abstract
The eukaryotic translation initiation factor 4F (eIF4F) consists of three polypeptides (eIF4A, eIF4G, and eIF4E) and is responsible for recruiting ribosomes to mRNA. eIF4E recognizes the mRNA 5′-cap structure (m7GpppN) and plays a pivotal role in control of translation initiation, which is the rate-limiting step in translation. Overexpression of eIF4E has a dramatic effect on cell growth and leads to oncogenic transformation. Therefore, an inhibitory agent to eIF4E, if any, might serve as a novel therapeutic against malignancies that are caused by aberrant translational control. Along these lines, we developed two RNA aptamers, aptamer 1 and aptamer 2, with high affinity for mammalian eIF4E by in vitro RNA selection-amplification. Aptamer 1 inhibits the cap binding to eIF4E more efficiently than the cap analog m7GpppN or aptamer 2. Consistently, aptamer 1 inhibits specifically cap-dependent in vitro translation while it does not inhibit cap-independent HCV IRES-directed translation initiation. The interaction between eIF4E and eIF4E-binding protein 1 (4E-BP1), however, was not inhibited by aptamer 1. Aptamer 1 is composed of 86 nucleotides, and the high affinity to eIF4E is affected by deletions at both termini. Moreover, relatively large areas in the aptamer 1 fold are protected by eIF4E as determined by ribonuclease footprinting. These findings indicate that aptamers can achieve high affinity to a specific target protein via global conformational recognition. The genetic mutation and affinity study of variant eIF4E proteins suggests that aptamer 1 binds to eIF4E adjacent to the entrance of the cap-binding slot and blocks the cap-binding pocket, thereby inhibiting translation initiation.
Keywords: eIF4E, cap binding, RNA aptamer, SELEX, translation initiation
INTRODUCTION
In eukaryotes, mRNAs are modified at their 5′-ends with a structure termed cap, m7GpppN, where N is any nucleotide (Shatkin 1976). The cap plays a key role in facilitating the binding of the ribosomal 40S subunit to the 5′-end of mRNA (Shatkin 1976) through the interaction with eukaryotic translation initiation factor 4F (eIF4F). eIF4F is composed of the three subunits eIF4E, eIF4A, and eIF4G. eIF4E recognizes the 7-methylguanosine-containing cap of mRNA, while eIF4A is an RNA-dependent ATPase and unwinds the secondary structure present in the 5′-untranslated region of mRNAs. eIF4G serves as a scaffold protein for binding to other eIFs including eIF3, which recruits the ribosomal 40S subunit. eIF4E is the least abundant factor of all eIFs (Duncan et al. 1987), and the recognition of the mRNA cap by eIF4E is the rate-limiting step of eukaryotic translation initiation. In fact, eIF4E is a major target for translational control by extracellular stimuli. eIF4E is phosphorylated on Ser209 (Flynn and Proud 1995; Joshi et al. 1995) by the eIF4E kinase Mnk1 through a MAP kinase signal transduction pathway (Fukunaga and Hunter 1997; Waskiewicz et al. 1997). Upon phosphorylation, eIF4E binds less efficiently to the capped mRNA (Scheper et al. 2002; Zuberek et al. 2003). The activity of eIF4E is also modulated by eIF4E binding proteins (4E-BPs) (Gingras et al. 2001), which interfere with the eIF4E–eIF4G interaction by occupying the same binding site on eIF4E as eIF4G. On the other hand, 4E-BP does not interfere with the eIF4E–cap interaction (Haghighat et al. 1995; Mader et al. 1995; Marcotrigiano et al. 1999).
The accurate control of translation initiation is important for cell growth. Overexpression of eIF4E is known to cause deregulated cell growth (De Benedetti and Rhoads 1990) and malignant transformation (Lazaris-Karatzas et al. 1990) of rodent and human cells. In fact, eIF4E levels are a prognostic indicator of clinical outcomes in a variety of human cancers including breast cancer as well as head and neck squamous cell carcinoma (Kerekatte et al. 1995; Nathan et al. 1997). Hence, it is conceivable that the growth-promoting and transforming properties of eIF4E are due to the increased translatability of certain, if not all, mRNAs that are important for growth control (for review, see Sonenberg and Gingras 1998). eIF4E overexpression does not uniformly stimulate translation for all mRNAs but, rather, a subset of transcripts is more sensitive to eIF4E levels (for review, see Sonenberg and Gingras 1998) such as those for c-Myc (Graff et al. 1995) and ornithine decarboxylase (ODC) (Shantz and Pegg 1994; Rousseau et al. 1996; Shantz et al. 1996). The recent studies have determined that deregulation of phosphorylation of eIF4E or 4E-BP1 in Akt signaling leads to tumorigenesis by the activation of eIF4F complex (Avdulov et al. 2004; Ruggero et al. 2004; Wendel et al. 2004).
Structural studies have shown that eIF4E resembles a cupped hand or baseball glove consisting of a single α/β domain composed of an eight-stranded, antiparallel curved β sheet, and backed on its convex surface by three long α-helices (Marcotrigiano et al. 1997; Matsuo et al. 1997). The concave basal surface contains a narrow cap-binding slot, where the side chains of two conserved tryptophans support the recognition of cap analog m7GDP (Marcotrigiano et al. 1997; Matsuo et al. 1997). The dorsal surface contains the eIF4G and 4E-BPs contact motif (YXXXXLΦ, where X is any amino acid and Φ is an aliphatic residue), where acidic and hydrophobic residues are phylogenetically conserved.
To create a novel molecule that inhibits or modulates the activity of eIF4E, we chose to generate RNA aptamers with high affinity for eIF4E by in vitro RNA selection-amplification. Using these RNAs, we investigated the sequence and conformational requirements in both the RNA aptamers and eIF4E for their high affinity interactions, and a possible constraint by the RNA on the cap-binding activity of eIF4E and translation initiation. Since a causal relationship between aberrant expression of initiation factors and malignant transformation has been documented (Clemens and Bommer 1999; Hershey and Miyamoto 2000; Dua et al. 2001; Avdulov et al. 2004; Ruggero et al. 2004; Wendel et al. 2004), RNA ligands that inhibit the eIF4E-dependent initiation step could serve as a molecular tool for investigating eIF4E-activated oncogenesis.
RESULTS
In vitro selection of eIF4E aptamers
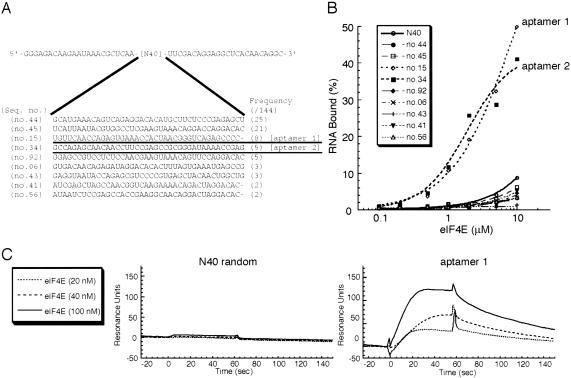
Affinity RNA selection experiments were performed using his-tagged mouse eIF4E (composed of 217 amino acids) and RNA pools of 40 random nucleotide positions (referred to as N40 RNA pool). The purified eIF4E preparation was fully active as it bound to the cap structure and 4E-BP1 (data not shown) as described previously (Altmann et al. 1988; Pause et al. 1994). In vitro selection was initiated using an N40 pool of 5 × 1014 different RNA molecules. To eliminate the matrix-binding sequences, the RNA pool was preincubated with Ni-NTA agarose resin, and unbound material was used for selection. RNA molecules bound to eIF4E (his-tagged) were captured by affinity precipitation with Ni-NTA agarose beads. The stringency of the selection was increased by decreasing the relative ratio of input protein to RNA. After 14 rounds of selection, 144 individual RNAs were cloned and sequenced, revealing nine unique aptamers that did not share any common motifs (Fig. 1A). When these RNA species were labeled with [α-32P]CTP and examined for their ability to bind to eIF4E in a nitrocellulose filter retention assay, only two RNAs, no. 15 and no. 34, bound efficiently to eIF4E (Fig. 1B). Thus, RNAs no. 15 and no. 34 were referred to as aptamer 1 and aptamer 2, respectively, and further characterized below. Both aptamers 1 and 2 bind to human eIF4E as efficiently as to mouse eIF4E, whose sequence only varies from the human sequence by four amino acid positions (data not shown). These aptamers did not bind to the other (control) proteins tested such as bovine serum albumin, yeast Saccharomyces cerevisiae eIF4E, and human eIF4A (data not shown).
FIGURE 1.
In vitro selected RNA sequences and their affinities for eIF4E. (A) Representative RNA sequences selected from randomized N40 RNA libraries. The sequence of the parental N40 RNA pool contains 5′ and 3′ constant sequences for primer annealing. After 14 rounds of selection, 144 individual clones were selected and nine nonhomologous sequences were identified. The frequency of each sequence in these selections is shown as numbers of each clone found in 144 independent isolates. (B) Nitrocellulose filter binding assays of selected RNAs and N40 random RNA (control) for wild-type eIF4A. Shown is the percentage of input [32P]-labeled RNA bound to the nitrocellulose filter. RNAs no. 15 and no. 34 showed efficient binding activity to eIF4E and are referred to as aptamer 1 and aptamer 2, respectively. (C) Sensorgrams of eIF4E binding to N40 random RNA (left panel, control) and aptamer 1 (right panel). Each eIF4E sample at the indicated concentrations were injected to flow cells immobilized with either aptamer 1 or N40.
Aptamer binding to eIF4E was monitored in real time with a BIAcore 2000 instrument based on the surface plasmon resonance (SPR) technique. 3′-Poly(A)-tailed RNAs were immobilized to the streptavidin sensor chip via 5′-biotinylated oligo(dT), and the formation of eIF4E-coupled complexes on this matrix was monitored as SPR signals. No positive signal was observed in a blank flow cell (data not shown). (Note that the background Resonance Units (RUs) of RNAs and 5′-biotinylated oligo(dT) immobilized on chips were subtracted in all the sensorgrams.) eIF4E was injected at a flow rate of 10 μL/min for 60 sec and dissociated for 300 sec by injecting a blank solution at the same flow rate. A set of sensorgrams for the eIF4E association with aptamer 1 and N40 random (control) is shown in Figure 1C, in which the eIF4E injection time was set as time zero. As expected, SPR signals on the aptamer 1 sensor chip appeared and increased in proportion to the 20, 40, and 100 nM eIF4E injections, while no positive signal was observed on an N40 random sensor chip. On the other hand, no SPR signals were observed for the aptamer 2 sensor chip (data not shown). We suggest that aptamer 2 underwent a conformational change upon poly(A) addition or immobilization to the sensor chip, leading to a loss of affinity for eIF4E. The association and dissociation rates estimated from the SPR profile for the eIF4E and aptamer 1 interaction are 9.36 ± 1.60 × 105 M−1 sec−1 and 1.05 ± 0.12 × 10−2 sec−1, respectively, and the apparent Kd is 11.2 nM.
Aptamer 1 interferes with the eIF4E–cap analog interaction
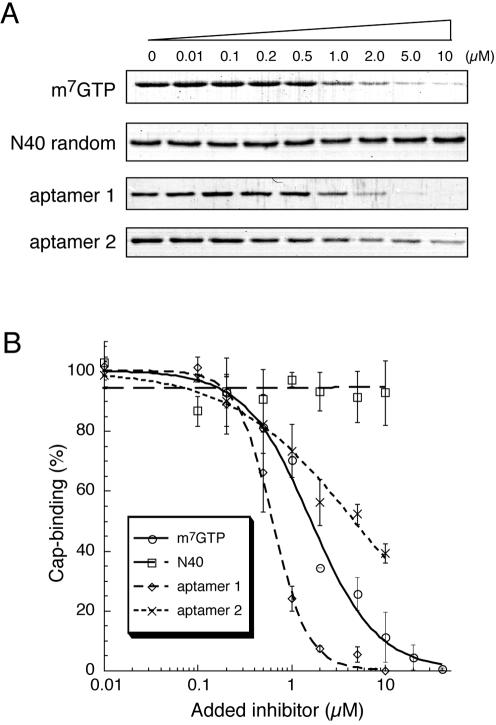
The potential effect of RNA aptamers on the interaction of eIF4E with the mRNA cap structure was investigated using the cap analog m7GTP (7-methylguanosine 5′-triphosphate) immobilized to Sepharose in a pull-down assay. eIF4E and m7GTP–Sepharose were incubated in the presence of RNA aptamers, N40 random RNA (negative control), or m7GTP (positive control) as competitors, and subjected to precipitation followed by repeated washing. Bound eIF4E was eluted with 10 mM m7GTP, separated by SDS-PAGE (polyacrylamide gel electrophoresis) and stained with Coomassie Brilliant Blue (Fig. 2A). As summarized in Figure 2B, the amount of eIF4E bound to m7GTP–Sepharose was reduced in proportion to the amount of aptamer 1, aptamer 2, and free m7GTP added, while unaffected in the presence of excess N40 random RNA. These results indicate that both aptamers inhibit the cap-binding activity of eIF4E. It is remarkable that the inhibitory effect of aptamer 1 is greater than those of aptamer 2 and free m7GTP cap analog. Hence, aptamer 1 was studied in detail below.
FIGURE 2.
Inhibition of the m7GTP–eIF4E interaction by RNA aptamers. (A) Pull-down assay of an eIF4E and m7GTP–Sepharose binary complex. eIF4E and m7GTP–Sepharose were mixed and challenged by increasing amounts of the indicated competitors—free m7GTP cap analog, aptamer 1, and aptamer 2. eIF4E remaining bound to m7GTP–Sepharose was detected by Coomassie staining after SDS-PAGE. (B) The intensity of eIF4E bands associated with m7GTP–Sepharose was quantified using NIH Image J, and the data are plotted using Kaleida Graph software as a percentage of the bound eIF4E in the absence of competitor: m7GTP (circle), N40 (square), aptamer 1 (diamond), aptamer 2 (cross).
Effect of aptamer 1 on eIF4E · 4E-BP1 binding
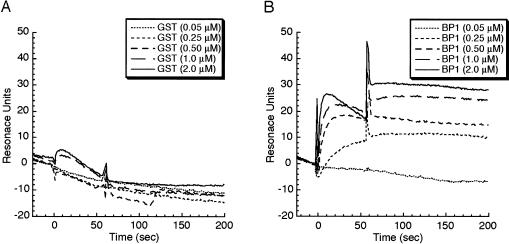
Next, we examined whether aptamer 1 interferes with eIF4E binding to 4E-BP1 in a glutathione-S-transferase (GST)-tagged 4E-BP1 coprecipitation assay using glutathione–Sepharose resin. When eIF4E and GST–4E-BP1 were incubated in the presence of excess aptamer 1, the level of eIF4E coprecipitable with GST–4E-BP1 was unaffected (data not shown), indicating that aptamer 1 does not interfere with the eIF4E · 4E-BP1 interaction. No interference was observed with aptamer 2 as well (data not shown). This was interpreted to indicate that eIF4E might be able to form a ternary complex with aptamer 1 and 4E-BP1. This was indeed the case when the ternary complex formation was examined by the SPR analysis. Aptamer 1 was immobilized to the sensor chip at a density of ~80 RUs of two flow cells. One flow cell was injected with 0.1 μM eIF4E for 60 sec (to form an aptamer–eIF4E complex, flow cell 2) while the other flow was not (as a single aptamer control, flow cell 1). Then, both flow cells were injected with 0.05, 0.25, 0.50, 1.0, and 2.0 μM solutions of GST–4E-BP1 or GST (control) for 60 sec. The SPR signal of flow cell 1 was subtracted from that of flow cell 2 to give rise to a net signal representing the specific interaction between 4E-BP1 and the aptamer-associated eIF4E (Fig. 3; note that GST or GST–4E-BP1 injection started at time 0 and the sharp peak that appeared at time 60 sec was due to a change of injecting solutions, which is referred to as an artificial “bulk effect”). The data indicated that GST alone does not interact with either aptamer or eIF4E (Fig. 3A), while GST–4E-BP1 is able to interact with the aptamer-associated eF4E, giving rise to SPR signals increased in proportion to the GST–4E-BP1 concentrations injected (Fig. 3B). These results led us to conclude that aptamer 1 does not interfere with the eIF4E · 4E-BP1 interaction and is capable of forming an aptamer · eIF4E · 4E-BP1 ternary complex. This probably means that the aptamer-binding site on eIF4E may be separated from the dorsal surface where 4E-BPs and eIF4G bind at the common binding motif YXXXXLΦ (where Φ means an aliphatic residue).
FIGURE 3.
Identification of the aptamer · eIF4E · 4E-BP1 ternary complex by SPR analysis. The aptamer 1 sensor chip was injected with 100 nM eIF4E for 60 sec to a signal of 80 RUs and then challenged with the indicated amounts of (A) GST and (B) GST-4E-BP1 at time 0 for 60 sec. A series of sensorgrams is normalized to represent the net interaction between (A) RNA and GST or (B) RNA and GST–4E-BP1. Experimental conditions and procedures are described in Materials and Methods.
Aptamer 1 inhibits cap-dependent translation in vitro
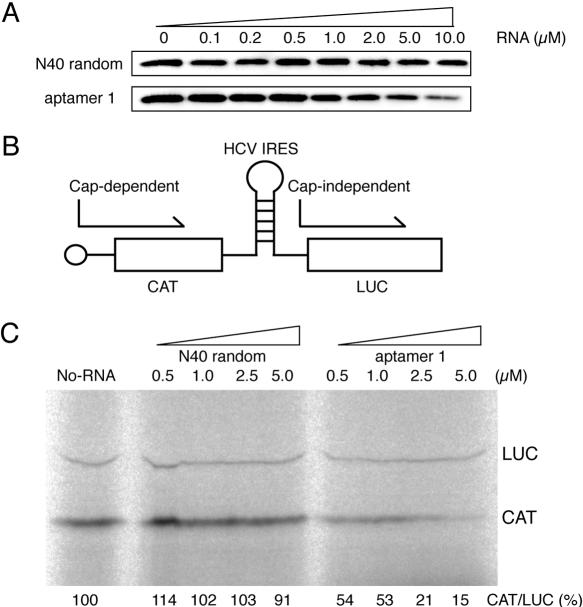
The effect of aptamer 1 on in vitro translation was examined using a 5′-capped chloramphenicol acetyltransferase (CAT) mRNA in a rabbit reticulocyte lysate (RRL) system. First, using the same procedure as shown in Figure 2 (i.e., coprecipitation of eIF4E–cap resin complex followed by anti-eIF4E immunostaining), it was confirmed that aptamer 1 is able to compete with the binding of RRL endogenous eIF4E to m7GTP–Sepharose (Fig. 4A). The binding and competition efficiency appeared to be similar to that for the recombinant eIF4E (see Fig. 2). Then, we examined the effect of aptamer 1 on in vitro translation. A bicistronic mRNA for which translation of the 5′-proximal CAT open reading frame is cap-dependent, whereas translation of the second luciferase (LUC) open reading frame is cap-independent as it is directed by the HCV internal ribosome entry site (IRES), was used (Fig. 4B). HCV IRES-directed translation initiation is independent of eIF4A, eIF4B, eIF4F, and eIF4H (Pestova et al. 1998). When translated in RRL in the presence of [35S]methionine and increasing amounts of N40 (control), efficient translation of both CAT and LUC was observed (Fig. 4C). In contrast, the addition of aptamer 1 inhibited cap-dependent CAT translation in a dose-dependent manner while it did not inhibit cap-independent LUC translation (Fig. 4C). It is noteworthy that a concentration of aptamer 1 in the micromolar range is sufficient to inhibit CAT synthesis in RRL that contains ~0.4 μM endogenous eIF4E (Rau et al. 1996). Aptamer 2 also exerted a weak but significant inhibition of cap-dependent translation (data not shown).
FIGURE 4.
Inhibition of cap-dependent in vitro translation by aptamer 1. (A) Pull-down of RRL-bearing endogenous eIF4E with m7GTP–Sepharose in the presence of N40 random and aptamer 1 RNAs. RRL and m7GTP–Sepharose were mixed, and the indicated amounts of N40 random RNA (control) and aptamer 1 were introduced as competitors. Pulled-down eIF4E was detected by immunostaining after SDS-PAGE as described in Materials and Methods. (B) Schematic diagram of capped CAT/HCV-IRES/LUC mRNA. (C) Translation products of capped CAT/HCV-IRES/LUC mRNA in RRL. Reaction mixtures were preincubated at 30°C for 3 min with increasing amounts (0.5, 1.0, 2.5, 5.0 μM) of N40 and aptamer 1 RNAs, followed by the addition of mRNA and [35S] methionine and further incubation for 60 min at 30°C. To stimulate cap-dependent translation as well as to avoid any nonspecific (inhibitory) effect of N40 (control) RNA on the in vitro translation, the reaction mix (25 μL) contained an increased amount (17.5 μL) of RRL, 100 mM potassium acetate, and 0.5 mM magnesium acetate. Products were analyzed by SDS-PAGE (15%) and fluorography. [35S]methionine incorporated into CAT and LUC is quantified using BAS-2000 PhosphorImager (Fuji Co.), and their relative values (CAT/LUC) are shown. The CAT/LUC ratio obtained in the absence of RNAs (left lane, buffer control) was set as 100%.
Structure and sequence requirements for aptamer 1
The aptamer 1 molecule used is 86 nt long and can be represented by at least three RNA secondary structures using the MFOLD program (Zuker 1989, 2003). The one shown in Figure 5B depicts the lowest free energy folding and is used in this study as the model structure for further studies. It contains five stems P1–P5 with 4-nt and 9-nt tails at both termini. Deletion analysis revealed that the 5′-tail (and P1 to some extent) is dispensable while P2 is essential for binding to eIF4E, and that a short 3′-tail preceding P5 is crucial for high affinity to eIF4E (data not shown).
FIGURE 5.
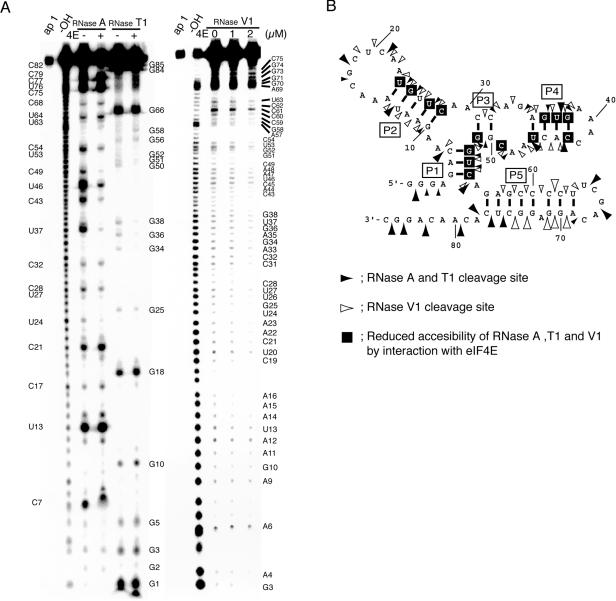
The MFOLD prediction and structural probing and footprinting of aptamer 1 by RNase digestion. (A) The 5′-end 32P-labeled aptamer 1 was digested with RNase A, T1 (left), and V1 (right) in the presence or absence of eIF4E, and the resulting digests were separated by electrophoresis on urea-denaturing gels as described in Materials and Methods. Undigested aptamer 1 (1/10 volume) and alkaline-digested ladders are also run (lanes ap1 and –OH, respectively). Signals generated by nuclease cleavage are assigned with nucleotide positions on both sides of the lanes. (B) The secondary structure of aptamer 1 examined by ribonuclease sensitivity and eIF4E protection assays. Solid arrowheads indicate the cleavage points with RNase A and T1, and open arrowheads indicate RNase V1 cleavage positions. The arrowhead size represents the degree of cleavage. The bases enclosed in black squares indicate sites protected by the addition of eIF4E from RNase A, T1, and V1 hydrolysis.
The apparent secondary structure of aptamer 1 and its eIF4E-binding site were investigated by probing the susceptibility of individual phosphodiester bonds with ribonucleases in the presence and absence of eIF4E. RNase T1 and RNase A selectively hydrolyze single-stranded RNA 3′ to G and U/C residues, respectively. RNase V1 hydrolyzes double-stranded regions. First, the 5′-end 32P-labeled aptamer 1 was partially digested by these RNases in the absence of eIF4E, and RNA digests were separated on a denaturing gel (Fig. 5A). As expected, single-stranded regions predicted by MFOLD were hydrolyzed efficiently by RNases A and T1 (Fig. 5A, left panel; Fig. 5B, solid arrowheads). However, RNases A and T1 were also, at least in part, accessible to several sites in the predicted stems. Probably, short stems P1–P4 may be occasionally unwound and become susceptible to RNases A and T1. Unexpectedly, RNase V1 hit nucleotides not only predicted to be in stems but also in some loop regions (Fig. 5A, right panel; Fig. 5B, open arrowheads). This RNase V1 sensitivity of loop regions cannot be explained by any MFOLD predictions (data not shown). These cleavage profiles suggest that aptamer 1 folds into several conformers in addition to the predicted MFOLD structure. In the presence of eIF4E, however, the intensity of RNase V1 cleavage in the putative loop regions greatly decreased (Fig. 5A, right). Of five stems predicted by MFOLD, P5 formed a relatively stable conformation that is sensitive to RNase V1 but is resistant to RNase T1 or RNase A (except for a few 3′-terminal nucleotides). These results are interpreted to indicate that although the MFOLD prediction may not be inconsistent with the main (or most common) structure of aptamer 1, it probably exists in several conformation states, and several flexible subdomains may undergo a conformational change upon binding to eIF4E, thereby stabilizing the predicted structure.
Moreover, it is important to point out that upon binding to eIF4E, the intensity of RNase A and T1 cleavage was decreased throughout the molecule at positions U24, G25, U27, C28, G34, G36–G38, C43, U46, and C49–G54. Likewise, the intensity of RNase V1 cleavage was also decreased throughout at positions A14–A16, C19, C21–C28, C31–G38, C43–C49, G51–C54, and A57–C60. Although this altered footprint profile may be in part due to a conformational change of aptamer 1 itself, it is reasonable to assume that most, if not all, of these nucleotides (i.e., U24, G25, U27, C28, G36–G38, C43, U46, C49, G51–C54) are protected from nuclease attacks upon binding to eIF4E (Fig. 6B, enclosed bases).
FIGURE 6.
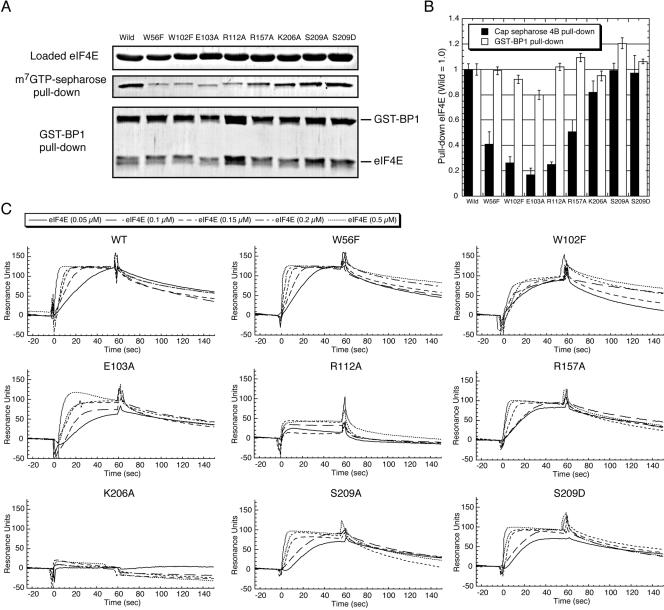
Characterization of variant eIF4E proteins changed at affected in functional amino acids. A set of amino acid changes was generated in eIF4E by site-directed mutagenesis, and the resulting variant proteins were purified and examined as described in Materials and Methods. (A) The activity of variant eIF4Es for binding to the m7GTP cap analog and GST–4E-BP1 was examined by respective pull-down assays. The same amount of variant eIF4E proteins (top panel) was applied to each pull-down assay, and those coprecipitated with m7GTP–Sepharose (middle panel) and GST–4E-BP1 (bottom panel) were eluted and detected by Coomassie staining after SDS-PAGE. (B) The intensity of each band derived from both pull-downs was measured by NIH image J application and used to estimate the relative efficiency (compared to wild-type eIF4E) of binding to m7GTP–Sepharose (closed box) and GST–4E-BP1 (open box). In the GST–BP1 pull-down, the ratio of variant eIF4E to 4E-BP1 in the staining was compared with that of wild-type eIF4E to 4E-BP1 (to normalize the efficiency of GST-4E-BP1 precipitation). (C) Sensorgrams of the interaction between aptamer 1 and increasing amount of variant eIF4Es.
Assembly of eIF4E variants
To understand the mechanism by which aptamer 1 hinders eIF4E from cap binding, we wish to determine the amino acids in eIF4E that are crucial for aptamer interaction. To this end, we constructed several eIF4E variants carrying single amino acid changes at positions that are known or predicted to be important for its function such as for binding to cap and phosphorylation. The available three-dimensional structure of eIF4E (Marcotrigiano et al. 1997; Niedzwiecka et al. 2002) helped us to assign or evaluate positions for site-directed mutagenesis. Trp56 and Trp102 are involved in the stacking interaction with 7-methylgua-nosine (Marcotrigiano et al. 1997; Matsuo et al. 1997) and their substitutions to Phe (W56F and W102F) render the cap-binding activity <50% (Altmann et al. 1988). Glu103 is involved in hydrogen bonding with the N1 and N2 of 7-methylguanosine, and its change to Ala (E103A) inhibits cap-binding by 80% (Morino et al. 1996). Thus, we prepared these known eIF4E variants (W56F, W102F, and E103A). Additionally, three novel variants—R112A, K157A, and K206A—were made since these positively charged amino acids are known to make a hydrogen-bond network with the three-phosphate (negative charge) chain in m7GpppG (Niedzwiecka et al. 2002). Moreover, the phosphorylation site Ser209 (Flynn and Proud 1995; Joshi et al. 1995) was changed to Ala and Asp (S209A and S209D) since these mutations have made indispensable contributions to uncover the crucial role of Ser209 phosphorylation in regulating mRNA cap binding and translation initiation (Niedzwiecka et al. 2002; Scheper et al. 2002). Acidic S209D variant is thought to mimic the putative salt bridge of phosphorylated Ser209 with Lys159, and the S209D variant binding affinity for capped mRNA, but not m7GTP (free from mRNA), is lower than wild type in SPR analysis (Scheper et al. 2002).
Characterization of variant eIF4E proteins
Variant eIF4E proteins were purified to homogeneity and examined for their activity to bind to the cap analog (m7GTP-resin) and GST–4E-BP1 by a pull-down assay (Fig. 6A). The estimated efficiency relative to wild-type eIF4E is summarized in Figure 6B. The data indicate that variant eIF4Es were as active as wild type in binding GST–4E-BP1 (Fig. 6B, white bar). This means that the variant eIF4Es fold into a functional conformation and the generated mutations do not affect the 4E-BP1 interaction site on eIF4E. On the other hand, cap-analog binding was hampered by the W56F, W102F, and E103A mutations to 40%, 25%, and 20% of the wild-type level, respectively (Fig. 6B, black bar). These results correlate well with previous results (Altmann et al. 1988; Morino et al. 1996). The efficiency of cap binding with novel variants R112A and R157A was also reduced to 25% and 45%, respectively, while that with K206A was only slightly affected, if at all (Fig. 6B). We confirmed that the two Ser209 variants, S209A and S209D, bind to the cap analog as efficiently as the wild type (Fig. 6B), as shown previously (Shibata et al. 1998; Scheper et al. 2002).
Specific amino acid changes in eIF4E affect aptamer binding
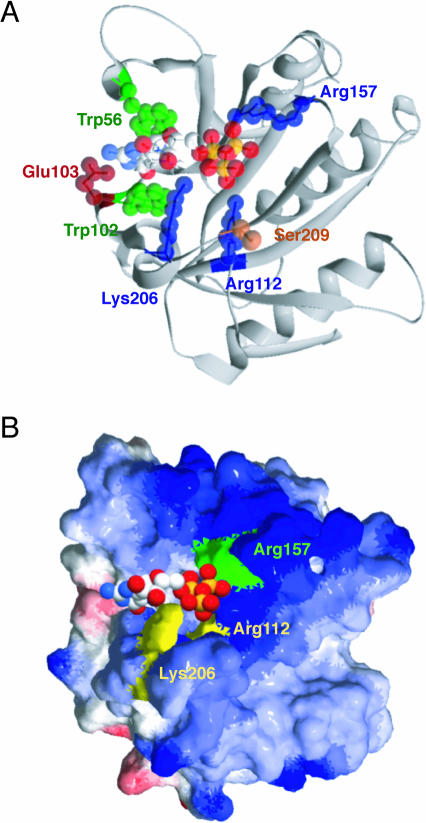
The binding efficiency of variant eIF4Es to aptamer 1 was examined by SPR analysis using the aptamer chip (Fig. 6C). The resulting sensorgram indicated that R112A and K206A showed either severe or complete loss of affinity to aptamer 1, respectively. Other variants did not show any appreciable defect in binding. Considering that W56F, W102F, and E103A are affected in cap binding (see Fig. 6B), the mode of binding of aptamer 1 is not necessarily the same as that of the cap analog. Nevertheless, it is evident that Arg112 and Lys206 play a crucial role in aptamer binding. These two basic residues are closely oriented on the eIF4E surface (Fig. 7) and interact with the third phosphate of the cap analog (Niedzwiecka et al. 2002). Although the same phosphate of cap also interacts with a distantly located Arg157, the R157A variant did not show any loss of affinity to the aptamer (see Fig. 6C). These findings suggest that aptamer 1 recognizes the entrance of the cap-binding slot of eIF4E via electrostatic interactions with the Arg112–Lys206 network in a manner that is similar to that of cap analog (m7GpppG) binding, with one key difference being that the aptamer does not require Arg157 for binding, while the cap analog does.
FIGURE 7.
Structure of the mouse eIF4E (amino acids 28–217) and m7GTP complex (Protein Data Bank accession code 1L8B) (Niedzwiecka et al. 2002). Structure modeling is performed by Swiss PDB Viewer application and rendered by POV-Ray (ver. 3.6). (A) Mutation sites are shown on the ribbon model of the eIF4E–m7GTP complex structure. Amino acids altered in this study are indicated as space-filling presentations showing (green) stacking, (red) acidic, (blue) basic, and (orange) phosphorylation residues. m7GTP is displayed as color-coded space filling—(white) carbon, (red) oxygen, (sky blue) nitrogen, (yellow) phosphorus—and located in the cap-binding slot in the cocrystal. (B) Surface electrostatic potential of the eIF4E–m7GTP complex structure. (Blue) positive (basic amino acid) and (red) negative (acidic amino acid) charges are shown. Arg 112 and Lys 206 residues required for aptamer 1 binding are yellow. Arg 157 that is not involved in aptamer 1 binding is green. m7GTP is shown as in A. The potentials were calculated with the program DELPHI.
Although the S209A and S209D substitutions may influence the phosphate bridging between Ser209–Lys159, in fact, these variants were not affected in binding to the aptamer (Fig. 6C), indicating that Ser209 is not involved in the aptamer interaction. This suggests that both phosphorylated and unphosphorylated forms of eIF4E can be equally recognized and inhibited by the aptamer. Furthermore, it is noteworthy that although S209D has been shown to bind much less efficiently to capped mRNA (Scheper et al. 2002), we failed to observe any loss of binding of S209D to the aptamer. This probably means that the mode of binding on eIF4E with aptamer 1 is different from that with capped mRNA. Taking these and other results into consideration, we suggest that while aptamer 1 and capped mRNA may have shared binding sites on eIF4E, the aptamer 1 binding probably occludes the cap-binding site, resulting in cap-dependent translation inhibition.
DISCUSSION
In this study, two species of high-affinity aptamers, aptamer 1 and aptamer 2, were generated for mammalian initiation factor eIF4E by in vitro RNA selection. The dissociation constant of aptamer 1, which was best characterized here, was 11.2 nM when determined by SPR. Aptamer 1 outcompetes m7GTP and aptamer 2 to bind to eIF4E, whereas it failed to interfere with the eIF4E · 4E-BP1 interaction. Consistently, aptamer 1 hinders cap-dependent in vitro translation in a RRL system, while it does not inhibit cap-independent (HCV IRES-directed) translation. By mutational analysis, it was determined that the 86-nt aptamer 1 can be truncated to 82 nt but may not be truncated beyond 76 nt (data not shown). Therefore, the overall structure and sequence of aptamer 1 seem to be required for high affinity binding to eIF4E. This prediction was, at least in part, confirmed by structural probing and footprinting of aptamer 1 with RNases T1, A, and V1 (see Fig. 5). In the native conformation, some of the regions predicted by MFOLD to form loops were cleaved, to some extent, by the double-stranded RNA specific nuclease V1 (probably because of forming alternate structures). However, the presence of eIF4E in the footprinting experiment reduced the amount of cleavage either by masking the cleavage site or altering the conformation. Therefore, we suggest that the RNA aptamer is in a more flexible and dynamic state in the free form than what is predicted by the MFOLD program, and is stabilized or undergoes a substantial conformational change upon interacting with eIF4E to fit the RNA structure to the target protein as an “induced fit.”
The binding site of aptamer 1 on eIF4E was estimated in part by introducing mutational changes for amino acids that are known or predicted to be important for cap binding or phosphorylation. Trp56, Trp102, and Glu103 were shown to interact directly with 7-methylguanosine residues in the crystal structure (Marcotrigiano et al. 1997), and, indeed, we confirmed here that these mutations affect cap analog binding. To our surprise, however, these variants interact as efficiently as wild-type eIF4E with aptamer 1. On the other hand, eIF4E variants changed at Arg112 or Lys206 were severely or completely inhibited in binding to aptamer 1, respectively. These residues, closely located on the eIF4E surface, interact with the cap phosphate chain via water solvent (Fig. 7B). The recent crystallographic study of eIF4E · m7GpppA complex has shown that the adenosine residue interacts with the Thr205–Lys206–Ser207 sequence pocket of the C-terminal flexible loop region, suggesting that the functional significance of this loop is to serve as the binding site for the second nucleoside of the cap structure (Tomoo et al. 2003). These findings are interpreted to indicate that aptamer 1 binds to eIF4E not by interacting with those residues that are directly involved in the 7-methylguanosine recognition, but, rather, those that are adjacent to the cap-binding site and are involved in the recognition of the downstream nucleoside(s) in the capped mRNA. Previous mutational and SPR analysis suggests that Ser209 is not directly involved in cap recognition but is responsible for mRNA capturing (Scheper et al. 2002). However, the Ser209 variants of eIF4E are not affected in the affinity to aptamer 1, suggesting that the aptamer binds to eIF4E in a way that may be in part similar to but mostly different from that with capped mRNA. Taking these results into consideration, we conclude that aptamer 1 binds to eIF4E around the entrance of the cap-binding slot, thereby blocking the cap-binding pocket.
Aptamer 1 does not affect the dorsal surface of eIF4E and is able to make a ternary complex with eIF4E and 4E-BP1 (probably with eIF4G as well). This feature might be useful as a specific inhibitor to knock out eIF4E in the complex without interrupting the association of dorsal-site-binding proteins. Aptamer 1 inhibits in vitro translation of the capped CAT mRNA at micromolar levels in the RRL translation system (see Fig. 4). This dose may be of one or two orders of magnitude higher than what is expected from the intracellular level of eIF4E, which should be the least abundant factor of all eIFs and the rate-limiting factor for eukaryotic translation initiation (Duncan et al. 1987). This apparent conflict can be explained in part by the fact that unlike in HeLa cell extracts (Duncan et al. 1987), eIF4E is highly phosphorylated and is not the limiting factor for translation in RRL (Rau et al. 1996). Furthermore, one can speculate that eIF4E is associated with other eIFs such as eIF4G in the RRL, which might affect susceptibility of eIF4E to aptamer 1. Therefore, translation inhibition would need much higher concentrations of inhibitors to inactivate either the initiation complex eIF4F or the 43S preinitiation ribosomal complex than that to inactivate a free eIF4E. In fact, the inhibition constant of the cap analogs obtained from in vitro translation in RRL was >100 times greater than its Kd (Cai et al. 1999). Although the efficacy of aptamer 1 is much greater than the cap analog, the observed similar increase in the inhibition effect compared with the Kd suggests that aptamer 1 interacts with both unbound and bound forms of eIF4E in the initiation complex (see Fig. 3).
It is noteworthy that while aptamer 1 needs to be >76 nt long, another RNA aptamer generated in this laboratory to human eIF4A helicase is 58 nt long, and truncated versions lose the high affinity binding to eIF4A (Oguro et al. 2003). In fact, NMR analysis shows that this 58-nt aptamer and its target protein eIF4A interact with each other at multiple sites that are widespread on the surfaces of both molecules (T. Sakamoto, G. Kawai, A. Oguro, T. Ohtsu, and Y. Nakamura, in prep.). Additionally, other RNA aptamers generated for eIF4G and eIF1A also require a relatively large mass (75 and 73 nt) for high affinity binding to their target proteins (S. Miyakawa, A. Oguro, T. Ohtsu, N. Sonenberg, J. Hershey, and Y. Nakamura, in prep.). Therefore, it might be argued that RNA aptamers to proteins without RNA recognition motifs or strong affinity to RNA (such as eIF4E, eIF4A, eIF4G, eIF1A, and others) can achieve specific high affinity to the target protein by capturing its global conformation. This is completely different from the pinpoint (i.e., epitope <10 amino acids) recognition of target protein by antibody. Previous, present, and ongoing studies (in this laboratory) of RNA aptamers to mammalian initiation factors contribute to strengthening the concept of conformational recognition of target proteins by RNA aptamer. For this reason, the RNA aptamer has promising potential to substitute for or complement the antibody as a new diagnostic or therapeutic tool that we refer to as “RNA antibody.” In view of the increasing awareness of relationships between aberrant expression of initiation factors and malignant transformation of mammalian cells, translation initiation might be a good target for anticancer therapeutics and diagnosis. RNA aptamers to these initiation factors might therefore serve in medical applications in the future.
MATERIALS AND METHODS
Protein expression and purification
Plasmids encoding wild-type or S209D mouse eIF4E and 4E-BP1 have been described previously (Morino et al. 2000). eIF4E variants were generated by the QuickChange Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s instruction using site-directed sequence primers:
W56F, 5′-AAAAATGATAAAAGCAAAACTTTTCAAGCAAACCTTCGATTGATC-3′;
W102F, 5′-CGGGATTGAGCCTATGTTTGAAGATGAGAAAAACA-3′;
E103A, 5′-GATTGAGCCTATGTGGGCAGATGAGAAAAACAAAC-3′;
R112A, 5′-AAACAAACGAGGAGGAGCATGGCTGATCACACTGA-3′;
R157A, 5′-AGCTGTTGTTAATGTTGCAGCTAAAGGCGATAAGA-3′;
K206A, 5′-AGACACAGCTACAGCAAGCGGCTCCACCA-3′; and
S209A, 5′-AGCTACAAAGAGCGGCGCAACCACTAAAAATAGGT-3′.
Recombinant his-tagged eIF4E was expressed in Escherichia coli BL21 (DE3), and the crude E. coli extract from 1 L of culture was applied to 0.5 mL of Ni-NTA agarose (QIAGEN) column equilibrated with buffer A (20 mM Tris-HCl at pH 7.6, 150 mM NaCl, 0.1 mM EDTA). After washing with buffer A + 20 mM imidazole, the protein was eluted with buffer A + 250 mM imidazole and dialyzed against buffer B (20 mM Tris-HCl at pH 7.6, 80 mM potassium acetate, 50% glycerol). The sample was then applied to a Resource S (Amersham Biosciences) column equilibrated with buffer C (20 mM Tris-HCl at pH 7.6, 80 mM potassium acetate, 2.5 mM magnesium acetate, 5% glycerol), and eluted by a 0%–100% buffer D gradient (20 mM Tris-HCl at pH 7.6, 1 M potassium acetate, 5% glycerol) by HPLC. Recombinant GST–4E-BP1 was expressed in E. coli BL21 (DE3), and the supernatants of cell extracts were applied to glutathione–Sepharose 4B (Amersham Biosciences) with buffer A + 40 mM reduced glutathione, dialyzed with buffer B, and purified on a Resource Q (Amersham Biosciences) column by a 0%–100% buffer D gradient elution. These proteins were dialyzed against buffer B, and stored at −20°C.
Selection-amplification of eIF4E aptamers
Preparation of random N40 RNAs and affinity RNA selection-amplification was carried out essentially as described previously (Oguro et al. 2003). Briefly, the initial selection pool involved 1014 different RNA molecules. RNAs were preincubated with 5 μL of Ni-NTA agarose resin in 100 μL of binding buffer (20 mM Tris-HCl at pH 7.6, 80 mM potassium acetate, 2.5 mM magnesium acetate, 1 mM dithiothreitol, 5% glycerol) for 30 min at room temperature. Unbound RNAs were then incubated with eIF4E for 30 min at room temperature, washed with 200 μL of binding buffer three times, and eluted with 50 μL of binding buffer + 250 mM imidazole. Selected RNAs were extracted by phenol/chloroform treatment, purified with Microcon YM-30 (Millipore), and reverse-transcribed using AMV reverse transcriptase (TaKaRa Co.). Reverse transcripts were then amplified by PCR using the appropriate primer and exTaq polymerase (TaKaRa Co.). The input RNA was in three molar excess to protein (i.e., 9 μM RNA to 3 μM protein) in the first round of selection, and gradually increased up to 60 molar excess to protein (12 μM RNA to 0.2 μM protein) by the 14th round of selection to increase the stringency of selection.
Filter retention assay
Filter binding assays were carried out as described previously (Oguro et al. 2003). After rounds 9 and 14, selected RNAs were labeled in an in vitro transcription reaction using [α-32P]GTP (800 Ci/mmol; Amersham Biosciences). The labeled RNA was incubated with eIF4E in 50 μL of binding buffer containing 10 μg/mL tRNA for 30 min at room temperature. The solution was filtered through a presoaked nitrocellulose membrane (0.45 μm pore size, type HA; Millipore) and washed three times with 1 mL of binding buffer, and the retained radioactivity was quantitated by scintillation counting. The data set was plotted using Kaleida Graph software (Synergy Software).
Surface plasmon resonance assay
The SPR assays were performed according to the same coupling method as described previously (Wood 1993; Van Ryk and Venkatesan 1999) using a BIAcore 2000 instrument (BIAcore AB). The aptamer templates were amplified and tagged at the 3′-end with dA16 by PCR using 5′-template primer (5′-CCGAAGCTTA ATACGACTCACTATAGGGAGACAAGAATAAACGCTCAA-3′) and dA16-tagging 3′-template primer (5′-TTTTTTTTTTTTTTTTGCCTGTTGTGAGCCTCCTGTCGAA-3′). 5′-Biotinylated dT16 oligomer was bound to the surface of the streptavidin sensor chip (BIAcore AB) of flow cells 1 and 2. Then 20 μg/mL 3′-A16-tagged RNA was immobilized to 150 RUs in flow cell 2 by complementary hybridization to the 5′-biotinylated dT16 oligomer in binding buffer at a flow rate of 20 μL/min at 25°C for 1 min by the NINJECT program (BIAcore AB). Unbound RNAs were washed in binding buffer at the same flow rate for 2 min. Purified eIF4E in binding buffer was passed through flow cells 1 and 2 of the sensor chip for 1 min at the KINJECT program (BIAcore AB), and bound proteins were gradually dissociated for 5 min. The data are obtained by subtracting the signals for 5′-biotinylated dT16 fixed on the sensor chip (flow cell 1) from the signal for RNA aptamer (flow cell 2), thereby showing the net interaction between RNA and protein (Figs. 1C, 6C).
To test the formation of the aptamer · eIF4E · 4E-BP1 complex, aptamer 1 was immobilized to both flow cells 1 and 2 of the sensor chip to ~150 RUs, and 100 nM eIF4E was injected only to flow cell 2 for 60 sec at 20 μL/min by NINJECT. Then, 0.05, 0.25, 0.50, 1.0, and 2.0 μM solutions of GST–4E-BP1 or GST (control) were injected to flow cells 1 and 2 for 60 sec at the same flow rate by KINJECT. The SPR signal of flow cell 1 was subtracted from that of flow cell 2 to eliminate the nonspecific interaction of aptamer with GST or with GST–4E-BP1 in the absence of eIF4E.
To regenerate the sensor chip, bound materials were completely removed by injecting 20 μL of 2 M urea at a flow rate of 20 μL/min. Close-fitting curves to the sensorgrams were calculated by global fitting curves (1:1 Langmuir binding) generated using BIAevaluation 3.0 software (BIAcore AB).
m7GTP–Sepharose pull-down assay
The indicated amount of RNA aptamers, 1 μM eIF4E, and 1 μL of m7GTP–Sepharose were incubated at 25°C for 30 min in 50 μL of binding buffer containing 10 μg/mL tRNA. The resin complex was washed three times with 200 μL of binding buffer, and the bound proteins were eluted with 20 μL of binding buffer containing 10 mM m7GTP (Sigma) after incubation for 60 min at room temperature. After centrifugation, 10 μL of the supernatant was applied to a 12% SDS-PAGE, and the intensity of eIF4E bands was quantified by Image J 1.30v (NIH) after staining with Coomassie Brilliant Blue G-250. For m7GTP–Sepharose pull-down of endogenous eIF4E from RRL, 15 μL of RRL, and 1 μL of m7GTP–Sepharose were mixed and incubated at 25°C for 30 min in 37.5 μL of solution of 100 mM potassium acetate and 5.0 mM magnesium acetate solution. The bound eIF4E was recovered, separated by 12% SDS-PAGE, and transferred to the polyvinylidene difluoride (PVDF) membrane. The membrane was soaked with blocking buffer (5% non-fat dry milk, 10 mM Tris-HCl at pH 7.5, 100 mM NaCl, 0.1% Tween-20) for 30 min at room temperature, and incubated with anti-eIF4E mouse polyclonal antibody (BD Biosciences) in buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.05% Tween-20 for 1 h at room temperature. After repeated washing, the membrane was incubated with anti-mouse IgG sheep secondary antibody HRP conjugate (Amersham Biosciences) for 1 h at room temperature. The signal was detected by ECL reagent (Amersham Biosciences) according to the manufacturer’s instruction and analyzed by LAS-1000 (Fujifilm). The linearity of detection was confirmed in each assay with an appropriate standard curve using recombinant eIF4E.
In vitro translation
In vitro translation was performed as described previously using RRL (Oguro et al. 2003). Capped-CAT/HCV-IRES/LUC mRNA was transcribed by using RiboMAX in vitro transcription systems (Promega). The in vitro translation mix was incubated at 30°C, and aliquots were withdrawn at the indicated time intervals and mixed with the SDS-PAGE sample buffer at 4°C.
RNA probing
The labeled RNA (~0.01 μM) was heated at 85°C, then slowly cooled to room temperature in binding buffer, and incubated with increasing amounts of RNase T1 or RNase A for 3 min at room temperature. The resulting digests were applied to a 9% PAGE in the presence of 7 M urea. Aptamer 1 was first dephosphorylated with 1 U of alkaline phosphatase (E. coli A19; TaKaRa Co.) at 37°C for 60 min. Then, the 5′-end was labeled with [γ-32P]ATP by T4 polynucleotide kinase (TaKaRa Co.). The resulting 32P-labeled aptamer 1 was subjected to 9%-acrylamide/7 M-urea gel electrophoresis, and the full-length aptamer 1 was purified by overnight elution from gel slices in 400 μL of elution buffer containing 0.5 M ammonium acetate, 1 mM EDTA (pH 8.0), and 0.1% (w/v) SDS. After ethanol precipitation, aptamer 1 was resuspended in binding buffer to ~30,000 cpm/μL. Alkaline digestion (for the control ladder) was performed in 10 μL of 50 mM sodium carbonate (pH 9.0), 1 mM EDTA for 3 min at 90°C. The labeled RNA was heated at 85°C and slowly cooled to room temperature in binding buffer, and incubated with RNase T1 (0.2 U; Sigma) and RNase A (4 × 10−5 mg/mL; Sigma) for 3 min at 37°C in the presence or absence of 15 μM eIF4E. RNase V1 partial digestion was conducted using 10−4 U of enzyme with 1.0 or 2.0 μM eIF4E, followed by incubation for 10 min. The resulting digests were subjected to a 9% PAGE in the presence of 7 M urea.
Acknowledgments
We thank Hiroaki Imataka for his experimental expertise and protein preparations. Colin Crist is thanked for critical reading of the manuscript and valuable comments. This work was supported in part by grants from The Ministry of Education, Sports, Culture, Science and Technology of Japan (MEXT); the Organization for Pharmaceutical Safety and Research (OPSR); and The Japan Health Sciences Foundation.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7108205.
REFERENCES
- Altmann, M., Edery, I., Trachsel, H., and Sonenberg, N. 1988. Site-directed mutagenesis of the tryptophan residues in yeast eukaryotic initiation factor 4E; effects on cap binding activity. J. Biol. Chem. 263: 17229–17232. [PubMed] [Google Scholar]
- Avdulov, S., Li, S., Michalek, V., Burrichter, D., Peterson, M., Periman, D.M., Manivel, J.C., Sonenberg, N., Yee, D., Bitterman, P.B., et al. 2004. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell 5: 553–563. [DOI] [PubMed] [Google Scholar]
- Cai, A., Jankowska-Anyszka, M., Centers, A., Chlebicka, L., Stepinski, J., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 1999. Assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry 38: 8538–8547. [DOI] [PubMed] [Google Scholar]
- Clemens, M.J. and Bommer, U.A. 1999. Translational control: The cancer connection. Int. J. Biochem. Cell. Biol. 31: 1–23. [DOI] [PubMed] [Google Scholar]
- De Benedetti, A. and Rhoads, R.E. 1990. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc. Natl. Acad. Sci. 87: 8212–8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua, K., Williams, T.M., and Beretta, L. 2001. Translational control of the proteome: Relevance to cancer. Proteomics 1: 1191–1199. [DOI] [PubMed] [Google Scholar]
- Duncan, R., Milburn, S.C., and Hershey, J.W.B. 1987. Regulated phosphorylation and low abundance of HeLa cell initiation factor 4F suggest a role in translational control. J. Biol. Chem. 262: 380–388. [PubMed] [Google Scholar]
- Flynn, A. and Proud, C.G. 1995. Ser209, but not Ser53, is the major site of phosphorylation in initiation factor eIF4E in serum treated Chinese hamster ovary cells. J. Biol. Chem. 270: 21684–21688. [DOI] [PubMed] [Google Scholar]
- Fukunaga, R. and Hunter, T. 1997. Mnk1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16: 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., Gygi, S.P., Niedzwiecka, A., Miron, M., Burley, S.K., Polakiewicz, R.D., Wyslouch-Cieszynska, A., Aebersold, R., and Sonenberg, N. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes & Dev. 15: 2852–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff, J.R., Boghaert, E.R., De Benedetti, A., Tudor, D.L, Zimmer, C.C., Chan, S.K., and Zimmer, S.G. 1995. Reduction of translation initiation factor 4E decreases the malignancy of ras-transformed cloned rat embryo fibroblasts. Int. J. Cancer 60: 255–263. [DOI] [PubMed] [Google Scholar]
- Haghighat, A., Mader, S., Pause, A., and Sonenberg, N. 1995. Repression of cap-dependent translation by 4E-binding protein 1: Competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14: 5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey, J.W.B. and Miyamoto, S. 2000. Translational control and cancer. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 637–654. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Joshi, B., Cai, A.L., Keiper, B.D., Minich, W.B., Mendez, R., Beach, C.M., Stepinski, J., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 1995. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser209. J. Biol. Chem. 270: 14597–14603. [DOI] [PubMed] [Google Scholar]
- Kerekatte, V., Smiley, K., Hu, B., Smith, A., Gelder, F., and De Benedetti, A. 1995. The proto-oncogene/translation factor eIF4E: A survey of its expression in breast carcinomas. Int. J. Cancer 64: 27–31. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas, A., Montine, K.S., and Sonenberg, N. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345: 544–547. [DOI] [PubMed] [Google Scholar]
- Mader, S., Lee, J., Pause, A., and Sonenberg, N. 1995. The translation initiation factor eIF4E binds to a common motif shared by the translation factor eIF-4g and the translational repressors, 4E-binding proteins. Mol. Cell. Biol. 15: 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.C., Sonenberg, N., and Burley, S.K. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell. 3: 707–716. [DOI] [PubMed] [Google Scholar]
- Matsuo, H., Li, H., McGuire, A.M., Fletcher, C.M., Gingras, A.C., Sonenberg, N., and Wagner, G. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4: 717–724. [DOI] [PubMed] [Google Scholar]
- Morino, S., Hazama, H., Ozaki, M., Teraoka, Y., Shibata, S., Doi, M., Ueda, H., Ishida, T., and Uesugi, S. 1996. Analysis of the mRNA cap-binding ability of human eukaryotic initiation factor-4E by use of recombinant wild-type and mutant forms. Eur. J. Biochem. 239: 597–601. [DOI] [PubMed] [Google Scholar]
- Morino, S., Imataka, H., Svitkin, Y.V., Pestova, T.V., and Sonenberg, N. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20: 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, C.A., Liu, L., Li, B., Nandy, I., Abreo, F., and De Benedetti, A. 1997. Detection of the proto-oncogene eIF4E in surgical margins may predict recurrence in head and neck cancer. Oncogene 15: 579–584. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka, A., Marcotrigiano, J., Stepinski, J., Jankowska-Anyszka, M., Wyslouch-Cieszynska, A., Dadlez, M., Gingras, A.C., Mak, P., Darzynkiewicz, E., Sonenberg, N., et al. 2002. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319: 615–635. [DOI] [PubMed] [Google Scholar]
- Oguro, A., Ohtsu, T., Svitkin, Y.V., Sonenberg, N., and Nakamura, Y. 2003. RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA 9: 394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause, A., Belsham, G.J., Gingras, A.C., Donzé, O., Lin, T.A., Lawrence Jr., J.C., and Sonenberg, N. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371: 762–767. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J., and Hellen, C.U. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 12: 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau, M., Ohlmann, T., Morley, S.J., and Pain, V.M. 1996. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J. Biol. Chem. 271: 8983–8990. [DOI] [PubMed] [Google Scholar]
- Rousseau, D., Kaspar, R., Rosenwald, I., Gehrke, L., and Sonenberg, N. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. 93: 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, D., Montanaro, L., Ma, L., Xu, W., Londei, P., Cordon-Cardo, C., and Pandolfi, P. 2004. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 10: 484–486. [DOI] [PubMed] [Google Scholar]
- Scheper, G.C., van Kollenburg, B., Hu, J., Luo, Y., Goss, D.J., and Proud, C.G. 2002. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem. 277: 3303–3309. [DOI] [PubMed] [Google Scholar]
- Shantz, L.M. and Pegg, A.E. 1994. Overproduction of ornithine decarboxylase caused by relief of translational repression is associated with neoplastic transformation. Cancer Res. 54: 2313–2316. [PubMed] [Google Scholar]
- Shantz, L.M., Hu, R.H., and Pegg, A.E. 1996. Regulation of ornithine decarboxylase in a transformed cell line that overexpresses translation initiation factor eIF-4E. Cancer Res. 56: 3265–3269. [PubMed] [Google Scholar]
- Shatkin, A. 1976. Capping of eukaryotic mRNAs. Cell 9: 645–653. [DOI] [PubMed] [Google Scholar]
- Shibata, S., Morino, S., Tomoo, K., In, Y., and Ishida, T. 1998. Effect of mRNA cap structure on eIF-4E phosphorylation and cap binding analyses using Ser209-mutated eIF-4Es. Biochem. Biophys. Res. Commun. 247: 213–216. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N. and Gingras, A.C. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10: 268–275. [DOI] [PubMed] [Google Scholar]
- Tomoo, K., Shen, X., Okabe, K., Nozoe, Y., Fukuhara, S., Morino, S., Sasaki, M., Taniguchi, T., Miyagawa, H., Kitamura, K., et al. 2003. Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J. Mol. Biol. 328: 365–383. [DOI] [PubMed] [Google Scholar]
- Van Ryk, D.I. and Venkatesan, S. 1999. Real-time kinetics of HIV-1 Rev–Rev response element interactions. Definition of minimal binding sites on RNA and protein and stoichiometric analysis. J. Biol. Chem. 274: 17452–17463. [DOI] [PubMed] [Google Scholar]
- Waskiewicz, A.J., Flynn, A., Proud, C.G., and Cooper, J.A. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16: 1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, H.-G., de Stanchina, E., Fridman, J.S., Malina, A., Ray, S., Kogan, S., Cordon-Cargo, C., Pelletier, J., and Lowe S.W. 2004. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428: 332–337. [DOI] [PubMed] [Google Scholar]
- Wood, S.J. 1993. DNA–DNA hybridization in real time using BIAcore. Microchem. J. 47: 330–337. [Google Scholar]
- Zuberek, J., Wyslouch-Cieszynska, A., Niedzwiecka, A., Dadlez, M., Stepinski, J., Augustyniak, W., Gingras, A.C., Zhang, Z., Burley, S.K., Sonenberg, N., et al. 2003. Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogs by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA 9: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244: 48–52. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]