Abstract
Mutations that generate premature translation-termination codons (PTCs) often result in production of alternatively spliced mRNAs. While in many cases, the PTC-causing mutation was found to affect splicing directly by disrupting an exonic splicing enhancer, induction of alternative splicing of TCR-βpre-mRNA has been reported to be specific for mutations that prematurely terminate the open reading frame. During testing of a cyto-nuclear feedback model that would have explained how cytoplasmic translation could influence nuclear splicing of TCR-βtranscripts, control experiments questioned the frame dependence of the nonsense-associated altered splicing (NAS) of TCR-βpre-mRNA. A subsequent detailed analysis of alternatively spliced TCR-βmRNA expressed from different minigene constructs with nonsense, silent, or frame-shift mutations at various positions revealed no correlation between truncation of the reading frame and production of alternatively spliced mRNA. Our study thus contradicts the previously reported PTC specificity of TCR-βNAS and points out the need for systematically testing the PTC specificity in other cases where NAS has been observed.
Keywords: premature translation-termination codon (PTC), nonsense-associated altered splicing (NAS), exonic splicing enhancer (ESE)
INTRODUCTION
Messenger RNAs whose open reading frame (ORF) is prematurely terminated by a nonsense mutation are recognized and specifically degraded by a quality control system called nonsense-mediated mRNA decay (NMD) (Maquat 2004). In addition to triggering NMD, nonsense mutations are also frequently found to alter the splicing pattern of the nonsense codon-containing precursor mRNA (pre-mRNA), an observation that is referred to as nonsense-associated altered splicing (NAS) (Hentze and Kulozik 1999). In many cases, splice site selection is altered so that the sequence comprising the offending premature translation-termination codon (PTC) is skipped from the spliced mRNA (Li and Wilkinson 1998; Valentine 1998; Hentze and Kulozik 1999; Maquat 2001; Cartegni et al. 2002). If the alternatively spliced transcript remains in-frame, it will encode for a protein that may in some cases save the protein’s function, although an internal part of the amino acid sequence is altered or deleted.
The term NAS implies that the alternative splicing event is dependent on the presence of a PTC and thus of premature termination of the ORF. However, in most cases of NAS that were thoroughly investigated, it was found that not only nonsense, but also some silent or missense mutations cause alternative splicing, and that these mutations disrupted an exonic splicing enhancer (ESE) critical for exon inclusion (Liu et al. 2001; Buchner et al. 2003; Pagani et al. 2003). Thus, in these cases, alternative splicing is a direct effect of mutating a splicing signal and does not depend on premature termination of the ORF.
In contrast, evidence for a link between splice site selection and maintenance of an ORF within an mRNA has also been published. For example, Li and colleagues recently showed that latent 5′splice sites present in introns of the CAD and IDUA gene are efficiently used when all in-frame termination codons between the normally selected 5′splice site and the latent 5′splice site are removed (Li et al. 2002). In our opinion, the most compelling evidence so far for frame-dependent NAS has been reported by Wang and colleagues, who showed that PTCs located in the VDJ exon of TCR-βgenes up-regulate usage of an alternative splice acceptor and splice donor site, resulting in exclusion of the offending PTC from the alternatively spliced mRNA (altSA/SD-mRNA) (Wang et al. 2002a,b). Up-regulation of altSA/SD-mRNA was PTC specific, since it was not observed with missense or silent mutations, and since it was suppressed in genes with a frame shift when the ORF was restored by an additional, compensating frame-shift mutation (Wang et al. 2002b). Furthermore, up-regulation of altSA/SD-mRNA was inhibited by a stem-loop structure in the 5′UTR, by expression of suppressor tRNA or by removal of the initiator AUG codons, indicating the requirement for frame scanning (Wang et al. 2002a,b). Finally, NAS of PTC+ TCR-βtranscripts was shown to require the NMD factor hUpf1/rent1, but not hUpf2/rent2, suggesting that NAS relies on a component of the nonsense surveillance machinery, but is not an indirect consequence of NMD (Mendell et al. 2002; Wang et al. 2002a).
Wang and colleagues proposed a model for frame-dependent NAS, according to which the PTC in the mature mRNA is recognized by nuclear translation at the site of transcription, affecting the local concentration of splicing factors or nucleases involved in general pre-mRNA turnover, and thus altering the production of spliced mRNAs from newly synthesized precursors in trans (Wang et al. 2002a,b). An alternative model to explain frame-dependent NAS of TCR-βtranscripts that does not involve nuclear translation was proposed by Dahlberg et al. (2003). Their so-called “cyto-nuclear feedback model” postulates that a shuttling splicing factor could signal the nucleus that a cytoplasmic mRNA contained a PTC.
We set out to test the cyto-nuclear feedback model by coexpressing a PTC- and a PTC+ TCR-βminigene in HeLa cells and found that NAS was allele specific in our experimental system. But more importantly, control experiments suggested that NAS was not frame dependent in our feedback reporter system. This led us to more systematically investigate the issue of frame dependence of TCR-βNAS, including the attempt to reproduce some of the data published by Wang and colleagues. In contrast to their results, our data provide no evidence for frame-dependent up-regulation of TCR-βaltSA/SD-mRNA.
RESULTS
Splicing of PTC-free TCR-βpre-mRNA is not affected in cells coexpressing both a PTC- and a PTC+ TCR-βgene
According to the pmodel proposed by Dahlberg et al. (2003), splicing of wild-type (PTC-) TCR-βpre-mRNA should be affected by coexpression of its PTC+ counterpart, resulting in increased levels of alternatively spliced PTC- mRNA (PTC- alt-mRNA). To test this, we selected polyclonal pools of HeLa cells that constitutively expressed either PTC-, PTC+, or PTC- and PTC+ TCR-βminigenes together. To be able to distinguish PTC- from PTC+ transcripts in the subsequent RNase protection assay (RPA), a short sequence tag was inserted into the 3′UTR of one of the constructs. Normally and alternatively spliced TCR-βmRNA levels were analyzed by RPA using a probe complementary to the final exon (including the tag) and the 3′part of the VDJ exon (Fig. 1A). This experimental setup allowed us to determine the relative levels of all four TCR-βmRNA species at once. To avoid potential effects on mRNA levels caused by promoters competing for a limiting transcription factor, each cell pool was transfected with the same amount of the expression plasmid. Thus, for cells expressing PTC-or the PTC+ TCR-βmRNA alone, the respective TCR-βexpression plasmid was cotransfected with an identical amount of the same expression vector expressing an unrelated gene. In addition, a third plasmid encoding the β-globin gene (Thermann et al. 1998) was included in each trans-fection for normalization.
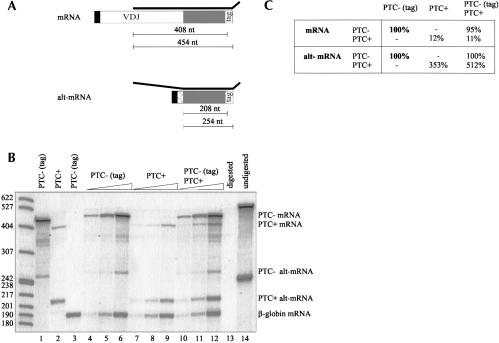
FIGURE 1.
TCR-βmRNA and alt-mRNA levels expressed from the wild-type allele are not affected upon coexpression with the nonsense allele. (A) Schematic illustration of the base pairing of the TCR-βprobe (thick black line) with the normally spliced (mRNA) and the alternatively spliced TCR-βtranscript (alt-mRNA). The leader exon (black box), the VDJ exon (white box), the alternative exon (diagonally striped box), and the final exon (gray box) of the TCR-βminigene are indicated. The alternative exon consists of the last 22 nt of the leader intron and the first 62 nt of the VDJ exon (Wang et al. 2002b). The box labeled with “tag” depicts the 46-nt-long tag inserted into to 3′UTR of the TCR-βminigene. The numbers below the illustrations indicate the lengths of the protected fragments for untagged and tagged mRNA and alt-mRNA, respectively. A T-to-A point mutation generates a PTC at amino acid 68 in construct “PTC+.” (B) RNase protection assay from total cellular RNA of HeLa cells stably expressing the indicated TCR-βconstructs. Lanes 4–12 contain both the TCR-βand the β-globin probe and increasing amounts of RNA (3, 9, and 27 μg). Lanes 1–3 are controls with only one probe (TCR-βin lanes 1 and 2, β-globin in lane 3) and 27 μg of the indicated RNA. Lanes 13 and 14 contain 9 μg RNA of untransfected HeLa cells, and no RNase was added in lane 14. (C) Quantification of the experiment shown in (B). Average values for TCR-βmRNA and alt-mRNA in each cell line were determined from lanes 4–6, 7–9, and 10–12, normalized to β-globin mRNA. Corrections to account for different amounts of radioactive label in protected fragments with different lengths were applied. mRNA and alt-mRNA levels from the cell line expressing only the wild-type allele (PTC- tag) were set to 100% (in bold).
As shown in Figure 1, B and C, the cells expressing the PTC+ TCR-βminigene produce about 3.5-fold more alt-mRNA compared to the cells expressing the PTC- construct (cf. lanes 7–9 and lanes 4–6). The cells expressing both constructs together produce fivefold more PTC+ than PTC- alt-mRNA (lanes 10–12). Thus, in contrast to the prediction made by the cyto-nuclear feedback model, alt-mRNA levels for the PTC- allele are not increased when the PTC+ TCR-βminigene is coexpressed. The difference in alt-mRNA levels between PTC+ and PTC- transcripts observed here is in the same range as reported previously from work with similar TCR-βconstructs (Wang et al. 2002a,b). In summary, this result is incompatible with a feedback mechanism that relies on a freely diffusible factor and shows that NAS is allele specific. On the other hand, the result would be consistent with the proposed nuclear scanning model, under the assumption that usage of these alternative splice sites was up-regulated as a result of truncation of the reading frame, as previously published (Wang et al. 2002b). However, allele-specific effects on splicing would also be observed if the nonsense mutation in the PTC+ construct had a direct effect on splicing, independent of truncating the ORF. It was therefore important to check for PTC specificity of TCR-βNAS in our experimental system.
NAS of the TCR-βfeedback reporter constructs is not PTC specific
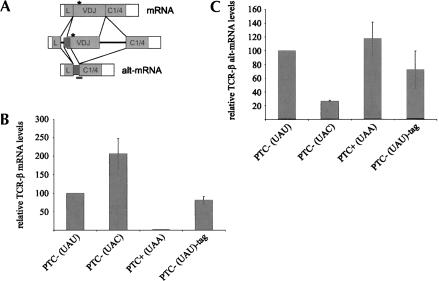
To test whether the increased levels of TCR-βalt-mRNA were specific for the T-to-A nonsense mutation at codon 68, we also introduced the silent T-to-C mutation at this position in the untagged TCR-βminigene [PTC- (UAC); Fig. 2]. The untagged wild-type [PTC- (UAU), PTC- (UAC), PTC+ (UAA) and the tagged wild-type [PTC- (UAU)-tag] constructs were transiently expressed in HeLa cells, and relative TCR-βmRNA and alt-mRNA levels were determined by real-time RT-PCR using splice variant-specific TaqMan probes. The probe to measure alt-mRNA spans the junction between the alternative splice donor site (altSD) in the VDJ exon and the 3′splice site of the final exon C1/4 (Fig. 2A); normally spliced mRNA was measured over the junction between the normal 5′splice site of the VDJ exon and the 3′splice site of the final exon (Buhler et al. 2002). A β-globin encoding plasmid (Thermann et al. 1998) was cotransfected for normalization. As shown in Figure 2B, the normally spliced PTC+ mRNA was reduced about 100-fold compared to the wild-type mRNA, irrespective of whether the wild-type mRNA was tagged or not. Surprisingly, the T-to-C mutation resulted in twofold more mRNA. The result of the alt-mRNA levels was unexpected in two respects. First, in contrast to the 3.5- to five-fold higher alt-mRNA levels from the PTC+ TCR-βminigene observed in the stably transfected cells (Fig. 1), this difference was less than twofold in the transient transfection experiment [Fig. 2C, cf. PTC+ (UAA) and PTC- (UAU)-tag], and compared to the untagged wild-type, there was no significant difference in alt-mRNA levels [cf. PTC+ (UAA) and PTC- (UAU)]. Secondly, the T-to-C mutation resulted in a fourfold reduction of alt-mRNA, suggesting that this mutation might have strengthened VDJ exon definition and thus promotes usage of the normal splice sites (twofold elevated mRNA; see above) and inhibits alt-mRNA production. Consistent with this possibility, analysis of the sequence comprising codon 68 with ESE-RESCUE (http://genes.mit.edu/chris/), a computational tool to predict exonic splicing enhancers (Fairbrother et al. 2002, 2004), found no ESE in the wild-type (UAU) and in the PTC+ (UAA) sequence, but predicted an ESE (CGTCGC, mutated base underlined) in the PTC-(UAC) construct. In summary, these results indicated that mutations of the third position of codon 68 in our TCR-βminigene did not alter splicing by a termination codon-dependent mechanism.
FIGURE 2.
High alt-mRNA levels of the TCR-βfeedback reporter constructs do not correlate with the presence of a PTC. (A) Schematic representation of the two major transcripts produced from the TCR-βpre-mRNA by alternative splicing. Exonic and intronic sequences are shown as boxes and lines, respectively. The dark box depicts the alternative exon, which comprises the last 22 nt of the leader intron and the first 62 nt of the VDJ exon. Alt-mRNA is produced by usage of the same alternative splice acceptor and splice donor sites as altSA/SD-mRNA (Wang et al. 2002a,b), but our minigenes contain as terminal exon a fusion between exons C1 and C4 instead of the full-length constant region of the TCR-βgene (Li et al. 1997) and a shortened JC intron (Stoecklin et al. 2001). The position of codon 68 is depicted by an asterisk, and the position of the TaqMan probe used to measure alt-mRNA is indicated by the short black line. (B,C) HeLa cells were cotransfected with plasmids expressing the indicated TCR-βminigenes and a plasmid expressing β-globin mRNA (Thermann et al. 1998), and relative levels of normally spliced (B) and alternatively spliced (C) TCR-βmRNA and β-globin mRNA were determined by real-time RT-PCR using TaqMan chemistry as described previously (Buhler et al. 2004). Relative TCR-βmRNA and alt-mRNA levels were normalized to the relative β-globin mRNA levels, and PTC- (UAU) was set as 100%. Average values and standard deviations of three PCR runs from one typical experiment are shown.
TCR-βalt-mRNA levels of constructs with frame shifts at codon 58 do not correlate with truncation of the ORF
Because we did not find evidence for PTC-specific up-regulation of alt-mRNA at codon 68, we decided to more systematically investigate the question of frame dependence of TCR-βNAS. To this end, we generated a set of frame-shift mutants by inserting or deleting one, two, or three nucleotides in a TCR-βminigene, giving constructs FS + 1, FS + 2, FS + 3 or FS − 1, FS − 2, FS − 3, respectively (Fig. 3A). No ESE is predicted in this region for any of the constructs by ESE-RESCUE (data not shown). The structure of these TCR-βminigenes is almost identical to the constructs used in Figures 1 and 2, except that the intron in the 5′UTR was deleted (see Materials and Methods). The frame-shift constructs were transiently expressed in HeLa cells and the relative levels of TCR-βmRNA and alt-mRNA were measured by real-time RT-PCR. As expected, in cells expressing the PTC-, in-frame constructs FS - 3 and FS + 3, TCR-βmRNA levels were 15- to 40-fold higher than in cells expressing frame-shift constructs with a PTC (Fig. 3B), reflecting the highly efficient NMD for TCR-βtranscripts. The highest level of alt-mRNA was obtained with the PTC+ construct FS - 1. If this increased alternative splicing were due to the premature termination of the ORF at codon 62, one would expect the same result with construct FS + 2, which has the same PTC. However, FS + 2 showed the lowest alt-mRNA level of all FS constructs. In fact, the second construct with an elevated alt-mRNA level was FS + 3, which has no PTC, while the other four constructs show relatively low alt-mRNA levels. In summary, we did not find any correlation between the relative amount of alt-mRNA and the presence or absence of a PTC in this set of constructs. We conclude that also in this second set of TCR-βconstructs, alt-mRNA production is not frame dependent.
FIGURE 3.
Testing a set of frame-shift TCR-β minigene constructs provides no evidence for frame-dependent up-regulation of alt-mRNA levels. (A) Sequence of the frame-shift constructs around codon 58 (GGG). Deletions are indicated with a β; inserted nucleotides are bold. On the right, the amino acid position at which the frame-shift mutation generates a PTC is indicated. Relative levels of TCR-β mRNA (B) and alt-mRNA (C) from transiently transfected HeLa cells were measured by real-time RT-PCR as described in Figure 2. The data is normalized to the relative mRNA levels of the neomycin resistance gene, which is encoded on the TCR-β expression plasmid, and the mRNA and alt-mRNA levels of construct FS + 3 were set as 100%. Average values and standard deviations from three PCR runs on cDNA from two independent transfections are shown.
No evidence for frame-dependent NAS with a set of TCR-βminigenes used by Wang and colleagues
In the light of the previously published evidence for frame-dependent NAS of TCR-βpre-mRNA, it was puzzling that both of our sets of TCR-βminigenes used to address this issue did not show any evidence for frame-dependent NAS. To further investigate this discrepancy, Miles Wilkinson kindly provided some of their TCR-β constructs that were reported to up-regulate alt-mRNA specifically in response to a PTC (Wang et al. 2002b). For easier comparison, the constructs are labeled with the same letters as in the previous publication (Wang et al. 2002b). Sequencing of the region between the translation initiator codon and the PTC in the C1 exon confirmed the identity of each construct (data not shown). Construct L (Wang et al. 2002b) is not informative and was not used, because the deletion of one base pair in the C1 exon shifts the alt-mRNA out of frame, which leads to degradation of this alt-mRNA by NMD. Constructs A, K, N, and M were transiently expressed in HeLa cells and relative levels of TCR-β mRNA and alt-mRNA were determined by real-time RT-PCR as before and normalized to the relative mRNA levels of the neomycin resistance gene, which is encoded on the same plasmid. The result of the mRNA levels was as expected, with 50- to 65-fold less mRNA from the PTC-containing construct K compared to the PTC- constructs K, N, and M (Fig. 4B). However, our result for the alt-mRNA levels is different from what has been reported by Wang et al. (2002b). Instead of the expected sevenfold up-regulation of alt-mRNA in response to a PTC, alt-mRNA increased only twofold with construct K in our experiments. And most importantly, this moderate up-regulation of alt-mRNA was not PTC specific, because the PTC- construct N also resulted in a threefold increased alt-mRNA level compared to construct A (Fig. 4C). Clearly, this result was contradictory to the result obtained by Wang and colleagues.
FIGURE 4.
No evidence for frame-dependent NAS with a set of TCR-β minigenes used by Wang and colleagues. (A) Schematic representation of the TCR-β minigenes used in these experiments. For easier comparison, the constructs were labeled as in the previous publication (Wang et al. 2002b). Construct K has an insertion of 10 base pairs (GCCCTCGAGG) at codon 83 of the parental TCR-β minigene A, introducing a frame-shift that generates a PTC at codon 151 in the C1 exon. Construct N has an almost identical insertion of nine base pairs (GCCCTCGAG) at codon 83 and remains in frame. Construct M is identical to construct K, except that the ORF was restored by deletion of one base pair of codon 133. After transient transfection of these constructs into HeLa cells, relative levels of TCR-β mRNA (B) and alt-mRNA (C) were determined by real-time RT-PCR and normalized to the relative mRNA levels of the neomycin resistance gene. Average values and standard deviations from four PCR runs of one typical experiment are shown, and the mRNA and alt-mRNA levels of construct A were set as 100%. (D,E) relative TCR-β mRNA and alt-mRNA levels of constructs A, K, N, and M in HEK293T and HT1080 cells. For normalization, a β-globin mRNA expressing plasmid was cotransfected. Average values and standard deviations from three PCR runs of one typical experiment are shown, and the mRNA and alt-mRNA levels of construct A were set as 100%.
We speculated that the different results could be due to differences in the cell lines used for the experiments, al though HeLa cells were used in both cases. To test if frame-dependent NAS could be observed in other cells, we repeated our experiment using the human embryonic kidney cell line HEK293T and the human fibrosarcoma cell line HT1080 (Fig. 4D,E). The relative TCR-β alt-mRNA levels measured in these cell lines after transfection was similar to the results obtained in HeLa cells (Fig. 4, cf. C and E) and showed again no evidence for PTC-specific up-regulation of alt-mRNA. In both cell lines, the alt-mRNA levels of construct K did not differ significantly from those of construct A, and the highest relative levels of alt-mRNA was obtained with the PTC- construct N.
Collectively, our data from transient expression of these TCR-β minigene constructs in three different human cell lines indicate that the level of alt-mRNA does not alter specifically in response to premature termination of the ORF.
Expression of TCR-β constructs integrated into the genome confirms data from transient transfections
In search of possible explanations for the discrepancy between our data and the data reported by Wang and colleagues, we also considered the possibility that high expression levels of the test genes could saturate the putative mechanism for frame-dependent NAS. From studying NMD of TCR-β minigenes, we know that transfecting the cells with large quantities of plasmid DNA can easily exhaust the capacity of the NMD system (O. Mühlemann, unpubl. observation). Although all transient transfection experiments shown here were done under conditions that do not saturate NMD (i.e., transfection of relatively small amounts of plasmid DNA), as can be seen by the efficient down-regulation of PTC+ mRNA levels (Figs. 2, 3, 4), the expression levels might still saturate the putative frame-dependent NAS mechanisms. Consistent with this idea, we observed 3.5- to 5-fold more alt-mRNA from the PTC+ feedback reporter construct compared to the PTC- construct when these constructs were stably transfected (Fig. 1), whereas this difference was less than twofold when the same constructs were transiently transfected (Fig. 2), which gives a higher rate of gene expression than stable transfections. Therefore, we selected polyclonal populations of HeLa cells that had integrated into the genome constructs A, K, N, or M and measured relative TCR-β mRNA and alt-mRNA levels as before. As shown in Figure 5A, the levels of normally spliced mRNA were the same as in the transient trans-fection (Fig. 4B), confirming that NMD is fully active under both conditions. Likewise, there was no significant difference in alt-mRNA levels between stable (Fig. 5B) and transient (Fig. 4C) transfections. Again, the PTC+ construct K gave only marginally more alt-mRNA than construct A, while the PTC- construct N showed twofold more alt-mRNA, which is inconsistent with frame-dependent regulation of alt-mRNA production.
FIGURE 5.
Expression of TCR-β constructs integrated into the ge-nome reveals no PTC-specific regulation of alt-mRNA levels. Poly-clonal populations of HeLa cells stably expressing constructs A, K, N, or M were selected by treating the cells with G418 for 22 d. Relative TCR-β mRNA (A) and alt-mRNA (B) levels were determined from total cellular RNA by real-time RT-PCR and normalized to the relative mRNA levels of the neomycin resistance gene. Average values and standard deviations from three duplicate PCR runs of one RNA preparation are shown.
Confirmation of real-time RT-PCR data by RNase protection assays
Finally, we wanted to rule out that the different results obtained by us and by the Wilkinson laboratory could be due to the different methods used to quantify RNA levels. Although there was no reason to assume that the RPA used by the Wilkinson laboratory or our real-time RT-PCR assay would produce incorrect results, it should be noted that our TaqMan assay for alt-mRNA will measure all alternatively spliced transcripts that have altSD spliced to the 3′splice site of the C1 exon, while the RPA probe allows specific quantification of the altSA/SD-mRNA. In fact, an alternative TCR-β transcript in which altSD is spliced to the 3′ splice site of the C1 exon, but which uses the normal 3′splice site of the VDJ exon instead of altSA has been described (Wang et al. 2002a). However, this splice form has a PTC and hence accumulates only at minute amounts in NMD-competent cells (Wang et al. 2002a). Thus, it should influence our real-time RT-PCR measurements of alt-mRNA only marginally.
To directly compare our real-time RT-PCR data with RPA data, we checked our RNA samples from the cells stably expressing constructs A, K, N, and M by RPA. The probe used for the RPAs and the expected size of the protected fragments for normally spliced mRNA, altSA/SD-mRNA, altSD-mRNA, and premRNA are shown in Figure 6A. Initial titration of probe and RNA ensured that our RPAs were performed with an excess of probe (data not shown), which is important to obtain quantitative results. As shown in Figure 6B, the RPAs confirmed the data obtained by real-time RT-PCR. Again, we could not observe a PTC-specific up-regulation of altSA/SD-mRNA. The level of altSA/SD-mRNA from construct K (PTC+) was only slightly higher than with construct A (wildtype PTC-). As in all experiments before, the highest level of altSA/SD- mRNA was obtained with construct N. Notably, a faint band of the size expected for altSD-mRNA could also be detected, and comparison of its intensity with the intensity of the band corresponding to altSA/SD-mRNA confirms that this minor splice form does not contribute significantly to our alt-mRNA levels determined by real-time RT-PCR. In conclusion, analysis of our RNA by RPA gives results very similar to analysis by real-time RT-PCR, excluding the possibility that the discrepancy between our results and the results reported by Wang and colleagues could be due to the different methods used.
FIGURE 6.
Confirmation of real-time RT-PCR results by RNase protection assays. (A) The RPA probe used in (B) is complementary to the last 58 nt of the leader intron and the first 154 nt of the VDJ exon of TCR-β pre-mRNA. The relevant splice sites and the alternative exon included in altSA/SD-mRNA are indicated, and the predicted sizes of protected fragments corresponding to the differently spliced TCR-β transcripts are listed. (B) Twenty-seven micrograms of total cellular RNA from cells expressing the indicated constructs were hybridized with 15 fmol 32P-labeled TCR-β and 15 fmol 32P-labeled neo probe (lanes 3–8) or with each probe alone as a control (lanes 1,2). RNA from untransfected HeLa cells was used as a specificity control of the assay (lane 7) and to show the probes when no RNases were added (lane 8). Protected fragments after RNase digestion were separated on a denaturing polyacrylamide gel and quantified by PhosphorImager scanning. The fragment sizes of the 32P-labeled DNA ladder are indicated on the left; the positions of the bands corresponding to the different TCR-β transcripts and to neo mRNA are shown on the right. The identity of the TCR-β-specific band at about 100 nt is not known. Relative amounts of TCR-β mRNA and altSA/SD-mRNA in lanes 3–6, normalized to neo mRNA and corrected for the number of 32P-labeled nucleotides, are shown below the gel.
DISCUSSION
At the outset of the study reported here, we wanted to test the cyto-nuclear feedback model (Dahlberg et al. 2003), which was proposed as an alternative to models that postulated the involvement of nuclear ORF scanning to explain the reported PTC-specific up-regulation of an alternatively spliced TCR-β transcript (Wang et al. 2002b). In contrast to the prediction made by the cyto-nuclear feedback model, the level of PTC- TCR-β alt-mRNA did not increase when the PTC+ TCR-β construct was coexpressed in the same cells, demonstrating that NAS in this experimental system was allele specific. While at first sight, this result appeared to support the proposed nuclear scanning models, we noted that allele-specific effects on splicing would also be observed if the nonsense mutation in the PTC+ construct had a direct effect on splicing, independent of truncating the ORF. It was therefore crucial to investigate whether TCR-β NAS was PTC specific in our experimental system.
Here, our study took an unexpected turn. All our experiments that systematically addressed the issue of frame-dependent up-regulation of TCR-β alt-mRNA indicated that there was no correlation between the presence of a PTC in a TCR-β construct and an elevated level of alt-mRNA. Even with the same constructs used previously to demonstrate the PTC specificity of TCR-β NAS, we could not find any evidence for frame-dependent production of alt-mRNA. In both sets of TCR-β minigenes designed to distinguish between frame-dependent NAS and direct effects of the mutations on splicing, there was one PTC- construct that yielded higher levels of alt-mRNA than the PTC+ constructs. Based on our results, we conclude that the different levels of TCR-β alt-mRNA observed with different TCR-β minigenes is unrelated to the presence of a PTC in these genes, but that instead the sequence alterations generated by point mutations or small insertions/deletions directly affect splice site selection. The reason for the discrepancy between our data and the data of Wang and colleagues remains elusive.
In addition to TCR-β, NAS has been observed in many other genes, but only in a few cases has it been systematically investigated whether it is frame dependent. In addition to the examples mentioned in the Introduction, we have recently identified an alternatively spliced immunoglobulin μ transcript in cells expressing Ig-μ minigenes with PTCs in the VDJ exon. Systematic analysis of different mutations at two positions in the Ig-μ minigene clearly showed that production of this alternative transcript was not PTC specific. Instead, the PTC-causing mutations happened to interfere with splicing directly (Buhler and Muhlemann 2005). Collectively, our data from the TCR-β and the Ig-μ minigene systems provide strong evidence against the intellectually appealing hypothesis that cells might possess mechanisms that would allow regulation of splice site selection in response to premature termination of the ORF.
Because it is quite difficult to unambiguously demonstrate frame dependency of NAS and rule out that the altered splicing was caused by mutation of splicing signals, it would be important to carefully reexamine the cases for which NAS has been claimed to be PTC specific before embarking on further projects that build on PTC specificity of the altered splicing. The fibrillin-1 gene is an example where such a reexamination of apparently PTC-specific NAS (Dietz et al. 1993; Dietz and Kendzior 1994) has revealed that the nonsense mutations disrupted an ESE (Caputi et al. 2002).
From the point of splicing regulation, our data fit well into a growing list of examples indicating that exon definition and splice site selection involve many cis-acting signals distributed across the pre-mRNA (Maniatis and Tasic 2002; Zheng 2004). These signals together constitute a still poorly understood second layer of information encoded by the RNA sequence, which determines the fate of the RNA. As for the ORF, the first layer of information, single nucleotide changes in many cases are apparently sufficient to alter this second layer information, resulting for example in altered splicing of the RNA.
MATERIALS AND METHODS
Plasmids
The TCR-β feedback reporter genes used in Figures 1 and 2 are based on constructs A (pβ333) and C (pβ337, PTC at amino acid position 68) described by Li et al. (1997), except that the JC intron was shortened to 357 nt by deletion of a 634-nt-long Eco0109I fragment. The T-to-C mutation in construct PTC- (UAC) was generated with the QuikChange Site-Direced Mutagenesis Kit (Stratagene). A 46-nt-long sequence tag (5′-GATCCCATTCGC CATTCAGGCTGCGCAACTGTTGGGAAGGGCGATG-3′) was inserted into the BamH1 site in the 3′UTR of constructs PTC+ (UAA) and PTC- (UAU) using a double-stranded oligo. To generate pBS-β433ΔJC-tag, the template for the TCR-β probe used for the RPA in Figure 1, the construct PTC- (UAU), from which the entire JC intron was deleted by fusion PCR, was cloned into Sal1–BamH1 of pBluescript II (Stratagene). To generate pBS-βglobin-exon2, the template for the β-globin probe, the Acc1–BamH1 fragment of pClneoβglobinwt, comprising β-globin exon 2, was cloned into Acc1–BamH1 of pBluescript II.
The frame-shift constructs used in Figure 3 are based on a modified version of the wild-type feedback reporter construct PTC- (UAU), in which the β-actin intron in the 5′UTR was deleted by replacing the Not1–Sal1 fragment with a double-stranded oligo (5′-GGCCGCGGCGGCGCCCTATAAAACCCA GCGGCGCGACGCGCCACCACCGCCGAGACCGGTACCG-3′), which shortened the 5′UTR to 61 nt. One, two, and three base pair insertions and deletions were generated generated with the QuikChange Site-Direced Mutagenesis Kit (Stratagene).
Plasmids encoding constructs A (pβ290), K (pβ291), N (pβ589), and M (pβ625) were obtained from Miles Wilkinson and are described elsewhere (Muhlemann et al. 2001). pBS-β433VDJ, the template for the RPA probe used in Figure 6, was generated by deleting the sequence between Nde1 and Not1 in pBS-β433-tag. pBS-neo, the template for the RPA probe detecting neomycin mRNA, was generated by inserting the neomycin resistance cassette of MSCVneoEB (Hawley et al. 1994) into Xho1–BamH1 of pBluescript II (Stratagene).
Plasmid sequences and sequences of the primers used for site-directed mutagenesis are available upon request.
Cell culture, transfection, and selection of stably transfected polyclonal cell pools
HeLa, HEK293T, and HT1080 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen), supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U/mL penicillin, and 100 μg/mL streptomycin (Amimed). Cells were trans-fected in six-well plates at about 60%–80% confluency using li-pofectamine (Invitrogen). For transient transfections 100–150 ng of TCR-β expressing plasmid DNA was used (Figs. 3, 4B,C) or 50 ng of TCR-β plasmid plus 50 ng of pClneoβglobinwt (Neu-Yilik et al. 2001; Figs. 2, 4D,E). To generate the cell lines that stably coexpressed the tagged PTC- (pβ433-tag) and the PTC+ (pβ434) TCR-β minigenes, 200 ng of each plasmid were transfected together with 100 ng pClneoβglobinwt. In parallel, 200 ng of pβ433-tag or pβ434 were transfected together with 200 ng pβhyb8-(Buhler et al. 2004) and 100 ng pClneoβglobinwt to generate cell lines that express only one allele of TCR-β. For generating the stably transfected cell lines used in Figures 5 and 6, 500 ng of pβ290, pβ291, pβ589, or pβ625 were used. Polyclonal populations of stably transfected HeLa cells were obtained by selection with 500 μg/ml G418 (Roche) for 2–3 weeks.
RNA isolation
Total cellular RNA was purified using the Absolutely RNA RT-PCR Miniprep Kit according to the manufacturer’s protocol (Stratagene).
RNase protection assay
The TCR-β probe illustrated in Figure 1A comprises the last 199 nt of the VDJ exon and the final exon until the end of the tag sequence plus an unspecific 5′-part originating from pBluescript II. The probe for detecting β-globin mRNA is complementary to the AvaII–BamHI fragment of exon 2 in pClneoβglobinwt and contains an unspecific 5′part originating from pBluescript II. The 32P-labeled probes were generated by in vitro transcription using T7 RNA polymerase and Nde1-digested pBS-β433ΔJC-tag or AvaII-digested pBS-βglobin-exon2 as template, respectively. Nineteen femtomoles of each gel-purified probe were hybridized with 3, 9, and 27 μg of total cellular RNA, digested with RNases A and T1, and separated on a denaturing polyacrylamide gel as previously described (Suter et al. 1999).
The TCR-β probe illustrated in Figure 6A comprises the last 58 nt of the leader intron and the first 154 nt of the VDJ exon plus an unspecific 5′part originating from pBluescript II. The 32P-labeled probe was generated by T7 in vitro transcription from the EcoR1-digested pBS-β433VDJ. The neo probe was generated by T7 in vitro transcription from the Sap1-digested pBS-neo. Fifteen fem-tomoles of each gel-purified probe were hybridized with 27 μg of total cellular RNA digested with RNases and analyzed as described above. Gels were exposed to PhosphorImager screens, scanned with a Storm 820 instrument (Amersham Biosciences), and quantified with Aida software. The relative TCR-β mRNA and alt-mRNA levels were normalized to the levels of β globin mRNA (Fig. 1) or neo mRNA (Fig. 6) and corrected for the different numbers of 32P-labeled nucleotides in TCR-β mRNA and alt-mRNA to allow comparisons between the relative amounts of these two RNA species.
Real-time RT-PCR
Reverse transcription and real-time PCR was as described by Buhler et al. (2004). TCR-β alt-mRNA was measured using 800 nM om120 (5′-CACCCAAAGTCCAAGAAGCAA-3′), 800 nM om121 (5′-GAGACCTTGGGTGGAGTCACA-3′), and 200 nM of TM120/121 (5′-FAM-TCTCAGATCCTCTTTCCTCCTGTTACT GCCA-TAMRA-3′). β-Globin mRNA was measured using 800 nM om111 (5′-GCTGCACTGTGACAAGCTGC-3′), 800 nM om112 (5′-AAAGTGATGGGCCAGCACAC-3′), and 200 nM TM111/112 (5′-FAM-TCCTGAGAACTTCAGGCTCCTGGGCA AC-TAMRA-3′).
Neomycin mRNA was measured using 800 nM om101 (5′-TGTGACATAATTGGACAAACTACCTACA-3′), 800 nM om102 (5′-CATTCCACCACTGCTCCCA-3′), and 200 nM TM101/102 (5′-FAM-AGATTTAAAGCTCTAAGATTCCAACCTATGGAACT GATGA-TAMRA-3′). Primers and TaqMan probe to measure TCR-β mRNA are described by Buhler et al. (2002). Real-time PCR reactions were run on the GeneAmp 5700 or 7000 Sequence Detection System (Applied Biosystems) using the standard thermal profile.
Acknowledgments
We are grateful to Miles Wilkinson for providing plasmids and for stimulating discussions. This work was financed by the Swiss National Research Foundation (grants 3100-61720.00 and 3100A0-102159), the Kanton Bern, the Novartis Foundation for Biomedical Research, and the Helmut Horten Foundation.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7182905.
REFERENCES
- Buchner, D.A., Trudeau, M., and Meisler, M.H. 2003. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science 301: 967–969. [DOI] [PubMed] [Google Scholar]
- Buhler, M. and Muhlemann, O. 2005. Alternative splicing induced by nonsense mutations in the immunoglobulin μ VDJ exon is independent of truncation of the open reading frame. RNA (this issue) [DOI] [PMC free article] [PubMed]
- Buhler, M., Wilkinson, M.F., and Muhlemann, O. 2002. Intranuclear degradation of nonsense codon-containing mRNA. EMBO Rep. 3:646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler, M., Paillusson, A., and Muhlemann, O. 2004. Efficient down-regulation of immunoglobulin mu mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res. 32: 3304–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi, M., Kendzior Jr., R.J, and Beemon, K.L. 2002. A nonsense mutation in the fibrillin-1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes & Dev. 16: 1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285–298. [DOI] [PubMed] [Google Scholar]
- Dahlberg, J.E., Lund, E., and Goodwin, E.B. 2003. Nuclear translation: What is the evidence? RNA 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz, H.C. and Kendzior Jr., R.J. 1994. Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat. Genet. 8: 183–188. [DOI] [PubMed] [Google Scholar]
- Dietz, H.C., Valle, D., Francomano, C.A., Kendzior Jr., R.J., Pyeritz, R.E., and Cutting, G.R. 1993. The skipping of constitutive exons in vivo induced by nonsense mutations. Science 259: 680–683. [DOI] [PubMed] [Google Scholar]
- Fairbrother, W.G., Yeh, R.F., Sharp, P.A., and Burge, C.B. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Fairbrother, W.G., Yeo, G.W., Yeh, R., Goldstein, P., Mawson, M., Sharp, P.A., and Burge, C.B. 2004. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 32: W187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, R.G., Lieu, F.H., Fong, A.Z., and Hawley, T.S. 1994. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1:136–138. [PubMed] [Google Scholar]
- Hentze, M.W. and Kulozik, A.E. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96: 307–310. [DOI] [PubMed] [Google Scholar]
- Li, S. and Wilkinson, M.F. 1998. Nonsense surveillance in lymphocytes? Immunity 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Li, S., Leonard, D., and Wilkinson MF. 1997. T cell receptor (TCR) minigene mRNA expression regulated by nonsense codons: A nuclear-associated translation-like mechanism J. Exp. Med. 185: 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Wachtel, C., Miriami, E., Yahalom, G., Friedlander, G., Sharon, G., Sperling, R., and Sperling, J. 2002. Stop codons affect 5′splice site selection by surveillance of splicing. Proc. Natl. Acad. Sci. 99: 5277–5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.X., Cartegni, L., Zhang, M.Q., and Krainer, A.R. 2001. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat. Genet. 27: 55–58. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. and Tasic, B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236–243. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. 2001. The power of point mutations. Nat. Genet. 27: 5–6. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5: 89–99. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T., ap Rhys, C.M., and Dietz, H.C. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419–422. [DOI] [PubMed] [Google Scholar]
- Muhlemann, O., Mock-Casagrande, C.S., Wang, J., Li, S., Custodio, N., Carmo-Fonseca, M., Wilkinson, M.F., and Moore, M.J. 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell 8: 33–43. [DOI] [PubMed] [Google Scholar]
- Neu-Yilik, G., Gehring, N.H., Thermann, R., Frede, U., Hentze, M.W., and Kulozik, A.E. 2001. Splicing and 3′end formation in the definition of nonsense-mediated decay-competent human β-globin mRNPs. EMBO J. 20: 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani, F., Buratti, E., Stuani, C., and Baralle, F.E. 2003. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9.J. Biol. Chem. 278: 26580–26588. [DOI] [PubMed] [Google Scholar]
- Stoecklin, G., Stoeckle, P., Lu, M., Muehlemann, O., and Moroni, C. 2001. Cellular mutants define a common mRNA degradation pathway targeting cytokine AUrich elements. RNA 7: 1578–1588. [PMC free article] [PubMed] [Google Scholar]
- Suter, D., Tomasini, R., Reber, U., Gorman, L., Kole, R., and Schumperli, D. 1999. Doubletarget antisense U7 snRNAs promote efficient skipping of an aberrant exon in three human β-thalassemic mutations. Hum. Mol. Genet. 8: 2415–2423. [DOI] [PubMed] [Google Scholar]
- Thermann, R., Neu-Yilik, G., Deters, A., Frede, U., Wehr, K., Hagemeier, C., Hentze, M.W., and Kulozik, A.E. 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 17: 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine, C.R. 1998. The association of nonsense codons with exon skipping. Mutat. Res. 411: 87–117. [DOI] [PubMed] [Google Scholar]
- Wang, J., Chang, Y.F., Hamilton, J.I., and Wilkinson, M.F. 2002a. Nonsense-associated altered splicing: A frame-dependent response distinct from nonsense-mediated decay. Mol. Cell 10: 951–957. [DOI] [PubMed] [Google Scholar]
- Wang, J., Hamilton, J.I., Carter, M.S., Li, S., and Wilkinson, M.F. 2002b. Alternatively spliced TCR mRNA induced by disruption of reading frame. Science 297: 108–110. [DOI] [PubMed] [Google Scholar]
- Zheng, Z.M. 2004. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 11: 278–294. [DOI] [PMC free article] [PubMed] [Google Scholar]