Abstract
Comparisons of full-length cDNAs and genomic DNAs available for Arabidopsis thaliana described here indicate that some adjacent loci are transcribed into extremely long RNAs spanning two annotated genes. Once expressed, some of these transcripts are post-transcriptionally spliced within their coding and intergenic sequences to generate bicistronic transcripts containing two complete open reading frames. Others are spliced to generate monocistronic transcripts coding for fusion proteins with sequences derived from both loci. RT-PCR of several P450 transcripts in this collection indicates that these extended transcripts exist side by side with shorter monocistronic transcripts derived from the individual loci in each pair. The existence of these unusual transcripts highlights variations in the processes of transcription and splicing that could not possibly have been predicted in the algorithms used for genome annotation and splice site predictions.
Keywords: bicistronic transcription units, Arabidopsis thaliana, pre-mRNA splicing, genome annotation
INTRODUCTION
Dogmatically, the transcription of eukaryotic genes has been prototyped as occurring in monocistronic units (containing a single open reading frame [ORF]) independent of adjacent genes. But, as individual transcripts and/or cDNAs have been characterized, cases of bicistronic or polycistronic transcription units (containing two or more ORFs) have been identified in a handful of organisms. In the animal kingdom, organisms containing these complex units include Caenorhabditis elegans, where ~25% of all genes exist in polycistronic units; Drosophila, where the stoned and Adh loci exist in bicistronic units with 55 nt and 298 nt separating their ORFs; and rat, where CREB and I-CREB loci exist in bicistronic units with 7 or 118 nt separating ORFs (Blumenthal 1998). In the plant kingdom, only the glutamyl kinase (GK) and glutamyl phosphate reductase (GPR) loci in tomato are known to exist in a biscistronic unit with 5 nt separating their ORFs (Garcia-Rios et al. 1997). Apart from C. elegans, where all polycistronic transcripts are processed to monocistronic transcripts by the unusual process of trans-splicing (Blumenthal 1995), it has remained unclear how many genes in other organisms might exist within bicistronic and/or polycistronic transcription units. Recent annotations of Drosophila cDNAs have suggested that additional dicistronic transcripts exist in this species, some of which have very short intergenic spacers (Misra et al. 2002), but no studies have yet determined the extent to which these are represented relative to monocistronic transcripts.
For a number of reasons, Arabidopsis thaliana has emerged as a model plant for studies in classical and molecular genetics, developmental biology, physiology, and biochemistry. Concerted efforts in this species have focused on developing and integrating genetic linkage maps, cytological maps, and physical maps for this genome, sequencing large EST and cDNA collections and, ultimately, on defining functions for more than 25,000 genes annotated in its genomic DNA sequence. In an initial effort aimed at verifying the transcripts derived from cytochrome P450 monooxygenase (P450) transcription units, we compared the collection of RIKEN full-length P450 cDNAs (Seki et al. 2002; http://rarge.gsc.riken.go.jp/) with the available annotated Arabidopsis genomic sequences only to discover a set of six transcripts spanning more than one P450 locus. In subsequent efforts aimed at defining the range of loci transcribed as part of polycistronic transcription units in this model plant, we surveyed the entire RIKEN collection of Arabidopsis full-length cDNA clones constructed with mRNA from normal as well as chemically and environmentally stressed Arabidopsis tissues for cDNAs spanning a minimum of two adjacent loci. These comparisons provide the first genome-wide description of the complex transcript processing events that occur on transcripts extending through two adjacent transcription units.
RESULTS
Identification of extended P450 transcripts
In an effort aimed at improving annotation of the 272 P450 genes present within the Arabidopsis genome that are now detailed at two Web sites (http://arabidopsis-P450.biotec.uiuc.edu; http://www.biobase.dk/P450/), we aligned each P450 gene with all sequences available through high-throughput cloning and sequencing efforts compiled at the dbEST (http://www.ncbi.nlm.nih.gov/Genbank/index.html), RIKEN (http://rarge.gsc.riken.go.jp/), and CERES (ftp://ftp.tigr.org/pub/data/a_thaliana/ceres/) databases as well as the validated and provisional REFSEQ sequences (Pruitt et al. 2002) becoming available through the GenBank database. These comparisons identified a number of extended P450 cDNAs extending through two adjacent transcription units that are detailed in Figures 1–6.
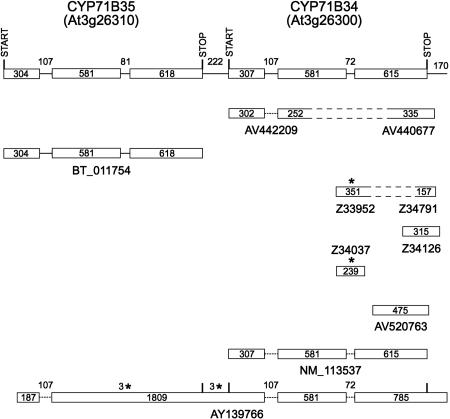
FIGURE 1.
CYP71B35 and CYP71B34 transcripts. The gene models for these loci are designated with the predicted start and stop codons for each open reading frame above the current TAIR gene models (top line) and termination codons in-frame with the P450 open reading frame shown with asterisks. GenBank accession numbers for ESTs and full-length cDNAs are shown below each diagram. REFSEQ NM_113537 is provisional.
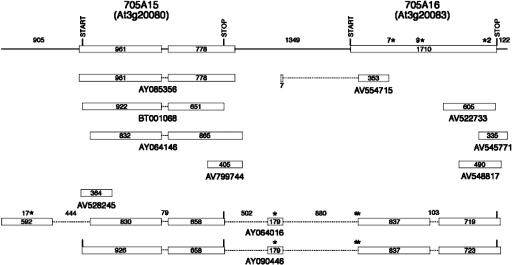
FIGURE 2.
CYP705A15 and CYP705A16 transcripts. The gene models for these loci are designated with the predicted start and stop codons for each open reading frame above the current TAIR gene models (top line) and termination codons in-frame with the P450 open reading frame designated with asterisks. GenBank accession numbers for ESTs and full-length cDNAs are shown below each diagram.
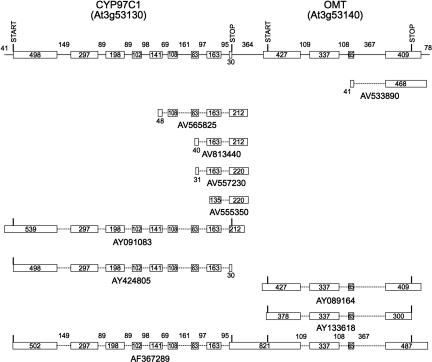
FIGURE 3.
CYP97C1 and OMT transcripts. The gene models for these loci are designated with the predicted start and stop codons for each open reading frame above the current TAIR gene model (top line). GenBank accession numbers for ESTs and full-length cDNAs are shown below each diagram.
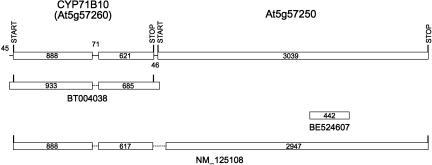
FIGURE 4.
CYP71B10 and At5g57250 transcripts. The gene models for these loci are designated with the predicted start and stop codons for each open reading frame above the current TAIR gene model (top line). GenBank accession numbers for ESTs and full-length cDNAs are shown below each diagram. REFSEQ NM_125108 is provisional.
FIGURE 5.
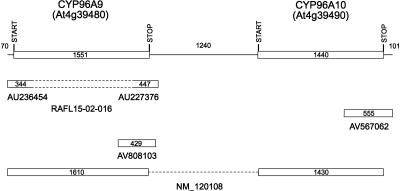
CYP96A9 and CYP96A10 transcripts. The gene models for these loci are designated with the predicted start and stop codons for each open reading frame above the current TAIR gene model (top line). GenBank accession numbers for ESTs and full-length cDNAs are shown below each diagram. REFSEQ NM_120108
FIGURE 6.
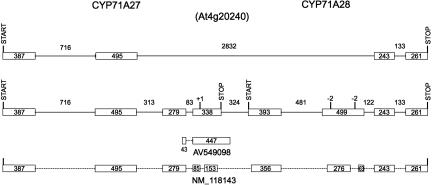
CYP71A27 and CYP71A28 transcripts. The gene models for these loci are designated with the predicted start and stop codons for each open reading frame above the previous TAIR gene model (top line). One nucleotide addition and two nucleotide deletions introduced to generate the gene model available at http://www.biobase.dk/P450/Arab_cyps/(middle line) are designated with +1 and −2, respectively. The single EST available for this locus is shown below the diagram. REFSEQ NM_118143 is provisional.
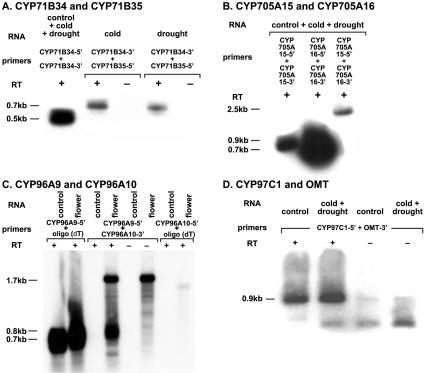
For the first of these locus sets, the existence of two full-length CYP71B35 cDNAs indicate that this locus generates two types of transcripts extending either through the CYP71B35 gene or through the CYP71B35 gene and the downstream CYP71B34 gene (Fig. 1). The first of these transcripts, represented by cDNA BT011754, encodes the predicted CYP71B35 protein. The second of these, represented by the longer AY139766 cDNA, lacks the first 117 nt of the CYP71B35 coding sequence, retains the second intron of the CYP71B35 gene, and includes a correctly spliced CYP71B34 gene. As a result of these changes, this transcript has potential to code for a truncated CYP71B35 protein of 238 amino acids with seven amino acid variations at its C terminus and a full-length CYP71B34 protein. The existence of a full-length CYP71B34 cDNA (AV442209 and AV440677 representing the 5′ and 3′ ends of APZ06C03) and a provisional REFSEQ (NM_113537) indicate that monocistronic transcripts coding for full-length CYP71B34 protein also exist. RT-PCR gel blot analysis with a CYP71B35-specific (71B35-5′) and CYP71B34-specific (71B34-3′) primer generates the 860-bp product expected from transcripts extending through the intergenic region (IGR) between these genes (Fig. 7A, below).
FIGURE 7.
RT-PCR analysis. Total RNAs isolated from the aboveground tissues of unstressed 1-mo-old plants (control), cold and drought stressed 7-d-old seedlings (cold + drought), unstressed 7-d-old seedling shoots or roots, 3-wk-old rosettes, and 1-mo-old leaves, stems, or flowers were RT-PCR amplified in one-step or two-step reactions as outlined in Materials and Methods. The RT-PCR products were electrophoresed on 1.0% agarose gels, blotted to Hybond-N nylon membranes, and probed with 32P-labeled gene-specific fragments.
For the second locus set, alignments of five available cDNAs indicate that there are two different types of bicistronic transcripts spanning the CYP705A15 and CYP705A16 loci as well as several monocistronic transcripts spanning only the CYP705A15 locus (Fig. 2). One type of bicistronic transcript represented in the AY090446 cDNA encodes the full-length CYP705A15 protein and contains a significant part of the CYP705A16 open reading frame but, due to unpredicted splicing events in the IGR, not its predicted in-frame translation start. The other type of bicistronic transcript represented in the AY064016 cDNA encodes significant portions of both P450 ORFs frames which are deleted for 28 and 31 N-terminal amino acids of the CYP705A15 and CYP705A16 proteins, respectively, because of the presence of long 5′ UTR sequences and unpredicted splicing events in the 5′ UTR and IGR. One EST (AV554715) confirms these unpredicted splicing events in the IGR preceding the CYP705A16 coding sequences. RT-PCR amplification with a CYP705A15-specific (705A15-5′) and a CYP705A16-specific (705A16-3′) primer results in 2.4–2.5-kb product (Fig. 7B, below) expected from transcripts that span and splice the CYP705A15/CYP705A16 intergenic region. Sequencing of the CYP705A16 RT-PCR product with primers from within the predicted exons has shown that a single intron of 103 nt is spliced from these transcripts.
For the third locus set, analysis of several full-length CYP97C1 cDNAs indicates that this locus generates two types of transcripts that extend either just through the CYP97C1 gene or through the CYP97C1 gene and downstream O-methyl transferase (OMT) gene (Fig. 3). The first of these transcripts represented by the AY091083 and AY424805 cDNAs is predicted to code for the CYP97C1 protein of 539 amino acids. The second of these transcripts, represented by the AF367289 cDNA, codes for the CYP97C1 protein and the full-length OMT protein and retains the entire IGR separating these adjacent loci. Alignments of independent OMT cDNAs (AY089164, AY133618) indicate that monocistronic transcripts coding for full-length OMT protein also exist. RT-PCR gel blot analyses with CYP97C1-specific (97C1-5′) and OMT-specific (OMT-3′) primers result in the 900-bp product expected from transcripts that span the CYP97C1 and OMT intergenic region (Fig. 7D, below).
Analysis of a full-length CYP71B10 cDNA and a provisional REFSEQ indicates that this locus generates two types of transcripts. The first, represented by the BT004038 cDNA, extends through the entire CYP71B10 gene (Fig. 4) and, after splicing of its single intron, codes for a full-length protein of 512 amino acids. The second of these, represented by the provisional NM_125108 sequence, extends through the CYP71B10 locus and adjacent At5g57250 locus and, after splicing of two introns, codes for a fusion protein of 1483 amino acids. Because splicing of the second intron in this transcript occurs immediately upstream of the stop codon in the CYP71B10 coding sequence and downstream of the start codon in the At5g57250 coding sequence, the fusion protein contains the entire CYP71B10 coding sequence fused to eight pentatricopeptide repeat (PPR) motifs (Small and Peeters 2000) of the predicted At5g57250 sequence.
In the fifth locus set, analysis of full-length CYP96A9 cDNA (AU236454 and AU227376 representing 5′ and 3′ ends of RAFL15-02-O16) and provisional REFSEQ sequences indicates that there are two types of transcripts generated from this locus. The first of these, represented by the RAFL15-02-016 cDNA, extends through the CYP96A9 gene (Fig. 5) and codes for a full-length CYP96A9 protein (516 amino acids). The second of these, represented by the NM_120108 REFSEQ, splices the CYP96A9 coding sequence in-frame to the CYP96A10 coding sequence at positions that delete the last two amino acids from the predicted CYP96A9 protein and the first four amino acids from the predicted CYP96A10 protein. The dimeric P450 protein predicted from this transcript contains two heme-binding domains and represents the first example of an unusual class of dimeric P450s not known to exist in other organisms. No monocistronic cDNAs have been identified for the CYP96A10 locus. RT-PCR analyses with CYP96A9-specific (96A9-5′), CYP96A10-specific (96A10-5′), and oligo(dT)17 primers conducted to determine whether polyadenylated transcripts for these individual P450 loci exist result in a 700-bp CYP96A9 product (Fig. 7C, below) that is indicative of transcripts extending ~250 nt downstream from the CYP96A9 stop codon and a 1700-bp CYP96A10 product that is indicative of transcripts extending 100 nt downstream from the CYP96A10 stop codon. RT-PCR gel blot analyses with CYP96A9-specific (96A9-5′) and CYP96A10-specific (96A10-3′) primers generate a CYP96A9-CYP96A10 product of ~800 bp expected from transcripts splicing the CYP96A9 and CYP96A10 coding sequences is provisional. into an ORF for the dimeric P450 outlined above. Clearly, both P450 loci are represented in short as well as long monocistronic transcripts.
For the sixth locus set, analysis of a full-length REFSEQ sequence spanning the CYP71A27 and CYP71A28 loci has begun to resolve several models for the organization of these loci. The original TAIR (The Arabidopsis Information Resource) gene model for these loci combined both loci into one that paired the first two exons of the CYP71A27 locus with the last two exons of the CYP71A28 locus and included an extremely long (2.8 kb) intron (Fig. 6, top line). The subsequent manually annotated gene model (Fig. 6, middle line) (http://www.biobase.dk/P450/Arab_cyps/) predicted the existence of two open reading frames for P450s if a single nucleotide was added to the CYP71A27 ORF (designated with +1 in Fig. 6) and 4 nt were deleted from the CYP71A28 ORF (designated with two −2 in Fig. 6). The newest gene model that is derived from provisional REFSEQ NM_118143 sequence (Fig. 6, bottom line) indicates that, as in the two cases of several P450s cited above, a variety of unpredictable splicing events fuse the CYP71A27 coding sequence in-frame with the CYP71A28 coding sequence to create another dimeric P450 protein that is predicted to lack the last 15 amino acids of the previously predicted CYP71A27 protein and the first 12 amino acids of the predicted CYP71A28 protein. RT-PCR gel blots supporting the existence of these transcripts are shown in Figure 7.
Genome-wide identification of extended Arabidopsis transcripts
Having demonstrated that several bicistronic and fused monocistronic P450 transcripts existed, BLAST programs (Altschul et al. 1990) were used to align all 13,181 FL-cDNA clones from the current RIKEN release (http://rarge.gsc.riken.go.jp/) with all annotated genes in the current Arabidopsis genome (ATH1.seq_cm_gz; ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/) to determine the total number of Arabidopsis loci generating extended transcripts. Processing of the BLAST results removed clones that aligned with nonadjacent loci, adjacent loci on opposing DNA strands, or with only a segment of an adjacent locus. The remaining data set contained a series of 60 FL-cDNA clones that aligned continuously to two adjacent annotated loci transcribed in the same direction. Because two sets of these 60 combined loci are each represented by two independent full-length cDNAs, these extended transcripts encompass a final count of 58 combined loci corresponding to 0.198% of all annotated Arabidopsis loci (if the combined locus is counted as one gene) or 0.395% of all loci (if both genes in the combined locus are counted as two genes).
Analysis of this collection of transcripts with respect to their chromosomal position indicated that they are randomly distributed across the Arabidopsis genome with 11, 8, 15, 10, and 14 paired loci distributed across the five chromosomes (Table 1). Analysis of this collection of transcripts with respect to their origin indicated that 44 are derived from libraries created from environmentally stressed plants (see Table 1 legend) and 16 are derived from libraries created from unstressed plants.
TABLE 1.
Sets of adjacent Arabidopsis loci expressing elongated transcripts
| Two ORFs | One ORF | ||||
| RIKEN clone | Category | Locus | RIKEN clone | Category | Locus |
| RAFL05-11-B17* | A | At4g00030/At4g00040 | RAFL05-02-O09 | P | At1g33470/At1g33475 |
| RAFL09-22-H13* | A | At2g25610/At2g25620 | RAFL06-69-H17 | P | At2g35710/At2g35715 |
| RAFL09-40-B19 | A | At3g06510/At3g06520 | RAFL07-15-G11* | P | At5g35980/At5g35990 |
| RAFL09-54-M24 | A | At2g24940/At2g24945 | RAFL09-16-G02 | P | At1g23430/At1g23440 |
| RAFL09-63-M16* | A | At2g25610/At2g25620 | RAFL09-44-D01 | P | At4g29490/At4g29500 |
| RAFL11-04-B08 | A | At4g34530/At4g34540 | RAFL09-60-B14 | P | At1g60200/At1g60210 |
| RAFL16-76-N11 | A | At5g53480/At5g53485 | RAFL09-61-C04 | P | At4g31080/At4g31090 |
| RAFL09-60-E22 | B | At3g45640/At3g45650 | RAFL09-81-M20* | P | At5g54120/At5g54130 |
| RAFL19-62-F22 | B | At3g48565/At3g48570 | RAFL15-41-F10 | P | At4g20120/At4g20130 |
| RAFL08-16-I18 | C | At3g26420/At3g26430 | RAFL16-67-A17 | P | At5g20530/At5g20540 |
| RAFL09-13-P07 | D | At3g53130/At3g53140 | RAFL16-69-P16 | P | At5g65350/At5g65360 |
| RAFL08-09-I21* | E | At1g23870/At1g23880 | RAFL19-84-B16 | P | At4g33330/At4g33340 |
| RAFL21-32-H16* | F | At4g29850/At4g29860 | RAFL05-11-G13 | Q | At2g39070/At2g39080 |
| RAFL16-98-M19* | G | At3g13700/At3g13710 | RAFL06-74-D12 | Q | At1g26280/At1g26300 |
| RAFL09-29-J06 | H | At5g57140/At5g57150 | RAFL07-15-F13* | Q | At2g40835/At2g40840 |
| RAFL09-67-C10 | I | At4g10955/At4g10960 | RAFL09-24-E16* | Q | At1g09980/At1g09990 |
| RAFL09-69-N10 | J | At3g20080/At3g20083 | RAFL14-80-M23 | Q | At1g62850/At1g62855 |
| RAFL14-83-D16 | K | At5g50970/At5g50980 | RAFL16-01-C03 | Q | At5g63420/At5g63430 |
| RAFL16-97-D24* | K | At3g59560/At3g59570 | RAFL09-56-O16 | R | At5g55030/At5g55040 |
| RAFL08-13-H11* | L | At3g20080/At3g20083 | RAFL05-09-I21 | S | At5g48250/At5g48260 |
| RAFL09-72-F15* | M | At2g31330/At2g31340 | RAFL06-10-K24* | S | At2g03390/At2g03400 |
| RAFL15-09-P04* | N | At3g03350/At3g03360 | RAFL09-32-C08* | S | At5g04300/At5g04310 |
| RAFL05-21-L13 | O | At4g35870/At4g35880 | RAFL15-09-A22* | S | At2g31110/At2g31120 |
| RAFL07-17-D11* | O | At3g26300/At3g26310 | RAFL16-73-D01* | S | At1g31700/At1g31710 |
| RAFL08-11-J20* | O | At1g01770/At1g01780 | RAFL09-85-I15* | T | At3g48900/At3g48910 |
| RAFL08-19-A05 | O | At4g20830/At4g20840 | RAFL11-13-C20 | U | At5g12160/At5g12170 |
| RAFL09-34-K18 | O | At1g11350/At1g11360 | RAFL08-08-P22 | V | At3g16855/At3g16857 |
| RAFL16-96-P09 | O | At1g73060/At1g73070 | RAFL08-17-N09* | V | At3g52640/At3g52650 |
| RAFL19-66-K06 | O | At5g23230/At5g23240 | RAFL21-17-J10 | V | At3g08830/At3g08840 |
| RAFL09-21-B11* | W | At3g24460/At3g24470 | |||
| RAFL07-12-J11 | W | At5g62590/At5g62600 |
Asterisks designate transcripts containing additional ATG codons in the 5′ UTR of the first ORF that potentially initiate on internal ATG codons. Bold type designates adjacent loci whose ability to generate bicistronic transcripts or fused monocistronic transcripts has been confirmed by RT-PCR gel blot analysis of Arabidopsis RNAs shown in Figure 7. Transcripts derived from enviromentally stressed plants include five and seven clones in the RAFL5 and RAFL8 libraries constructed with plants treated with dehydration, 3 and 21 clones in the RAFL6 and RAFL9 libraries constructed with plants at different developmental stages as well as plants treated with dehydration and cold, four in the RAFL7 library constructed with plants treated with cold, two in the RAFL11 library constructed with plants at different developmental stages as well as plants treated with dehydration, cold, salt, heat, UV, or ABA, and two in the RAFL21 library constructed with plants treated with environmental stresses (heat, UV) and various chemical treatments (ABA, auxin, ethylene, JA, SA, GA, cytokinin, BTH) (Seki et al. 2002). Transcripts derived from unstressed plants include two in the RAFL14 library constructed with root tissue, three and three in the RAFL15 and RAFL19 libraries constructed with silique and flower tissues, and eight in the RAFL16 library constructed with dark-grown plants (Seki et al. 2002).
Bicistronic transcripts with two predicted ORFs
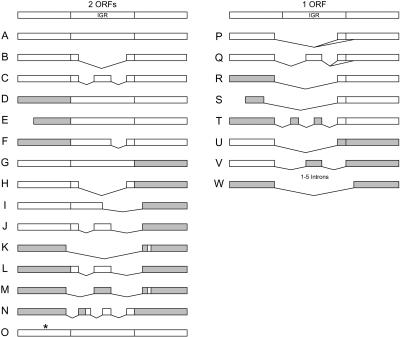
Further examination of the 60 clones distinguished 15 categories of bicistronic transcripts (Fig. 8, left) and 8 categories of monocistronic transcripts (Fig. 8, right) that shared varying degrees of overlap with existing gene models. The first group of 10 cDNAs contains sets of accurately annotated bicistronic transcripts potentially coding for two different proteins with each at least 45 amino acids in length. In each of these cases (categories A–C), all introns are spliced from each ORF as annotated in the TAIR models (correct annotations as of Dec. 2003 are designated with white boxes in Fig. 8) indicating that these do not represent partially processed or truncated transcripts derived from these adjacent loci but, rather, real bicistronic transcripts capable of coding for multiple proteins. The annotated functions for loci included in this set of bicistronic transcripts include many proteins with no obvious functional relationship with one another (see Supplemental Table 1 at www.life.uiuc.edu/csb/faculty/publications/bicistronic_supp_tables.html).
FIGURE 8.
Classes of bicistronic and monocistronic transcripts derived from adjacent loci. Diagrams for two adjacent loci are depicted with their intergenic region (IGR). White boxes indicate that each ORF is as annotated in current gene models for the individual loci with all introns spliced as predicted. Gray boxes indicate that the ORF is altered from predictions due to splicing of introns at alternate positions within the coding region of the transcript, to N-terminal extension of coding sequences due to transcript initiation at sites upstream from the predicted translation start site, or to N-terminal truncation of coding sequences due to placement of the 5′ end of the transcript downstream from the predicted translation start site. Vertical lines designate the translation start and stop codons used in each ORF. The numbers of cDNAs in each category are as follows: A (7), B (2), C–J (1 each), K (2), L–N (1 each), O (7), P (12), Q (6), R (1), S (5), T (1), U (1), V (3), W (2).
Alignments with the available Arabidopsis genomic sequences indicate that, in this first group of bicistronic loci, the lengths of the intergenic regions (IGR) between the stop codon of the first ORF and the start codon of the second ORF vary from 194 to 1994 nt in genomic DNA and from 23 to 1994 nt in the corresponding bicistronic transcripts. Based on the presence or absence of splicing events occurring in their intergenic regions, these 10 bicistronic transcripts have been further subdivided into categories A–C. Seven cases (category A) include the entire length of the IGR (RAFL16-76-N11 in this collection has a previously undetected exon in its 5′ UTR), two cases (category B) excise a single large intron (465 or 554 nt) from the IGR, creating a short noncoding spacer from the 3′ UTR of the first gene and the 5′ UTR of the second gene, and one case (category C) excises a small intron (108 nt) from the IGR, allowing a long IGR-derived sequence (847 nt) to exist between the two predicted ORFs. For one set of combined loci (At2g25610/At2g25620), two independent cDNAs in category A have been identified that both include the entire IGR.
Bicistronic transcripts with at least one reannotated ORF
Of the original 60 clones, a second group of 19 cDNAs contains sets of bicistronic transcripts in which one or both of the ORFs differ from the predicted gene model. In the clones represented by categories D–J, one ORF is the same as predicted in the current TAIR gene model and the second ORF deviates from the gene model (designated with gray boxes in Fig. 8). In the clones represented by categories K–N, both ORFs differ from those predicted in the TAIR gene models. In the clones represented by category O, both ORFs are the same as predicted in the current TAIR models but inclusion of an intron in the transcript is predicted to prematurely truncate the encoded protein (designated by the asterisk in Fig. 8).
These deviations from predictions arise for a number of reasons. In categories D, F, K, M, and N, differences from the gene model for the first ORF result from splicing of alternate introns within the coding region that are not predicted in the TAIR gene model. In the clones represented by categories E and L, transcript initiation at a site within the predicted ORF or incomplete reverse transcription of the first cDNA strand results in truncated ORFs that code for the C-terminal portion of the predicted ORF. Deviations from the gene model for the second ORF result from the splicing of alternate introns within the coding region (categories G, H, J, K, M, and N) or splicing of the preceding ORF or IGR sequences to the second ORF at a point downstream from its predicted start codon (categories I, J, L, and M). In cases where the next start codon exists in the same reading frame as the predicted downstream ORF (category I), the second ORF is predicted to code for an N-terminally truncated protein similar to the originally predicted protein. In cases where the next start codon exists in a reading frame different from the originally predicted downstream ORF (categories J, L, and M), the second ORF is predicted to code for a protein different from that previously predicted.
Among the 12 clones represented by categories D–N, individual transcripts can be subdivided into three cases that include the entire length of the IGR and nine cases that excise one to three introns from the IGR. When the first ORF utilizes its predicted stop codon (categories D–J and L), these splicing events within the IGR include varying amounts of noncoding sequence upstream from the start site of the second ORF (either as originally predicted or downstream from it). When the first ORF does not utilize its predicted stop codon (categories K, M, and N), splicing within the IGR fuses codons derived from the IGR or downstream locus in-frame with those derived from the upstream locus. For one set of combined loci (At3g20080/At3g20083), two independent cDNAs in categories J and L have been identified that represent bicistronic transcripts with different splicing events in the first ORF and the same splicing events in the IGR region and second ORF. The protein sequences derived from the loci included in this set of reannotated bicistronic transcripts (see Supplemental Table 1 at www.life.uiuc.edu/csb/faculty/publications/bicistronic_supp_tables.html) include combinations of various unrelated proteins as well as the one shown in Figure 3 encoding P450 and O-methyltransferase proteins potentially mediating sequential steps in a chemical detoxification process. Three other bicistronic transcripts encode the closely related and obviously duplicated P450 proteins shown in Figures 1 and 2.
Alignments of the loci represented in bicistronic transcripts with the annotated full-length cDNAs indicate that most of these loci (13/18) in the first group and half of the loci (19/36) in the second group are represented by at least one monocistronic cDNA derived from each of the two ORFs as predicted in the current gene models. Depending on which locus they align with, some of these monocistronic transcripts are as predicted in the current set of TAIR models and others agree with the revised gene model based on mapping of these bicistronic transcripts.
Monocistronic transcripts fusing two adjacent ORFs
A third group of 31 cDNAs contains sets that encode true fusion proteins with codons derived from two adjacent and independently annotated loci. In the first set of 18 clones represented by categories P and Q, splicing occurs between two accurately predicted ORFs. The splice sites combining these nearly full-length ORFs often occur just upstream from the stop codon of the first ORF and downstream from the start codon of the second ORF. In the second set of 11 clones represented by categories R–V, splicing occurs between a predicted ORF and an ORF that differs from predictions. In two cases (categories R and T), changes in the first ORF due to alternate introns coupled with alternative splicing upstream from the stop codon of the first ORF merge a reannotated first ORF with the full length of the predicted ORF for the second locus. In five cases (category S), the first ORF is truncated at its N terminus (due possibly to transcription initiation at an alternate site or incomplete reverse transcription) and at its C terminus (due to alternate splicing) and fused to nearly the full length of the second ORF; in two of these cases, the truncations are so severe that only the second ORF has potential to yield an extended translation product. In four cases (categories U and V), splicing upstream from the stop codon of the first ORF to a reading frame different from that predicted for the second locus yields transcripts coding for an extended fusion protein containing nearly the full length of the first ORF and an unpredicted ORF derived from the second locus. In the third set of two clones represented by category W, splicing occurs between two ORFs different from predictions generating a fusion protein containing entirely an unpredicted sequence. In the first two sets of clones generating a single ORF, variations in the numbers, sizes, and positions of introns spliced from the IGR region (as described above for the bicistronic transcripts) cause different IGR-encoded sequences to be included between the sequences derived from the predicted and reannotated reading frames. Seven of these bicistronic transcripts are represented by multiple cDNAs in RIKEN and non-RIKEN collections1. Annotations associated with loci in this group of fused monocistronic transcripts are detailed in Supplemental Table 2 www.life.uiuc.edu/csb/faculty/publications/bicistronic/supp_tables.html.
Many of the loci represented in these sets of bicistronic and monocistronic transcripts utilize the first translation start codon present in the transcript. But, because of variations in transcription start sites and/or inclusion of additional exons near the 5′ end of these transcripts, some in the first and second groups of bicistronic transcripts (12/29) and the third group of fused monocistronic transcripts (11/31) contain one or more translation start codons in their 5′ UTRs.
Splice sites on intergenic introns
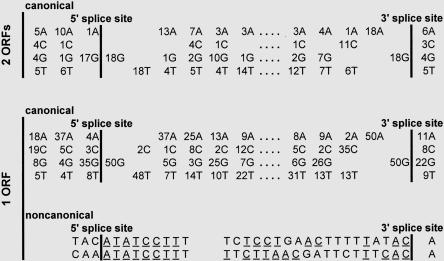
Examination of the 5′ and 3′ splice sites on the intergenic introns in the bicistronic transcripts versus the fused monocistronic transcripts indicates that 66 have canonical/GT…AG/dinucleotides at either end of the intron and two have noncanonical/GC…AG/dinucleotides (Table 2). The more extended consensus sequences spanning these junctions contain consensus nucleotides typical of many U2 snRNA-dependent plant introns (Simpson and Filipowicz 1996; Schuler 1998). But, as diagrammed in Figure 9 for examples in bicistronic categories C and F and monocistronic categories P and Q, individual splice sites have varying numbers of contiguous nucleotides corresponding to the optimal (T/C)(G/A)G/GT(G/A)(G/A)GT-5′ splice site consensus sequence and the optimal TGCAG/G-3′ splice site consensus sequence; matches to these are designated with underlines in Figure 9. An additional two introns in the set of fused monocistronic transcripts have noncanonical /AT…AC/dinucleotides and an extended set of consensus nucleotides typical of U12 snRNA-dependent introns (Patel and Steitz 2003; Zhu and Brendel 2003); matches to these consensus sequences are designated with underdots in Figure 9 and Table 2.
TABLE 2.
Survey of splice sites in intergenic regions
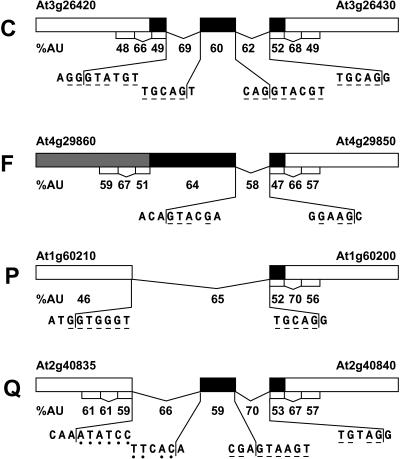
FIGURE 9.
Intron junctions in intergenic regions of bicistronic transcripts.
In the four examples shown in Figure 9, comparisons of the adenosine plus uridine (A + U) content of introns and exons within intergenic regions indicate that the transitions in A + U content are less dramatic than for most of the introns and exons within the upstream and downstream coding sequences. For example, the intergenic introns in bicistronic transcript C are 2%–10% and 9%–20% richer in A + U than their adjacent exonic sequences compared to adjacent introns in the coding sequence that are 17%–18% (upstream) and 16%–19% (downstream) richer in A + U content. Similarly, the intergenic intron in bicistronic transcript F is 8%–11% richer in A + U content than its adjacent exonic sequence compared to adjacent introns in the coding sequence that are 8%–16% and 9%–19% richer in A + U content. The lower transitions in A + U content seen for intergenic introns do no reflect the fact that this group of introns has lower A + U content but rather that their exons, which are derived from AU-rich intergenic regions, have higher A + U contents than exons in typical coding regions. This point is reiterated in the intergenic introns in monocistronic transcripts P and Q, which have the long intergenic intron in transcript P excising all AU-rich intergenic sequences from the adjacent AU-poor ORFs and the long intergenic introns in transcript Q excising only part of the AU-rich intergenic sequence. These less obvious and variable transitions in AU content suggest that intergenic introns cannot be computationally distinguished from intergenic exons based solely on their AU content compared to adjacent sequences.
DISCUSSION
These alignments have identified some very surprising transcription units with potential to code for multiple proteins. Among the P450s in this collection, the first and clearest example of this is the CYP97C1/OMT transcript that contains two complete ORFs separated by 364 nt. The second example is the CYP705A15/CYP705A16 transcript discussed above that contains a complete first ORF and a close-to-full-length second ORF lacking only a translation start site. The third example is the CYP71B35/CYP71B34 transcript that contains an incomplete first ORF and complete second ORF. RT-PCR amplifications across these combined loci indicate quite conclusively that these bicistronic transcripts exist in normal and stressed Arabidopsis tissues. Recent compilations of Drosophila cDNAs have identified a number of bicistronic transcripts (Misra et al. 2002) organized in a fashion similar to those that we have identified in Arabidopsis but it is as yet unclear the level to which these are expressed in vivo.
Whereas most eukaryotic transcripts are presumed to utilize their first AUG as a translation start site (Futterer and Hohn 1996), a growing number of examples exist of Drosophila, rat, and tomato bicistronic transcripts containing two complete ORFs that are translated variously by ribosome skipping and internal ribosome initiation mechanisms (Blumenthal 1998). Other examples exist of C. elegans polycistronic transcripts containing complete ORFs and N-terminally truncated ORFs that are translated after trans-splicing of translation start leader sequences onto the 5′ end of truncated ORFs (Blumenthal 1998). In an attempt to assess whether highly structured RNA duplexes characteristic of internal ribosome entry sites (IRES) exist in the longer intergenic regions of the combined CYP97C1/OMT locus and the combined CYP71B35/CYP71B34 locus, 298 of 413 bases in the IGR of CYP97C1/OMT and all bases in the IGR of CYP71B35/CYP71B34 have been modeled using M-fold programs (Zuker 2003). Both of these transcripts are predicted to fold into structures with multiple hairpins characteristic of IRES (not shown) that have potential to facilitate translation initiation on these downstream ORFs.
In several other surprising instances, transcripts from adjacent loci fuse ORFs at unpredictable splice sites to create monomeric transcripts containing functional domains for two functionally distinct types of proteins (e.g., P450 and PPR repeat proteins) or functionally redundant proteins (e.g., dimeric P450 proteins). While the sum of this evidence might suggest that pairs of adjacent loci in the single ORF group should be collapsed to single loci encoding these long monocistronic transcripts, alignments with ESTs and shorter cDNAs indicate that four second ORFs2 represented in these 31 pairs of adjacent loci are represented by one shorter monocistronic cDNA that exists as predicted for the second locus. Thus, like the bicistronic transcripts described previously, these reannotated monocistronic fusion transcripts confound the process of genome annotation. The long post-transcriptionally fused versions of their sequences, which might serve to decrease the complexity of annotated loci in the Arabidopsis genome by merging short annotated domains into longer ORFs3 and hypothetical open reading frames into recognizable proteins4, exist in conjunction with short independently transcribed versions of their sequences, which encode shorter monomeric proteins. RT-PCR analyses of several P450 loci have indicated that both long and short monocistronic transcripts derived from paired P450 loci coexist in RNA pools (i.e., they are not mutually exclusive). The new bicistronic and monocistronic models described here emphasize the need for future gene annotations to take into account the production of both bicistronic and monocistronic transcripts from some sets of “combined” loci and the production of alternate monocistronic transcripts from other sets of “combined” loci. Although helping to explain the transcription units of the most closely spaced genes, these alignments have now clouded definition of transcription units and genetic loci in the Arabidopsis genome. Future appreciations of microarray and oligonucleotide array data sets must take into account the fact that at least some loci are represented in both monocistronic and bicistronic transcripts capable of hybridizing with array elements designed to be specific for one or another locus. Importantly, these transcript models begin to explain the unusually high proportion (23%) of transcriptional units spanning IGR sequences recently detected in Arabidopsis whole genome arrays (Yamada et al. 2003). The extent to which these downstream ORFs are translated clearly merits further investigation so that the full complexity of the Arabidopsis proteome can be appreciated.
MATERIALS AND METHODS
Alignments
P450 ESTs and full-length cDNAs were identified by stand-alone BLAST (blastall) alignments (Altschul et al. 1990) of P450 loci with all ESTs available on the GenBank database (http://www.ncbi.nlm.nih.gov/dbEST/index.html) and full-length cDNAs available on the RIKEN (http://rarge.gsc.riken.go.jp/) and GenBank (http://www.ncbi.nlm.nih.gov/Genbank/index.html) databases. Clones that aligned with P450 loci were scanned for alignments with existing TAIR gene models and exceptional ones were searched for open reading frames containing the P450 signature motif FXXGXRXCXG that is conserved in most P450s.
Full-length cDNAs spanning at least two adjacent loci in the Arabidopsis genome were identified by stand-alone BLAST (blast-all) alignments of the current data set of genes from TAIR (ATH1.seq_cm_gz; ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/) with full-length cDNAs available from the RIKEN (http://rarge.gsc.riken.go.jp) and GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) databases. BLAST results were then processed to eliminate clones that aligned with nonadjacent loci and adjacent loci on opposite strands of the DNA. Finally, the set of transcripts derived from two adjacent loci on the same DNA strand was visually inspected and manually adjusted for splice junctions at the ends of BLAST alignments for adjacent exons.
Plant materials and RNA isolation
A. thaliana (Columbia ecotype) seeds were surface sterilized with 70% ethanol for 30 sec, 12% Clorox bleach for 20 min, and washed four times with sterile water prior to plating on half-strength MS-agar media (MS salts plus B5 vitamins; Sigma) (pH 5.7) supplemented with 1% sucrose. Seven-day-old seedlings were grown at a temperature of 21°C with a 16 h light (120 μmol sec−1 m−2 of cool-white fluorescent)/8 h dark cycle and harvested directly from these MS-agar plates. One-month-old plants were grown on these plates for 1 wk, transferred to soil, and grown for an additional 3 wk under the same temperature and light conditions. Individual tissues were harvested from 7-d-old seedlings and 1-mo-old plants, frozen in liquid nitrogen, and stored at −80°C. For drought stress, 7-d-old seedlings were removed from the agar and desiccated in plastic dishes at 22°C for 15 min. For cold treatment, plates with 7-d-old seedlings were transferred to a 4°C growth chamber for either 3 or 27 h. Cold-stressed RNAs from 3-and 27-h time points were combined for RT-PCR analysis.
For all tissue samples except 7-d-old roots, total RNA was isolated using TRIzol reagent (Invitrogen). For this, 1 g of tissue was ground in a mortar and pestle with 12 mL TRIzol, transferred to a 15-mL Falcon tube, extracted with 3 mL chloroform, incubated for 5 min at room temperature, and centrifuged at 5,000g at 4°C for 15 min. The supernatant was transferred to a fresh tube and nucleic acids were precipitated by adding a half volume of isopropanol and a half volume of 0.8 M sodium citrate, 1.2 M NaCl, gently inverting the mixture, incubating at room temperature for 10 min, and centrifuging at 10,000g at 4°C for 10 min. Nucleic acids were redissolved in 500 μL RNase-free water (Invitrogen), reprecipitated with one-tenth volume of 3 M sodium acetate (pH 5.0) and two volumes of ethanol for 20 min at −20°C, centrifuged at 13,000g for 10 min at 4°C, dried, resuspended in 300 μL sterile RNase-free water, and stored at −80°C.
For 7-d-old roots, total RNA was extracted from 200 mg of root tissue by grinding it for 1 min with a Beadbeater (Biospec Products) in 1 mL buffer containing equal volumes of phenol:chloroform (1:1) and 100 mM LiCl, 100 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% SDS. After centrifugation for 2 min in an Eppendorf centrifuge, the aqueous phase was re-extracted twice with an equal volume of phenol:chloroform (1:1) and the nucleic acids were precipitated by adding 0.1 volumes 3 M sodium acetate and 2 volumes ethanol. After precipitation, residual DNA was removed by treating each sample with 5 U RQ1 DNase (Promega) in the presence of 20 U RNasin and the manufacturer’s buffer for 60 min at 37°C, re-extracting with an equal volume of phenol:chloroform (1:1) and precipitating with 0.3 M sodium acetate and ethanol.
RT-PCR gel blot analysis
For RT-PCR gel blot analysis of most P450 transcripts, ~0.5–1.0 μg total RNA isolated from different tissues were used for one step RT-PCR amplification in 50 μL reactions containing 50 mM KCl, 10 mM Tris-HCl (pH 8.4), 200 μM each dNTP, 200 μg/mL gelatin, 40 pmol each primer, 4 U AMV reverse transcriptase (Promega), 20 U RNasin, and 2.5 U Taq polymerase (Gibco-BRL). First strand cDNAs were synthesized for 40 min at 42°C, and subsequently PCR amplified for 30 cycles with each cycle consisting of denaturation at 95°C for 1 min, annealing at 62°C for 1 min, and extension at 72°C for 2.5 min, followed by a final extension step of 72°C for 10 min. For the RT-PCR gel blot analysis of the CYP96A9 and CYP96A10 transcripts, 30 μg total RNA were reverse transcribed for 3.5 h at 42°C in a first-step 20-μL reaction containing 300 U Superscript II RNase H− reverse transcriptase (Invitrogen) and 30 pmol oligo (dT)17. Five microliters of these RT products were PCR amplified in a second-step 50-μL reaction containing 50 mM KCl, 10 mM Tris-HCl (pH 8.4), 200 μM each dNTP, 200 μg/mL gelatin, 40 pmol each gene-specific primer, and 2.5 U Taq polymerase (Gibco-BRL) for 35 cycles of denaturation at 94°C for 45 sec, annealing at 62°C for 45 sec, extension at 72°C for 2 min, with a final extension step 10 min at 72°C. The gene-specific primer sequences and their positions relative to the start codon (number in the parentheses) are as follows:
CYP71B34-5′ (71) tcggaggaattatcaacggac;
CYP71B34-3′ (515) acagttaaagccagcaatgtc;
CYP71B35-5′ (1479) CCAGAGATGAGAAGATCGTG;
CYP705A15-5′ (687) gcgaacatgttgcgtgcggg;
CYP705A15-3′ (1552) tcatgaactccggttcagtg;
CYP705A16-5′ (772) tggattcgacgagctactagag;
CYP705A16-3′ (1473) cgctatatttgttccaggacat;
CYP97C1-5′ (1402) ataacatccatcgttcttccg;
OMT-3′ (230) gctggtgagcatccgaagt;
CYP96A9-5′ (1104) GGTGTATTTACACGGCGCGG;
CYP96A10-5′ (−50) GCAATTTGGTGTATCTCAAAAGG; and
CYP96A10-3′ (420) CTCGGGATTCATCATCATGC.
The oligo(dT)17 primer was CGGAATTCTTTTTTTTTTTTTTTTT.
PCR products were fractionated on 1.0% agarose gels containing 1× Trisborate-EDTA (TBE) buffer, transferred to Hybond-N (Amersham Pharmacia-Biotech), and probed with random hexamer 32P-labeled probes corresponding to individual P450 cD-NAs. Blots were prehybridized in 50 mM Na2HPO4 (pH 7.2), 0.5% SDS, 5× SSC, 5× Denhardt’s, 50% formamide for at least 4 h at 42°C and hybridized for 12–16 h at 42°C with 32P-labeled probe added directly to the prehybridization solution. The blots were washed twice for 15 min at 42°C with 2× SSC, 0.1% SDS and twice for 15 min at 62°C with 0.2× SSC, 0.1% SDS and autoradiographed.
Acknowledgments
We thank Ms. Kathryn Palacio for help with RT-PCR analyses. This compilation was funded by National Science Foundation grant MCB 0115068.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7114505.
Footnotes
In two cases, both transcripts are RIKEN clones (RAFL07-15-G11 equivalent to RAFL05-14-C13; RAFL09-44-D01 equivalent to RAFL09-70-M08). In four cases, the two transcripts are RIKEN and non-RIKEN clones (RAFL09-60-B14 equivalent to AV528973 and AV523321 and AV530163 and AV524280; RAFL11-13-C20 equivalent to AY084340; RAFL15-09-A22 equivalent to AY087549; RAFL16-01-C03 equivalent to AV529139 and AV523459).
RAFL06-69-H17, RAFL14-80-M23, RAFL16-69-P16, RAFL21-17-J10.
Functions annotated for 13 combined loci are extended and the same as one or both of the previously annotated functions for RAFL06-69-H17, RAFL06-10-K24, RAFL07-12-J11, RAFL07-15-F13, RAFL07-15-G11, RAFL09-56-O16, RAFL19-84-B16, RAFL21-17-J10, RAFL05-02-O09, RAFL08-08-P22, RAFL09-60-B14, RAFL16-73-D01, and RAFL16-69-P16.
Functional annotations have now been made for five combined loci including RAFL08-17-N09 (nicastrin precursor), RAFL09-16-G02 (pyrrolodone carboxyl peptidase-like protein), RAFL15-41-F10 (ribulose-1,5-bisphosphate carboxylase large subunit N-methyltransferase), RAFL09-24-E16 (ZW18 protein), RAFL09-85-I15 (single-strand DNA endonuclease 1).
REFERENCES
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T. 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11: 132–136. [DOI] [PubMed] [Google Scholar]
- ———. 1998. Gene clusters and polycistronic transcription in eukaryotes. BioEssays. 20: 480–487. [DOI] [PubMed] [Google Scholar]
- Futterer, J. and Hohn, T. 1996. Translation in plants—Rules and exceptions. Plant Mol. Biol. 32: 159–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ríos, M., Fujita, T., LaRosa, P.C., Locy, R.D., Clithero, J.M., Bressan, R.A., and Csonka, L.N. 1997. Cloning of a polycistronic cDNA from tomato encoding γ-glutamyl kinase and γ-glutamyl phosphate reductase. Proc. Natl. Acad. Sci. 94: 8249–8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, S., Crosby, M.A., Mungall, C.J., Matthews, B.B., Campbell, K.S., Hradecky, P., Huang, Y., Kaminker, J.S., Millburn, G.H., Prochnik, S.E., et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 3: 0083.1–0083.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A.A. and Steitz, J.A. 2003. Splicing double: Insights from the second spliceosome. Nature 4: 960–970. [DOI] [PubMed] [Google Scholar]
- Pruitt, K., Tatusova, T., and Ostell, J. 2002. The Reference Sequence (RefSeq) Project, chapter 18. In The NCBI Handbook. National Library of Medicine, National Center for Biotechnology Information, Bethesda, MD. http://ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books
- Schuler, M.A. 1998. Plant pre-mRNA splicing. In A look beyond transcription: Mechanisms determining mRNA stability and translation in plants (eds. J.N. Bailey-Serres and D.R. Gallie), pp. 1–19. ASPP Publications, Rockville, MD.
- Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A., Akiyama, K., Oono, Y., et al. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G. and Filipowicz, W. 1996. Splicing of precursors to messenger RNA in higher plants: Mechanism, regulation and sub-nuclear organization of the spliceosomal machinery. Plant Mol. Biol. 32: 1–41. [DOI] [PubMed] [Google Scholar]
- Small, I.D. and Peeters, N. 2000. The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25: 46–47. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Lim, J., Dale, J.M., Chen, H., Shinn, P., Palm, C.J., Southwick, A.M., Wu, H.C., Kim, C., Nguyen, M., et al. 2003. Empirical analysis of transcriptional activity in the Arabidopsis gene. Science 302: 842–846. [DOI] [PubMed] [Google Scholar]
- Zhu, W. and Brendel, V. 2003. Identification, characterization and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome. Nucleic Acids Res. 31: 4561–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]