Abstract
Sensitive profile searches and fold recognition were used to predict the structures of two yeast RNase P/MRP proteins. Rpp1p, which is one of the subunits common to eukaryotes and archaea, is predicted to adopt the seven-stranded TIM-barrel fold found in PHP phosphoesterases. Pop3p, initially thought to be one of the RNase P/MRP subunits unique to yeast, has been assigned the L7Ae/L30e fold. This RNA-binding fold is also present in human RNase P subunit Rpp38, raising the possibility that Pop3p and Rpp38 are functional homologs.
Keywords: RNase P, homology modeling, metal-dependent hydrolase, L7Ae, kink-turn
INTRODUCTION
Ribonuclease P (RNase P) is the ribonucleoprotein enzyme that cleaves the leader sequence of precursor tRNAs to generate mature tRNAs (for review, see Xiao et al. 2002). Bacterial RNase P has an essential RNA subunit, M1 RNA, and a single protein subunit (Guerrier-Takada et al. 1983). M1 RNA can recognize and cleave precursor tRNAs under high ionic strength conditions in the absence of proteins (for review, see Frank and Pace 1998; Xiao et al. 2002). Eukaryotic RNase P also has a single RNA species, H1 RNA, and a variable number of protein subunits (Chamberlain et al. 1998; Jiang et al. 2001). Although the evidence supports RNA-based catalysis in eukaryotic RNase P (Thomas et al. 2000), the catalytic activity of naked H1 RNA is yet to be shown in vitro, suggesting that it requires proteins for proper folding or for substrate binding and catalysis (Frank et al. 2000; Xiao et al. 2002). The purification of archaeal RNase P revealed multiple proteins similar to eukaryotes (Hall and Brown 2002). In contrast, the archaeal RNA subunits of RNase P are more similar to bacteria, and some of them, though not all, show low levels of in vitro catalytic activity without proteins (Pannucci et al. 1999). The RNase P functions in eukaryotes have been divided between several enzymes that operate in different cellular compartments (Xiao et al. 2002). Among those, mostly nucleolar enzyme RNase MRP plays an important role in pre-rRNA processing and shows considerable subunit overlap with RNase P (Chamberlain et al. 1998).
A diminished ability of RNA subunits of eukaryotic RNase P to bind and cut their RNA substrates is possibly the result of some of their functions being relegated to proteins. The protein subunits also may be needed for H1 RNA folding and to help maintain its precise subcellular localization (Xiao et al. 2002). Many of the identified protein subunits have been studied both in eukaryotes and archaea (Dichtl and Tollervey 1997; Stolc and Altman 1997; Chamberlain et al. 1998; Stolc et al. 1998; Jarrous et al. 1999; Jiang et al. 2001; Hall and Brown 2002), yet their roles in RNase P/MRP structure and catalytic mechanism are still not well understood. Recent comparative genomics and sequence profile analyses showed that a chromosomal protein, Alba, shares structural similarity to RNase P/MRP subunits Pop7p/Rpp20 and Rpp25 (Aravind et al. 2003). In an attempt to provide similar structural/functional assignments for other RNase P/MRP subunits, we used sequence profile methods, fold recognition, model building, and evaluation (Sippl 1993; Altschul et al. 1997; Sanchez and Sali 1998; Eswar et al. 2003; Ginalski et al. 2003) for all RNase P/MRP subunits of Saccharomyces cerevisiae. The results of this study provide clues about the functions of RNase P/MRP subunits Rpp1p and Pop3p.
RESULTS AND DISCUSSION
Rpp1p of S. cerevisiae is essential for tRNA and rRNA maturation (Stolc and Altman 1997). Orthologs of Rpp1p have been identified in higher eukaryotes and archaea (Koonin et al. 2001; Hall and Brown 2002). Iterative profile searches with PSI-BLAST (Altschul et al. 1997) using Rpp1p as a query identified putative phosphoesterases containing the PHP domain (first occurrence in third iteration, E = 3 × 10−5) (Aravind and Koonin 1998). This finding was confirmed by the 3D-Jury meta-server (Ginalski et al. 2003), which ranked the PHP-containing protein YcdX of Escherichia coli in the top six structural matches to Rpp1p, all of which scored better than the threshold value of 50 (the highest score was 109). YcdX contains a trinuclear zinc cluster inside the deep cleft of an unusual seven-stranded β-barrel capped with helix α8 (Teplyakov et al. 2003). Although the exact function of YcdX is unknown, it likely belongs to metal-dependent phosphohydrolases based on the function of a characterized member of the same superfamily (Morona et al. 2002; Teplyakov et al. 2003). Another group of proteins that had three structures scoring in a 50–65 range were eight-stranded TIM-barrel metallohydrolases that catalyze the hydrolytic cleavage of amide and ester bonds (Nagano et al. 2002). According to the SCOP database (Andreeva et al. 2004), these proteins are close structural relatives of PHP proteins as both groups of proteins coordinate metals and are capped by an α-helix at the C terminus. The decision to proceed further with PHP proteins as best matches was based on three considerations: (1) the sequence similarity of Rpp1p to PHP proteins noted above; (2) the secondary structure prediction (Jones 1999) of Rpp1p accounted for seven instead of eight β-strands, which favors seven-stranded PHP proteins; and (3) 3D-Jury scores for YcdX were much higher than those of TIM-barrel metallohydrolases.
Manually adjusted 3D-Jury alignment of Rpp1p and YcdX (Fig. 1A) was used to generate the homology model of Rpp1p by satisfying spatial restraints (Fig. 1B; Eswar et al. 2003). Energy-based structure quality assessment (Sippl 1993; Sanchez and Sali 1998) showed that the overall fold of this model is correct (pG = 0.99), further supporting the choice of PHP proteins as closest structural homologs of Rpp1p. The colors in Figure 1B are based on residue conservation among eukaryotic orthologs of Rpp1p and suggest their relative functional importance; the most conserved residues are shown in red and the least conserved in blue (Pei and Grishin 2001). Two interesting things about the residue conservation are worth noting. First, Rpp1p and its orthologs lack several histidine residues that coordinate Zn2+ in YcdX (see Supplemental Fig. 1 at http://www.homepage.montana.edu/~mdlakic/Rpp1p_YcdX_Suppl_Fig1.html). This is not unexpected as the types of metals and residues used for their binding vary in TIM-barrel proteins; for example, Mg2+ and Mn2+ are typically coordinated by acidic residues (Nagano et al. 2002). At least one group of PHP-containing phosphatases is known to be manganese-dependent (Morona et al. 2002), highlighting the versatility of this fold in coordinating metals other than zinc. Second, many of the conserved residues in Rpp1p occupy similar positions as the metal-binding residues of YcdX and cluster inside the barrel surrounding putative metal ions (blue spheres in Fig. 1A). Most of the metal-binding residues in TIM-barrel enzymes are found in strands 1, 5, 6, and 7 (Nagano et al. 2002). This holds true for three highly conserved acidic residues of Rpp1p, D4, D127, and E157 (marked with red triangles in Fig. 1A and Supplemental Fig. 1), which are found in strands 1, 5, and 6, respectively. Considering that the majority of TIM-barrel proteins are enzymes (Nagano et al. 2002), the structural similarity of Rpp1p to the zinc-binding PHP proteins suggests a possible enzymatic function for Rpp1p that involves metal ions. Although at present there are no data indicating that Rpp1p binds metal ions, it is well known that RNase P requires Mg2+ (for review, see Xiao et al. 2002), and coordination of Mg2+ by conserved acidic residues is a common theme among nucleases (Dlakić 2000).
FIGURE 1.
3D model of Rpp1p shows structural similarity to Ph1877p (PDB code 1V77). (A) The initial alignment of Rpp1p and YcdX was obtained from the 3D-Jury meta-server (Ginalski et al. 2003). Slight manual adjustments were made to position insertions and deletions within loop regions. The residues are color-coded according to the following scheme: (red letters) negative residues; (green shade) aliphatic; (blue letters) positive; (light-blue letters) aromatic; (orange letters) charged; (purple) small; (red shade) polar; (cyan shade) big; (dark-blue shade) hydrophobic. Red triangles beneath the alignment denote conserved acidic residues of Rpp1p and are equivalent to red triangles in Supplemental Figure 1 (see text for details). (B) The model of Rpp1p was colored by sequence conservation so that most conserved residues are in red and the least conserved ones are in blue (Pei and Grishin 2001). The color palette between red and blue indicates intermediate levels of sequence conservation. The positions of ions in the putative trinuclear metal cluster (blue spheres inside the β-barrel) have been assigned from the template structure (Teplyakov et al. 2003). (C) The Ph1877p structure (Takagi et al. 2004) was aligned to the Rpp1p model using CE (Shindyalov and Bourne 1998). The optimal alignment spans 192 residues with an overall RMSD of 3.57 Å. Helices are colored differently from β-strands and loops to facilitate the comparison of secondary structure elements with Figure 1B.
While this manuscript was under consideration, the crystal structure of the Pyrococcus horikoshii homolog of Rpp1p, Ph1877p, has been reported (Takagi et al. 2004). This study showed that Ph1877p is, indeed, a TIM-barrel protein with seven instead of canonical eight β-strands. The Ph1877p structure aligns with the Rpp1p model over 192 residues with an overall root-mean-square deviation of 3.57 Å. Furthermore, this alignment has a Z-score of 6.0, which is indicative of a relationship at the level of protein family (Shindyalov and Bourne 1998). To evaluate the quality of comparative models made from target–template alignments with <30% identity, one has to consider that for these difficult cases only 50%–80% of residues are expected to be within 3.5 Å of their correct positions (Marti-Renom et al. 2000; Baker and Sali 2001). Given that Rpp1p and Ph1877p have only 14% sequence identity and that the template sequence (YcdX of E. coli) has only 11%–16% sequence identity to either one of them, the Rpp1p model can be considered reliable. The superposition of Ph1877p structure (Fig. 1C) and the Rpp1p model (Fig. 1B) shows the overall agreement between most of the secondary structure elements, although their lengths and spatial orientations are not always identical. Interestingly, the Ph1877p structure also aligns well with eight-stranded TIM-barrel metallohydrolases (Takagi et al. 2004), providing an explanation for the initial difficulty in choosing the correct template for modeling. Finally, it should be noted that no metal ions were found inside the barrel of Ph1877p. This could be due to the fact that no divalent metals were used in crystallization (Takagi et al. 2004), or because Rpp1p homologs adopt this unusual variant of the TIM-barrel fold for reasons other than metal coordination, for example, for RNA binding (Jiang et al. 2001; Takagi et al. 2004).
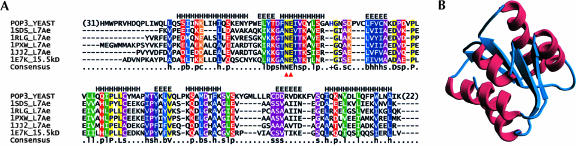
Pop3p is essential for both pre-tRNA and pre-rRNA processing in S. cerevisiae (Dichtl and Tollervey 1997). It has no orthologs outside of related fungal species. Iterative database searches with PSI-BLAST identified Pop3p relatives among proteins containing the L7Ae/L30e domain (first occurrence in fourth iteration, E = 7 × 10−4). Human RNase P subunit Rpp38 (hRpp38), which is known to have the L7Ae/L30e domain (Jarrous 2002), was among the identified proteins (seventh iteration, E = 7 × 10−10). Since both Pop3p and hRpp38 are known to bind RNA (Brusca et al. 2001; Jiang et al. 2001), it is tempting to speculate that they are functional homologs. The PSI-BLAST finding was supported by 3D-Jury predictions (Ginalski et al. 2003), as all of the top 20 structural matches had the L7Ae/L30e domain (six of them scored higher than the threshold of 50). This universal agreement of 3D-Jury structural templates was also evident in close correlation between the predicted secondary structure of Pop3p and the structural consensus of L7Ae/L30e proteins (Fig. 2A), and provided greater confidence for the overall Pop3p prediction. In a final step, the homology model of Pop3p was built (pG = 1) (Fig. 2B) using archaeal RNA-binding protein L7Ae as a template (Hamma and Ferre-D’Amare 2004).
FIGURE 2.
Multiple alignment of Pop3p with L7Ae/L30e proteins and 3D model of Pop3p. (A) Sequence names correspond to PDB codes of solved structures containing L7Ae/L30e proteins. Red triangles beneath the consensus line mark Asn–Glu residues that directly recognize the tandem of sheared G • A base pairs of the kink-turn (Vidovic et al. 2000; Klein et al. 2001; Hamma and Ferre-D’Amare 2004; Moore et al. 2004). Secondary structure assignments above the alignment (H for α-helices, E for β-strands) were derived from the consensus of L7Ae and 15.5kD structures (Vidovic et al. 2000; Klein et al. 2001; Hamma and Ferre-D’Amare 2004; Moore et al. 2004). The initial alignment by PCMA (Pei et al. 2003) was manually adjusted based on secondary structure prediction. Shaded residues are conserved in all sequences. Lowercase letters on the consensus line have the following meaning: (h) hydrophobic; (b) big; (p) polar; (s) small; (c) charged; (a) aromatic; (+) positively charged; (−) negatively charged; (l) aliphatic. Capital letters on the consensus line and columns shaded in yellow indicate single-residue conservation. (B) Structures of a spliceosomal 15.5-kDa protein (Vidovic et al. 2000) and the archaeal RNA-binding protein L7Ae (Hamma and Ferre-D’Amare 2004) were used as templates, and in both cases the produced models were of good quality (Sippl 1993; Sanchez and Sali 1998). The Pop3p model shown is based on the archaeal L7Ae structure.
Proteins containing the L7Ae/L30e fold often bind a conserved RNA secondary structure motif called the kink-turn (K-turn) (Klein et al. 2001). This general type of protein–RNA interaction is found in ribosomes, spliceosomal RNPs, and C/D-box snoRNA RNPs (Vidovic et al. 2000; Klein et al. 2001; Rozhdestvensky et al. 2003). Two key residues of L7Ae/L30e proteins that interact with tandem sheared G • A base pairs of the K-turn (Vidovic et al. 2000; Klein et al. 2001; Hamma and Ferre-D’Amare 2004; Moore et al. 2004) are conserved in Pop3p (indicated by red triangles in Fig. 2A). Given this remarkable conservation of interactions between L7Ae/L30e proteins and K-turn motifs and the prediction that human RNase MRP RNA subunit contains the K-turn (Klein et al. 2001), it seems likely that Pop3p and hRpp38 both recognize K-turn motifs. Finally, an analysis of the ribosome structure suggested that L7Ae also forms a conserved protein–protein interface with L15e (Klein et al. 2001). This type of interaction is unlikely for Pop3p because there is no obvious structural homolog of L15e among RNase P/MRP subunits and the key residues that contribute to the L7Ae/L15e interface are not conserved in Pop3p.
In conclusion, the successful prediction of Rpp1p structure validates the approach that combines remote homology detection with model building. It also inspires confidence in Pop3p prediction and suggests further experiments to test whether Pop3p and hRpp38 recognize an RNA secondary structure motif formed by the RNA subunit of RNase P/MRP, its substrate RNAs, or the combination of both (Brusca et al. 2001).
MATERIALS AND METHODS
Iterative profile searches were done with PSI-BLAST (Altschul et al. 1997) using composition-based statistics and a conservative threshold for inclusion into profile (E = 0.001). This stringent threshold was used in order to eliminate any spurious hits to unrelated proteins. The searches were iterated until convergence.
Protein structures were predicted using the 3D-Jury meta-server (http://bioinfo.pl/Meta; Ginalski et al. 2003). This server takes as input models generated by a set of prediction servers, compares them with each other, and generates a similarity score. The model is more likely to be correct if two or more independent methods generate similar predictions (Ginalski et al. 2003). Final scores >50 are considered significant, though the top 20 matches are always shown regardless of whether they score above the threshold or not. The server also provides target–template alignments that can be used for subsequent model building.
Comparative modeling was done by satisfying spatial restraints as implemented in MODELLER (http://salilab.org/modeller/modeller.html; Eswar et al. 2003). The quality of models critically depends on correct target–template alignments (Marti-Renom et al. 2000). For alignments with low target–template sequence identity (typically below 30%), it is often necessary to edit the alignments by accounting for secondary structure predictions. When dealing with difficult cases of low sequence identity, and because model evaluation is more reliable than alignment evaluation (Marti-Renom et al. 2000), the 3D models were evaluated and their flawed sections were rebuilt by adjusting the target–template alignment. The cycle of model building and evaluation was repeated, including the consideration of multiple templates if available.
Models were evaluated by Prosa II empirical mean-force potentials devised to detect faulty parts of structural models (Sippl 1993). In addition to energy graphs that visually identify suspicious parts of the structure, Prosa II provides a Z-score that indicates whether the fold is native-like or not. The Z-score was used to estimate the probability (pG) on a 0–1 scale that the model is based on a correct alignment with an appropriate template (http://sanchezlab.org/servers/pg/; Sanchez and Sali 1998). This means that the model may not be correct in all the details even when its pG value is very close to 1. More detailed description of practical aspects of remote homology detection, model building, and evaluation procedures are provided in a step-by-step example at http://www.homepage.montana.edu/~mdlakic/modeling/methods.html.
Acknowledgments
I am grateful to Elisa Caffarelli, Alessandro Fatica, and David Tollervey for comments and to Dave Engelke for discussions about Rpp1p. This work was supported by NIH Grant P20 RR16455 from the BRIN Program of the National Center for Research Resources.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7128905.
REFERENCES
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva, A., Howorth, D., Brenner, S.E., Hubbard, T.J., Chothia, C., and Murzin, A.G. 2004. SCOP database in 2004: Refinements integrate structure and sequence family data. Nucleic Acids Res. 32: D226–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L. and Koonin, E.V. 1998. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 26: 3746–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., Iyer, L.M., and Anantharaman, V. 2003. The two faces of Alba: The evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 4: R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D. and Sali, A. 2001. Protein structure prediction and structural genomics. Science 294: 93–96. [DOI] [PubMed] [Google Scholar]
- Brusca, E.M., True, H.L., and Celander, D.W. 2001. Novel RNA-binding properties of Pop3p support a role for eukaryotic RNase P protein subunits in substrate recognition. J. Biol. Chem. 276: 42543–42548. [DOI] [PubMed] [Google Scholar]
- Chamberlain, J.R., Lee, Y., Lane, W.S., and Engelke, D.R. 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes & Dev. 12: 1678–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl, B. and Tollervey, D. 1997. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 16: 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakić, M. 2000. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci. 25: 272–273. [DOI] [PubMed] [Google Scholar]
- Eswar, N., John, B., Mirkovic, N., Fiser, A., Ilyin, V.A., Pieper, U., Stuart, A.C., Marti-Renom, M.A., Madhusudhan, M.S., Yerkovich, B., et al. 2003. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 31: 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D.N. and Pace, N.R. 1998. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67: 153–180. [DOI] [PubMed] [Google Scholar]
- Frank, D.N., Adamidi, C., Ehringer, M.A., Pitulle, C., and Pace, N.R. 2000. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA 6: 1895–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski, K., Elofsson, A., Fischer, D., and Rychlewski, L. 2003. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics 19: 1015–1018. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N., and Altman, S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. and Brown, J.W. 2002. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA 8: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma, T. and Ferre-D’Amare, A.R. 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 12: 893–903. [DOI] [PubMed] [Google Scholar]
- Jarrous, N. 2002. Human ribonuclease P: Subunits, function, and intranuclear localization. RNA 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous, N., Eder, P.S., Wesolowski, D., and Altman, S. 1999. Rpp14 and Rpp29, two protein subunits of human ribonuclease P. RNA 5: 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, T., Guerrier-Takada, C., and Altman, S. 2001. Protein–RNA interactions in the subunits of human nuclear RNase P. RNA 7: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195–202. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V., Wolf, Y.I., and Aravind, L. 2001. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 11: 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom, M.A., Stuart, A.C., Fiser, A., Sanchez, R., Melo, F., and Sali, A. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29: 291–325. [DOI] [PubMed] [Google Scholar]
- Moore, T., Zhang, Y., Fenley, M.O., and Li, H. 2004. Molecular basis of box C/D RNA–protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure 12: 807–818. [DOI] [PubMed] [Google Scholar]
- Morona, J.K., Morona, R., Miller, D.C., and Paton, J.C. 2002. Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J. Bacteriol. 184: 577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano, N., Orengo, C.A., and Thornton, J.M. 2002. One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 321: 741–765. [DOI] [PubMed] [Google Scholar]
- Pannucci, J.A., Haas, E.S., Hall, T.A., Harris, J.K., and Brown, J.W. 1999. RNase P RNAs from some Archaea are catalytically active. Proc. Natl Acad. Sci. 96: 7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, J. and Grishin, N.V. 2001. AL2CO: Calculation of positional conservation in a protein sequence alignment. Bioinformatics 17: 700–712. [DOI] [PubMed] [Google Scholar]
- Pei, J., Sadreyev, R., and Grishin, N.V. 2003. PCMA: Fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics 19: 427–428. [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky, T.S., Tang, T.H., Tchirkova, I.V., Brosius, J., Bachellerie, J.P., and Huttenhofer, A. 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 31: 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, R. and Sali, A. 1998. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl Acad. Sci. 95: 13597–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov, I.N. and Bourne, P.E. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11: 739–747. [DOI] [PubMed] [Google Scholar]
- Sippl, M.J. 1993. Recognition of errors in three-dimensional structures of proteins. Proteins 17: 355–362. [DOI] [PubMed] [Google Scholar]
- Stolc, V. and Altman, S. 1997. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes & Dev. 11: 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc, V., Katz, A., and Altman, S. 1998. Rpp2, an essential protein subunit of nuclear RNase P, is required for processing of precursor tRNAs and 35S precursor rRNA in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. 95: 6716–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, H., Watanabe, M., Kakuta, Y., Kamachi, R., Numata, T., Tanaka, I., and Kimura, M. 2004. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 319: 787–794. [DOI] [PubMed] [Google Scholar]
- Teplyakov, A., Obmolova, G., Khil, P.P., Howard, A.J., Camerini-Otero, R.D., and Gilliland, G.L. 2003. Crystal structure of the Escherichia coli YcdX protein reveals a trinuclear zinc active site. Proteins 51: 315–318. [DOI] [PubMed] [Google Scholar]
- Thomas, B.C., Chamberlain, J., Engelke, D.R., and Gegenheimer, P. 2000. Evidence for an RNA-based catalytic mechanism in eukaryotic nuclear ribonuclease P. RNA 6: 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic, I., Nottrott, S., Hartmuth, K., Luhrmann, R., and Ficner, R. 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell 6: 1331–1342. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Scott, F., Fierke, C.A., and Engelke, D.R. 2002. Eukaryotic ribonuclease P: A plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 71: 165–189. [DOI] [PMC free article] [PubMed] [Google Scholar]