Abstract
A recent re-examination of the role of the helices surrounding the conserved core of the hammerhead ribozyme has identified putative loop–loop interactions between stems I and II in native hammerhead sequences. These extended hammerhead sequences are more active at low concentrations of divalent cations than are minimal hammerheads. The loop–loop interactions are proposed to stabilize a more active conformation of the conserved core. Here, a kinetic and thermodynamic characterization of an extended hammerhead sequence derived from Schistosoma mansoni is performed. Biphasic kinetics are observed, suggesting the presence of at least two conformers, one cleaving with a fast rate and the other with a slow rate. Replacing loop II with a poly(U) sequence designed to eliminate the interaction between the two loops results in greatly diminished activity, suggesting that the loop–loop interactions do aid in forming a more active conformation. Previous studies with minimal hammerheads have shown deleterious effects of Rp-phosphorothioate substitutions at the cleavage site and 5′ to A9, both of which could be rescued with Cd2+. Here, phosphorothioate modifications at the cleavage site and 5′ to A9 were made in the schistosome-derived sequence. In Mg2+, both phosphorothioate substitutions decreased the overall fraction cleaved without significantly affecting the observed rate of cleavage. The addition of Cd2+ rescued cleavage in both cases, suggesting that these are still putative metal binding sites in this native sequence.
Keywords: RNA catalysis, hammerhead, ribozyme, metal ions, RNA ligation
INTRODUCTION
Since their discovery by Cech and Altman, ribozymes have been dissected to find their minimal active form and to determine their catalytic mechanisms (Doudna and Cech 2002). Of the nucleolytic ribozymes, the hammerhead ribozyme (HHRz) is the smallest member (Takagi et al. 2001; DeRose 2002; Lilley 2003). The HHRz performs a reversible site-specific phosphodiester bond cleavage reaction that results in a 5′-OH and a 2′, 3′-cyclic phosphate. Originally found in several single-stranded circular plant pathogenic RNAs, including the tobacco ringspot virus satellite RNA, the lucerne transient streak virus, and the avocado sun-blotch viroid (Hutchins et al. 1986; Prody et al. 1986; Forster and Symons 1987), HHRzs have also been found in cave crickets (Rojas et al. 2000), salamanders (Zhang and Epstein 1996), and schistosomes (Ferbeyre et al. 1998). Hammerhead and hammerhead-like sequences have also been found in a variety of other genomes (Ferbeyre et al. 2000), and in vitro evolution studies indicate that these highly divergent organisms may have settled on this small ribozyme as the simplest solution to a biochemical problem (Salehi-Ashtiani and Szostak 2001).
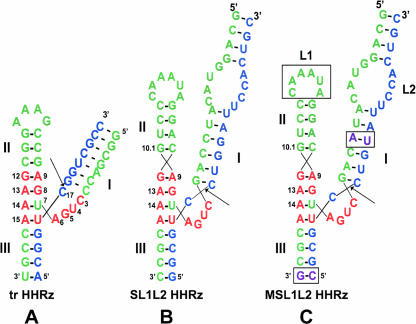
The minimal HHRz consists of three base-paired stems surrounding a conserved core of nucleotides (Forster and Symons 1987; Uhlenbeck 1987; Haseloff and Gerlach 1988). In early studies, the helical stems of the HHRz were truncated to facilitate in vitro studies resulting in a minimal sequence of fewer than 50 nt (Fig. 1A). Crystal structures of minimal HHRzs have shown that stem III coaxially stacks on stem II, forming a y-shaped structure (Pley et al. 1994; Scott et al. 1995). A recent re-examination of the role of the helices surrounding the conserved core of the HHRz has indicated important loop–loop interactions that appear to be conserved in native HHRz sequences (Khvorova et al. 2003). When native or minimal HHRzs were placed in the 3′ untranslated region of a reporter gene, only the native HHRzs were active in vivo. Comparison of the difference between the native and minimal HHRzs suggests that the presence of loops in both stems I and II was the only major structural difference. Loop swapping studies in constructs examined in vitro gave additional support for the importance of these loop–loop interactions.
FIGURE 1.
(A) A minimal hammerhead ribozyme. (B) SL1L2 sequence derived from Schistosoma mansoni. (C) Modified SL1L2 (MSL1L2) sequence. Nucleotides in green are the enzyme strand and those in blue are the substrate strand. Red residues are components of the conserved core. Residues boxed and shown in purple were modified in SL1L2 to make MSL1L2. The boxed nucleotides of loop 1 (L1) were changed to U nucleotides in the U Loop MSL1L2. The arrow identifies the site of cleavage.
At physiological ionic strengths, divalent metal ions are thought to aid in the folding and possibly the catalytic mechanism of the HHRz (DeRose 2003). In comparison with minimal HHRzs, native HHRz sequences containing putative loop–loop interactions have higher observed rates and lower divalent ion requirements (De la Pena et al. 2003; Khvorova et al. 2003; Canny et al. 2004). The lower divalent ion requirements may be related to the ability of these loop–loop interactions to stabilize a correctly folded ribozyme core. Consistent with this hypothesis, fluorescence resonance energy transfer (FRET) experiments on an extended HHRz from Schistosoma mansoni, a species of digenetic trematodes that parasitize many animal species, suggests that folding occurs in a single step in the micromolar concentration range of Mg2+ (Penedo et al. 2004).
To further understand the influence of interacting loops on the characteristics of HHRzs, we have studied the activity and stabilization of constructs derived from S. mansoni (Fig. 1B,C; Ferbeyre et al. 1998). At moderate ionic strengths, the sequences studied here appear to exist in at least two conformers, one of which exhibits very fast cleavage rates at low Mg2+ concentrations. The effects of loop I modification and selective thiophosphate substitutions were examined, and an observed ligation rate was determined. Observed results are consistent with a model in which loop–loop interactions stabilize a more active conformation.
RESULTS
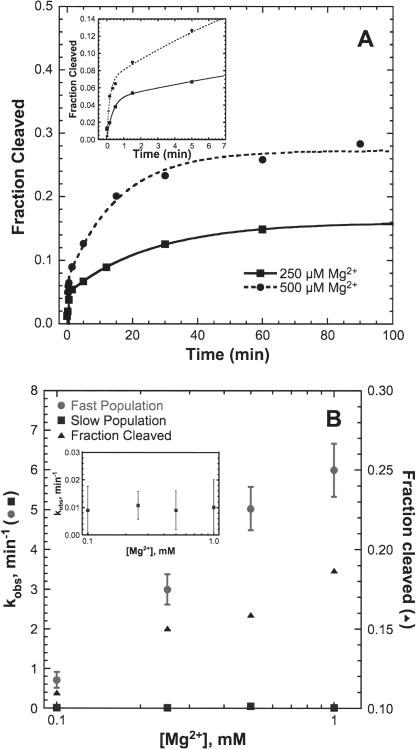
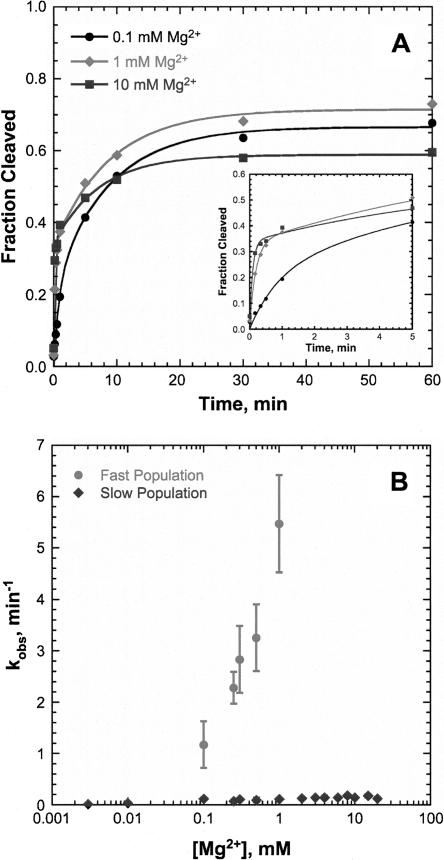
An extended hammerhead construct derived from S. mansoni (SL1L2) with a 43-nt enzyme strand and a 20-nt substrate strand was used to explore HHRzs with putative loop–loop interactions between stems I and II (Fig. 1B). Kinetic studies were performed at 20°C in 5 mM HEPES (pH 7.0), 100 mM Na+, and varying concentrations of Mg2+. Typical single-turnover kinetic traces of SL1L2 are shown in Figure 2A. At Mg2+ concentrations > 100 μM, a biphasic rate equation (Equation 2, below) is necessary to fit the kinetic data for the SL1L2 HHRz. At 1 mM Mg2+, the fast phase kobs of SL1L2 of 5.5 min−1 is > 40-fold faster than the kobs of the minimal HHRz (Table 1; Hunsicker and DeRose 2000). The kobs of the fast phase increases with increasing Mg2+ concentration, but the slow phase kobs does not (Fig. 2B and inset). The fraction cleaved of the SL1L2 HHRz is low and is dependent on Mg2+ concentration. In a background of 100 mM Na+, a maximum fraction cleaved of < 0.3 is observed with up to 2 mM Mg2+. This correlation between maximal fraction cleaved and divalent cation concentration is not observed for the minimal HHRz, which generally has a maximum fraction cleaved of 0.6–0.8 at all concentrations of Mg2+ (Hunsicker and DeRose 2000).
FIGURE 2.
Single-turnover kinetic study of the SL1L2 HHRz. (A) SL1L2 rate curves at 250 (solid line) and 500 (dashed line) μM Mg2+ in 5 mM HEPES (pH 7.0) and 100 mM Na+ fit to a biphasic rate equation (Equation 2). (Inset) Expansion of 0- to 5-min time points. (B) Fraction cleaved (▴) and kobs of slow phase (▪) and fast phase (•) as a function of Mg2+ concentration. (Inset) Expansion of the kobs of the slow phase.
TABLE 1.
Rates (kobs) for the minimal and extended hammerheads in divalent and monovalent cations
| kobs (min−1) | ||
| Cationa | Minimal hammerhead | Extended hammerhead |
| 0.01 mM Mg2+ | <0.001b | 0.036 |
| 1 mM Mg2+ | 0.125b | 5.5 |
| 10 mM Mg2+ | 0.25b | >6 |
| 0.01 mM Mn2+ | ~0.02b | 4.2 |
| 1 mM Mn2+ | 2.2b | >6 |
| 10 mM Mn2+ | 5b | >6 |
| 0.01 mM Co(NH3)63+ | <0.001c | 0.019 |
| 0.1 mM Co(NH3)63+ | <0.001c | 0.009 |
| 100 mM Co(NH3)63+ | 0.0071c | NDd |
| 1 M Na+ | NDd | 0.13 |
| 4 M Li+ | 0.18e | NDd |
Note: All extended hammerhead reactions were single-turnover with excess enzyme relative to substrate and were carried out in 5 mM HEPES (pH 7.0) at 20°C.
aAll experiments with multivalent cations were performed in a background of 100 mM Na+ except the rate determined in 100 mM Co(NH3)63+.
dExperiment not performed at that concentration of cation.
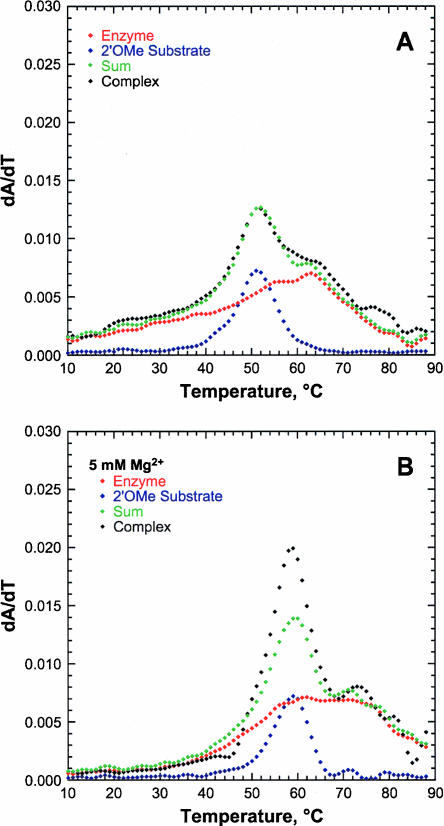
The low fraction cleaved of SL1L2 suggests that hybrid formation might be impeded in this sequence. Indeed, thermal denaturation studies of the SL1L2 substrate strand alone show evidence for a stable structure with a Tm of 51°C and a ΔH of 59.1 kcal/mol (Fig. 3A), which are consistent with a stable stem–loop that is predicted using the RNA-folding program Mfold (Mathews et al. 1999; Zuker 2003). The enzyme strand is predicted to have several conformations, but a common feature in all conformations is stem II, which is formed by the enzyme strand alone. UV thermal denaturation studies of the substrate strand, the enzyme strand, and the hybrid SL1L2 were performed in 5 mM HEPES (pH 7) and 100 mM Na+ in the absence and presence of 5 mM Mg2+ (Fig. 3). These studies indicate that both the enzyme and the substrate strands have stable secondary structures that could hinder hybrid formation and that are further stabilized by addition of Mg2+. The summation of the 260- nm spectra of the substrate strand and the enzyme strand overlaid on the spectrum of the combined strands are almost identical, suggesting that the latter spectrum derives mainly from the melting of the individual secondary structures of the enzyme and substrate strands and that little hybrid is formed.
FIGURE 3.
UV thermal denaturation study of the SL1L2 HHRz. UV thermal denaturation melting profiles observed at 260 nm for the enzyme strand (red), 2′OMe substrate strand (blue), sum of substrate and enzyme strand (green), and complex (black) in 5 mM HEPES (pH 7.0), 100 mM Na+ in the absence of divalent cations (A) and the presence of 5 mM Mg2+ (B).
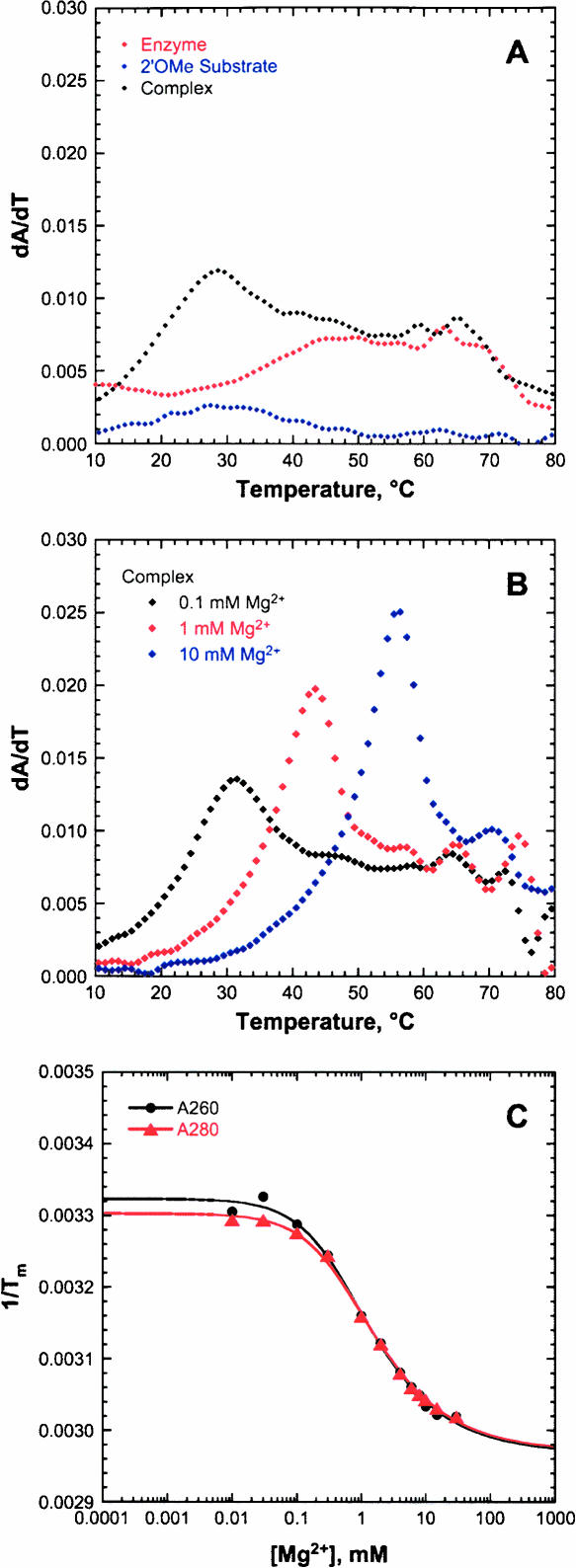
In an attempt to destabilize competing structures and enhance hybrid formation, SL1L2 was altered slightly. Using Mfold predictions, the boxed residues highlighted in purple in the stems were changed, generating the MSL1L2 sequence (Fig. 1C). The melt profile of the altered MSL1L2 substrate strand shows little evidence for significant secondary structure, with a single broad transition with a Tm of 29°C (Fig. 4A) which is 20°C lower than that of the SL1L2 substrate strand. The melt profile of the enzyme strand still shows some residual structure as expected due to formation of stem II (Fig. 1C). The MSL1L2 complex shows a new transition at 27°C in the absence of Mg2+ that we believe is due to complex formation. This transition increases in Tm and sharpness with increasing concentration of Mg2+ (Fig. 4B). Fitting the change in Tm for the first transition of the MSL1L2 hybrid as a function of Mg2+ (Equation 3, below) gives an apparent Kd of 1.6 ± 0.1 mM for A260 and 1.4 ± 0.2 mM for A280 (Fig. 4C), which reflects stabilization of the structure corresponding to this unfolding transition. These apparent Mg2+ Kd values likely reflect stabilization of base-paired regions of the structure by nonspecifically bound ions (Draper 2004).
FIGURE 4.
UV thermal denaturation study of the MSL1L2 HHRz. (A) UV thermal denaturation melting profiles at 260 nm for enzyme strand (red), 2′OMe substrate strand (blue), and complex (black) in 5 mM HEPES (pH 7.0) and 100 mM Na+. (B) Melting profiles at 260 nm for complex with addition of 0.1 (black), 1 (red), and 10 (blue) mM Mg2+. (C) Plot of 1/Tm as a function of Mg2+ concentration for the first transition of the complex at 260 nm (black) and at 280 nm (red) fit to Equation 3. Apparent Kds of 1.6 ± 0.1 mM for A260 and 1.4 ± 0.2 mM for A280 are observed for Mg2+ stabilization of structure monitored in this transition.
These studies indicate that the small changes made to SL1L2 decreased the stability of alternative conformations of the individual strands resulting in an increase in the amount of hybrid formed. Consistent with this, the MSL1L2 hammerhead showed a total fraction cleaved above 0.6 for all concentrations of Mg2+ (Fig. 5A). As with SL1L2, kinetic traces of MSL1L2 were best fit using a biphasic rate equation except at concentrations of Mg2+ below 100 μM, where a monophasic rate equation (Equation 1, below) was sufficient. The fast phase kobs of MSL1L2 increases significantly with increasing metal concentration, while the slow phase kobs does not (Fig. 5B).
FIGURE 5.
Single-turnover kinetic studies of the MSL1L2 HHRz. (A) Rate curves of MSL1L2 in 0.1 (•), 1 (♦), and 10 (▪) mM Mg2+ in 5 mM HEPES (pH 7.0) and 100 mM Na+ fit to a biphasic rate equation (Equation 2). (Inset) Expansion from 0- to 5-min time points. (B) kobs for fast (•) and slow (♦) populations as a function of Mg2+ concentration.
Activity studies in other supporting cations all show much faster rates for MSL1L2 over those of the minimal HHRz (Table 1). Activity studies of MSL1L2 in 0.01 mM Mn2+ result in a kobs for the fast population that is 200-fold greater than that observed for the minimal HHRz (Hunsicker and DeRose 2000). The kobs for MSL1L2 in 1 M Na + is similar to that seen for the minimal HHRz in 4 M Li+ (O’Rear et al. 2001). This indicates that MSL1L2 requires lower concentrations of cations for activity supported by either divalent or monovalent ions. MSL1L2 activity was also observed in 0.01 mM Co(NH3)63+ with a kobs of 0.09 min−1 (Table 1), whereas no activity was observed for the minimal HHRz at concentrations of Co(NH3)63+ < 10 mM (Horton and DeRose 2000; Curtis and Bartel 2001). This result suggests that inner sphere cation coordination is not necessary for activity of the extended HHRz sequence.
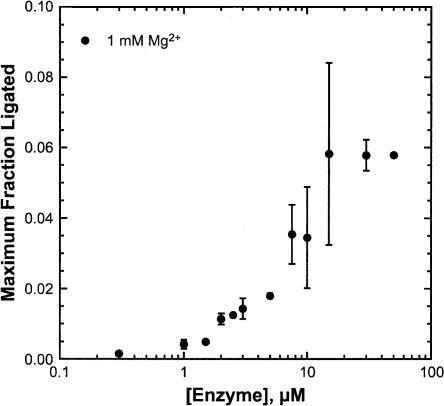
Biphasic kinetic traces indicate the presence of multiple processes. Ligation studies were performed to determine whether the reverse reaction could be contributing to the biphasic kinetics. For MSL1L2 in 5 mM HEPES (pH 7.0), 100 mM Na+, and 1 mM Mg2+, values of Keqint ~ 17 and klig of ~ 0.32 min−1 are determined (Fig. 6). A previous study of a minimal HHRz using the same method resulted in values of Keqint ~ 130 and klig ~ 0.008 min−1 (Hertel et al. 1994). While ligation is more favored than in the minimal HHRz, cleavage is still favored > 10-fold over ligation in MSL1L2, and compared to kcleavage, klig is not significant.
FIGURE 6.
Ligation study of the MSL1L2 HHRz. Plot of maximum fraction ligated as a function of enzyme strand concentration in 5 mM HEPES (pH 7.0), 100 mM Na+, and 1 mM Mg2+.
An alternative method to test the influence of ligation on the observed kinetics is to perform a chase experiment. In this experiment, excess unlabeled 3′ or 5′ product strand is added to an ongoing reaction in order to compete with labeled enzyme-bound products, preventing possible religation (Zamel et al. 2004). If the reaction has reached an equilibrium between cleavage and ligation, addition of cold product should result in a burst of new cleavage products. Here, when excess of either cleavage product was added to the MSL1L2 reaction, no additional burst phase was observed (data not shown). This is consistent with the low measured rate of ligation for MSL1L2. Taken together, these data suggest that the reverse reaction is not contributing to the biphasic kinetics and a likely scenario is that MSL1L2 exists in at least two conformations that result in slow and fast cleaving populations.
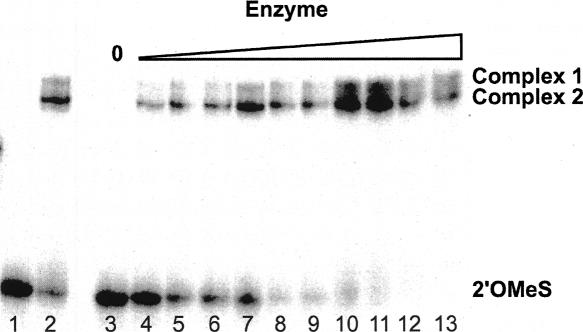
To further test the possibility that two species contribute to the biphasic kinetics, a native gel mobility assay was performed on the MSL1L2 complex (Fig. 7). Complex formation is readily observed, and an apparent Kd of 50 nM (0.1 M NaCl and 1 mM Mg2+) is estimated. Two bands are observed for the complex, one of which is consistently more populated for the MSL1L2 complex. This observation suggests that at least two conformations of MSL1L2 are present, supporting the model that multiple conformations contribute to the two kinetic phases observed for this ribozyme.
FIGURE 7.
Native gel mobility assay of MSL1L2 HHRz. The RNA was renatured in 5 mM HEPES (pH 7.0) and 100 mM NaCl, fast cooled on ice, and allowed to fold in the presence of 1 mM Mg2+ prior to fractionation. (Lane 1) 2.5 μM 2′OMe substrate strand only with trace 32P-end-labeled 2′OMe substrate; (lane 2) 3 μM enzyme annealed with 2.5 μM 2′OMe substrate and trace 5′ 32P-end-labeled 2′OMe substrate; (lane 3) control lane of 1 nM 5′ end-labeled 2′OMe substrate with no enzyme strand; (lanes 4–13) a titration of 1 nM 5′ end-labeled 2′OMe substrate with 10-fold or greater cold enzyme (up to 10 μM). Two conformations for the complex are observed for all concentrations of enzyme, with an apparent Kd for complex formation of 47 nM (in 1 mM Mg2+).
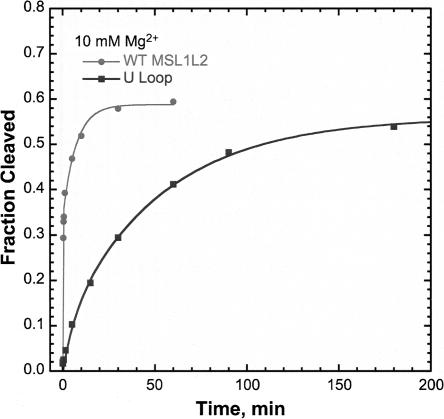
The importance of loop–loop interactions in the MSL1L2 ribozyme was examined by changing the boxed loop nucleotides in stem II to U residues (Fig. 1C). This loop 2 substitution resulted in significantly reduced cleavage rates measured at Mg2+ concentrations of up to 10 mM Mg2+. At 100 μM Mg2+, no activity is observed with this construct, which is consistent with the results of Penedo et al. (2004) and Canny et al. (2004), who both report that obliteration of loop I results in no detectable activity as measured in 100 μM Mg2+. As seen in kinetic studies performed at 20°C in 5 mM HEPES (pH 7.0), 100 mM Na+, and 10 mM Mg2+ (Fig. 8), the U loop MSL1L2 cleaves with a kobs of 0.02 min−1, which is slower than the slow population of wild-type MSL1L2. The U loop rate curves were best fit to a monophasic rate equation (Equation 1, below). The slow rate measured here for the U-loop substitution in stem 2 is within the range measured for some minimal hammerhead sequences in 10 mM Mg2+ (Hertel et al. 1994).
FIGURE 8.
Single-turnover kinetic study of the MSL1L2 U Loop HHRz. Kinetic traces of WT MSL1L2 (•) and U Loop (▪) in 5 mM HEPES (pH 7.0), 100 mM Na+, and 10 mM Mg2+. WT MSL1L2 data are fit to a biphasic equation (Equation 2) with a fast kobs of >6 min−1 and a slow kobs of 0.14 min−1. Data for U Loop MSL1L2 are fit to a monophasic rate equation (Equation 1), and a kobs of 0.02 min−1 is obtained.
In studies of the minimal hammerhead, a decrease in activity in Mg2+ due to phosphorothioate substitution and a subsequent rescue by the addition of Cd2+ has indicated potential metal binding to the Rp oxygen of the cleavage site phosphate and to the Rp oxygen of the phosphate 5′ to A9 (Dahm and Uhlenbeck 1991; Slim and Gait 1991; Wang et al. 1999). These studies have been repeated here with the MSL1L2 HHRz. In each case, activity studies of MSL1L2 sequences with a phosphorothioate either at the cleavage site or 5′ to A9 were performed on a mixture of the Rp and Sp diastereomers.
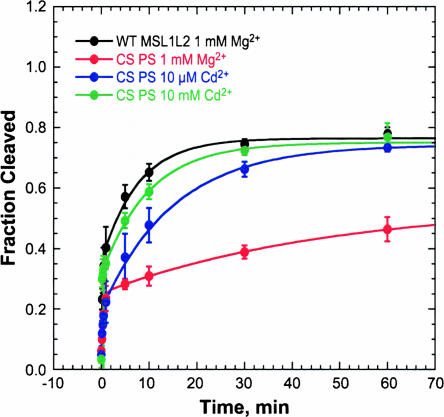
A phosphorothioate modification (PS) at the cleavage site of MSL1L2 results in a total fraction cleaved of ~ 0.45 in 1 mM Mg2+ (Fig. 9). This is a loss of approximately one-third of the total fraction cleaved in comparison with the unmodified MSL1L2. The kobs of the fast phase increases with Mg2+ concentration (data not shown) and has a value of 2.7 min−1 at 1 mM Mg2+, which is less than that of the unmodified MSL1L2 of 5.5 min−1. These results suggest that a fraction of the substrate strands, which are a mixture of cleavage-site Rp and Sp phosphorothioates, have reduced cleavage in the presence of Mg2+. In the case of the minimal HHRz, the Rp phosphorothioate does not cleave in Mg2+, but that activity can be rescued by Cd2+. Similarly, for MSL1L2, addition of 10 mM Cd2+ rescued cleavage to the level of the unmodified MSL1L2.
FIGURE 9.
Single-turnover kinetic study of MSL1L2 HHRz with a cleavage site phosphorothioate (CS PS). Rate curve of WT MSL1L2 (black) and of MSL1L2 with a mixture of Rp and Sp cleavage site phosphorothioate diastereomers in 1 mM Mg2+ (red), 10 μM Cd2+ (blue), and 10 mM Cd2+ (green) in 5 mM HEPES (pH 7.0) and 100 mM Na+. Data are fit to a biphasic rate equation (Equation 2). For WT MSL1L2 in 1 mM Mg2+, a fast kobs of 5.8 min−1 and a slow kobs of 0.14 min−1 are obtained. For the cleavage site PS in 1 mM Mg2+, a fast kobs of 2.7 min−1 and slow kobs of 0.02 min−1 are obtained. For the cleavage site PS in 10 μM Cd2+, a fast kobs of 4.5 min−1 and slow kobs of 0.08 min−1 are obtained while for the cleavage site PS in 10 mM Cd2+, a fast kobs of >6 min−1 and slow kobs of 0.10 min−1 are obtained.
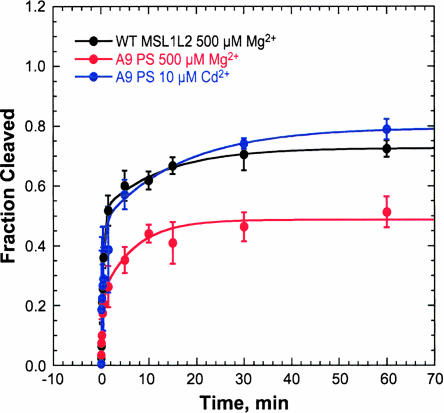
A phosphorothioate substitution 5′ to A9 also results in a decrease in the total fraction cleaved in the presence of Mg2+, giving a kobs of the fast phase of 3.3 min−1, similar to that of the unmodified MSL1L2 (Fig. 10). With the mixture of A9 PS diastereomers, approximately one-third of the sample is not cleaved in Mg2+ compared to the unmodified MSL1L2. In the presence of Cd2+, however, rate curves of the A9 phosphorothioate substituted MSL1L2 resemble those of the unmodified MSL1L2. Thus, the inactive A9 PS population of MSL1L2 can be rescued by Cd2+. Partial Mn2+ rescue of an A9 phosphorothioate substitution in a salamander extended HHRz has also been observed (Mitrasinovic and Epstein 1997).
FIGURE 10.
Kinetic study of the MSL1L2 HHRz with a phosphorothioate 5′ to A9 (A9 PS). Rate curve of WT MSL1L2 (black) and MSL1L2 with a mixture of Rp and Sp phosphorothioate diastereomers 5′ to A9 in 500 μM Mg2+ (red) and 10 μM Cd2+ (blue). Studies were performed in 5 mM HEPES (pH 7.0) and 100 mM Na+. Data are fit to a biphasic rate equation (Equation 2). In 500 μM Mg2+ a fast kobs of 2.3 min−1 and slow kobs of 0.08 min−1 are obtained for the WT MSL1L2 and a fast kobs of 3.2 min−1 and slow kobs of 0.15 min−1 are obtained for the A9 PS. In 10 μM Cd2+ a fast kobs of 2.2 min−1 and slow kobs of 0.06 min−1 are obtained.
DISCUSSION
For many years, HHRz research has focused on minimal hammerheads that were greatly truncated from native sequences (Uhlenbeck 2003). These HHRzs require millimolar concentrations of divalent cations for maximal activity, which are well above physiological concentrations. In addition, biochemical data on chemically modified HHRzs have often seemed incongruent with crystal structure data, where functional groups seemingly unimportant for activity in the crystal structure greatly reduced the activity observed in biochemical experiments (McKay 1996). A recent study showed that when a minimal HHRz was inserted in the 3′ untranslated region of a reporter gene, inactivation was not observed, suggesting that this minimal HHRz was not active in vivo (Khvorova et al. 2003). However, when the experiments were performed with native HHRzs, gene inactivation did occur. Further in vitro study of these native HHRzs indicated that they were highly active at cellular divalent cation concentrations and that loop–loop interactions were responsible for this increase in activity. Thus, referring back to nature’s original design has refreshed hammerhead research. Here, we explore a native HHRz construct derived from S. mansoni that is reasonably behaved and find high levels of activity at micromolar concentrations of divalent cations. Unlike the minimal HHRz, activity is observed at Mg2+ concentrations as low as 10 μM. At higher Mg2+ concentrations, a burst phase is measured with rates >10-fold over those observed in the minimal HHRzs (Table 1). Minimal HHRzs have notoriously slow kinetics, which has been proposed to be due to problematic conformational equilibria between inactive and active structures (Wang et al. 1999). This problem may be alleviated in the fast-cleaving form of the MSL1L2, bringing its rate closer to those of larger, more structured ribozymes. In the hepatitis delta virus (HDV) ribozyme, for example, the fast folding mutant G11C cleaves with a rate of 3.3 min−1 in 10 mM Mg2+ at pH 7.0 (Nakano et al. 2003), similar to the rate observed here for the fast cleaving phase.
Multiple conformations of the MSL1L2 hammerhead appear to be responsible for the observed biphasic kinetics. A native gel mobility study suggests the presence of at least two conformations. Alternative conformations have also been proposed to cause complications in the newt extended HHRz (Marusic et al. 1997). An observed rate of ligation that is much less than that of cleavage as well as a chase experiment indicate that increased ligation is not a significant contributor to the observed biphasic kinetics. However, the Keqint of the minimal hammerhead is almost 10 times larger than that of MSL1L2. An increase in ligation rate has also been observed in cross-linked minimal HHRzs (Stage-Zimmermann and Uhlenbeck 2001; Blount and Uhlenbeck 2002), and a recent crystal structure of a minimal hammerhead with cross-linked stems I and II suggests that switching between ligase and nuclease activity is determined by the helical orientation of stem I relative to stem II (Dunham et al. 2003). This suggests that the loop–loop interactions between stems I and II could stabilize the hammerhead in a near active conformation and ease the switch from cleavage to ligation.
FRET studies indicate that the folding of an extended hammerhead occurs in a single step in the presence of micromolar concentrations of Mg2+ (Penedo et al. 2004). In this study, however, as well as in a study by Pardi and coworkers of a similar sequence (Canny et al. 2004), higher concentrations of Mg2+ are found to be needed for maximal cleavage activity. These results parallel findings for the minimal HHRz, indicating functional importance for at least two types of cation interactions can be observed, one of which induces an initial folding step and the other of which enhances catalysis (for review, see DeRose 2003).
The increased activity at low concentrations of divalent cations of the MSL1L2 hammerhead over minimal HHRzs appears to result from loop–loop interactions. When these loop–loop interactions are disrupted, a large decrease in activity is observed (Fig. 7; De la Pena et al. 2003; Khvorova et al. 2003; Penedo et al. 2004). As has been pointed out (Khvorova et al. 2003; Penedo et al. 2004), the discovery of stabilizing elements in the HHRz parallels another small ribozyme. In the hairpin ribozyme, changing a two- or three-way junction to the native four-way junction increases catalytic activity in lower concentrations of Mg2+ (Esteban et al. 1997; Walter et al. 1999; Zhao et al. 2000), shifts the internal equilibrium toward ligation (Fedor 1999), and increases the rate of folding (Tan et al. 2003).
In the minimal HHRz, studies of phosphorothioate substitutions at the cleavage site and 5′ to A9 indicated potential metal binding to the pro-R oxygens of the phosphates at these locations (Dahm and Uhlenbeck 1991; Slim and Gait 1991; Wang et al. 1999). Here, initial studies with mixed Rp and Sp phosphorothioate diastereomers at the cleavage site or 5′ to A9 indicate that modification of these sites can also disrupt activity in the native HHRz. As is found in the minimal HHRz, Cd2+ rescues the activity of both phosphorothioates in the native sequence. The A9/G10.1 metal site has been corroborated by spectroscopic methods in the minimal HHRz (Morrissey et al. 1999a,b, 2000; Maderia et al. 2000). The importance of these two putative metal sites appears to be similar in both minimal and native hammerhead constructs.
MATERIALS AND METHODS
Oligonucleotides
RNA sequences in Figure 1B,C were purchased from Dharmacon, purified by denaturing gel electrophoresis on 20%/7 M urea polyacrylamide gels, and electroeluted from the gel slice. Sequences corresponding to P1 and P2 (5′ and 3′-OH) were also purchased from Dharmacon. Eluted transcripts were dialyzed against 5 mM triethanolamine (TEA; pH 7.0), 100 mM Na+, for at least 48 h with several reservoir changes, concentrated (Centricon-3, Millipore), ethanol precipitated, and resuspended into the same buffer to form a stock solution. The substrate strand used in the UV thermal denaturation and negative gel mobility studies had a 2′-O-methyl modification at the cleavage site.
Chemicals
All reagents used were molecular biology grade or higher. MgCl2 and MnCl2 metal stocks were purchased from Sigma. CdCl2 was purchased from Alfa Aesar. Co(NH3)63+ was purchased from Al-drich Chemical.
Measurements of hammerhead ribozyme activity
Activities and rate constants for the hammerhead ribozyme were determined using the 43-nt RNA enzyme and 5′-32P-labeled 20-nt substrate strand pictured in Figure 1B,C. End labeling was performed using T4 polynucleotide kinase (USB) and the radiolabeled transcripts were gel purified as described above. To form the hybrid hammerheads, the substrate (2.5 μM final concentration) and an excess of enzyme (3 μM final concentration) were combined in 5 mM HEPES (pH 7.0) and 100 mM Na+, heated to 90°C for 90 sec, and cooled on ice for 30 min. Cleavage reactions were initiated by addition of equal volumes of various concentrations of MgCl2 or MnCl2, and samples were incubated for a given time at 20°C. Reactions were terminated by the addition of an equal volume of 90% formamide, 40 mM EDTA, 0.6 mg/mL bromophenol blue, and 0.6 mg/mL xylene cyanol quench buffer and placed on dry ice. The substrate and product strands were separated by gel electrophoresis on 20%/7 M urea polyacrylamide gels. Labeled substrate and product strands were visualized and quantified by phosphor-imaging analysis (Fuji). Two equations were used to fit the data. Data where a monophasic fit was sufficient were analyzed using Equation 1:
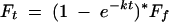 |
(1) |
where Ft is the fraction cleaved at time t, and Ff is the maximum fraction cleaved. Where Equation 1 was deemed insufficient to describe the data, the biphasic Equation 2 was used:
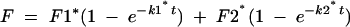 |
(2) |
where Fi and ki correspond to the fraction cleaved and rate constant for population (i). Due to the limitations of a manual quench protocol, k values above ~ 6 min−1 are not reported.
Ligation reactions
The 5′-32P-labeled P1 strand (5′-CGCGUC-3′) containing the appropriate 2′,3′-cyclic phosphate was generated by ribozyme-mediated cleavage of 5′-32P-labeled substrate strand and purified by gel electrophoresis on a 20%/7 M urea polyacrylamide gel. The RNA was eluted from the gel in 5 mM TEA (pH 7.0), with shaking overnight at 4°C, ethanol precipitated, and resuspended in an appropriate volume of 5 mM TEA (pH 7.0). The concentration was determined by scintillation counting. Strand P2 (5′-CUGUAUUCCACUGC-3′) was ordered from Dharmacon and purified as described above.
Single turnover ligation reactions were performed by combining varying concentrations of enzyme strand (1–50 μM), a 20% excess of strand P2, and 1 nM 5′-32P-labeled P1 in 5 mM HEPES (pH 7.0) and 100 mM Na+ in a final volume of 18 μL. The reactions were initiated by the addition of 2 μL of 10 mM MgCl2 to yield a final concentration of 1 mM Mg2+. Samples were taken at various times, quenched with formamide loading buffer, and placed on dry ice. Samples were fractionated on 20%/7 M urea polyacrylamide gels. The fraction of ligated 5′-32P-labeled substrate was measured as a function of time. The maximal fraction ligated was determined for each concentration of enzyme by fitting the observed data to the same double exponential equation used for the cleavage reactions (Equation 2) and totaling the fraction ligated from each phase. The value for klig was determined as was previously described for a minimal hammerhead sequence (Hertel et al. 1994). The internal equilibrium constant can be described as Keqint = [E · P1 · P2]/[E · S]. Assuming that Fmax,lig ≅ [E ·S], where Fmax,lig is the maximal fraction ligated, Keqint = 1/Fmax,lig. In this case, Fmax,lig = 0.058 (the average from measurements taken with 15 to 50 μM enzyme), which gives a Keqint of 17.2. As Keqint = kcleavage/klig and at 1 mM Mg2+ kcleavage = 5.5 min−1, a klig of 0.32 min−1 was calculated.
Chase experiments were also performed as an additional test on the effect of ligation on the observed cleavage rates. Reactions were allowed to proceed as described above for single-turnover cleavage kinetics until equilibrium was reached, usually for 30–60 min. Unlabeled cleavage product P1 (3′-OH) or P2 was then added in 50-fold excess of the enzyme strand to compete with enzyme-bound product strands, and additional time points were collected to search for a new burst of 5′-labeled cleavage product.
UV thermal denaturation studies
Substrate strands containing an inhibitory 2′-O-methyl at the cleavage site cytosine were used for thermal denaturation studies. Samples were prepared at final concentrations of 3 μM RNA and heated to 90°C for 90 sec and cooled on ice for 30 min. Bench cooling for 1 h did not change the results of the melt experiments. Melts were performed using a Varian Cary 300 UV-Vis spectrometer. Absorbance was recorded at 260 and 280 nm. The melting temperature (Tm) was determined by taking the first derivative of absorbance with respect to temperature (dA/dT). Data were fit using a nonlinear least squares fitting algorithm to a sequential folding model previously described (Theimer et al. 1998). Tm values at different concentrations of divalent cation were plotted as 1/Tm versus metal concentration and fit to Equation 3 (Laing et al. 1994):
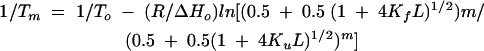 |
(3) |
where Tm is the melting temperature at a given metal concentration, Kf and Ku are the metal binding constants for the folded and unfolded forms of the RNA, L is the free metal concentration and m is the number of phosphates participating in a given unfolding event. ΔHo and To are the enthalpy and melting temperature of the transition in the absence of added metal.
Native gel mobility assay
For observing the mobility of the substrate strand alone 2.5 μM of cold 2′OMe substrate strand plus a trace amount of 5′ end-labeled 2′OMe substrate was renatured in 5 mM HEPES, pH 7.0, and 100 mM NaCl at 90°C for 90 sec and then placed on ice for 30 min before loading onto a nondenaturing gel. To prepare samples used for observing complex formation under the conditions used for kinetics, 3 μM of cold enzyme strand was mixed with 2.5 μM cold 2′OMe substrate plus a trace amount of 5′ end-labeled 2′OMe substrate and renatured as above. For the enzyme-substrate titration, 10-fold excess or greater of cold enzyme (up to 10 μM) was mixed with 1 nM cold 2′OMe substrate plus a trace amount of 5′ end-labeled 2′OMe substrate and annealed as above. After cooling on ice, Mg2+ was added to a final concentration of 1 mM and the samples were allowed to equilibrate at room temperature for 5 min. Glycerol was added to 10% final concentration and the samples were fractionated on a 20% (19:1 acrylamide/bis)/0.5× TBE native gel containing 1 mM Mg2+. The running buffer was 0.5× TBE and it also contained 1 mM Mg2+. The gels were run at 250 V for 8–10 h at 4°C to prevent excessive heating. Gels were dried and visualized using a phosphorimager system (Fuji).
Acknowledgments
This work was supported by the funding from the NIH (GM58096) and NSF (0111696) to V.J.D. and an NIH Training Grant fellowship to E.M.O.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7950605.
REFERENCES
- Blount, K.F. and Uhlenbeck, O.C. 2002. Internal equilibrium of the hammerhead ribozyme is altered by the length of certain covalent cross-links. Biochemistry 41: 6834–6841. [DOI] [PubMed] [Google Scholar]
- Canny, M.D., Jucker, F.M., Kellogg, E., Khvorova, A., Jayasena, S.D., and Pardi, A. 2004. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 126: 10848–10849. [DOI] [PubMed] [Google Scholar]
- Curtis, E.A. and Bartel, D.P. 2001. The hammerhead cleavage reaction in monovalent cations. RNA 7: 546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm, S.C. and Uhlenbeck, O.C. 1991. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry 30: 9464–9469. [DOI] [PubMed] [Google Scholar]
- De la Pena, M., Gago, S., and Flores, R. 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22: 5561–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose, V.J. 2002. Two decades of RNA catalysis. Chem. Biol. 9: 961–969. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Metal ion binding to catalytic RNA molecules. Curr. Opin. Struct. Biol. 13: 317–324. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A. and Cech, T.R. 2002. The chemical repertoire of natural ribozymes. Nature 418: 222–228. [DOI] [PubMed] [Google Scholar]
- Draper, D.E. 2004. A guide to ions and RNA structure. RNA 10: 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, C.M., Murray, J.B., and Scott, W.G. 2003. A helical twist-induced conformational switch activates cleavage in the hammerhead ribozyme. J. Mol. Biol. 332: 327–336. [DOI] [PubMed] [Google Scholar]
- Esteban, J.A., Banerjee, A.R., and Burke, J.M. 1997. Kinetic mechanism of the hairpin ribozyme. Identification and characterization of two nonexchangeable conformations. J. Biol. Chem. 272: 13629–13639. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. 1999. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry 38: 11040–11050. [DOI] [PubMed] [Google Scholar]
- Ferbeyre, G., Smith, J.M., and Cedergren R. 1998. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell. Biol. 18: 3880–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferbeyre, G., Bourdeau, V., Pageau, M., Miramontes, P., and Cedergren, R. 2000. Distribution of hammerhead and hammerhead-like RNA motifs through the GenBank. Genome Res. 10: 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, A.C. and Symons, R.H. 1987. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell 50: 9–16. [DOI] [PubMed] [Google Scholar]
- Haseloff, J. and Gerlach, W.L. 1988. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature 334: 585–591. [DOI] [PubMed] [Google Scholar]
- Hertel, K.J., Herschlag, D., and Uhlenbeck, O.C. 1994. A kinetic and thermodynamic framework for the hammerhead ribozyme reaction. Biochemistry 33: 3374–3385. [DOI] [PubMed] [Google Scholar]
- Horton, T.E. and DeRose, V.J. 2000. Cobalt hexammine inhibition of the hammerhead ribozyme. Biochemistry 39: 11408–11416. [DOI] [PubMed] [Google Scholar]
- Hunsicker, L.M. and DeRose, V.J. 2000. Activities and relative affinities of divalent metals in unmodified and phosphorothioate-substituted hammerhead ribozymes. J. Inorg. Biochem. 80: 271–281. [DOI] [PubMed] [Google Scholar]
- Hutchins, C.J., Rathjen, P.D., Forster, A.C., and Symons, R.H. 1986. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14: 3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., and Jayasena, S.D. 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10: 708–712. [DOI] [PubMed] [Google Scholar]
- Laing, L.G., Gluick, T.C., and Draper, D.E. 1994. Stabilization of RNA structure by Mg ions. Specific and non-specific effects. J. Mol. Biol. 237: 577–587. [DOI] [PubMed] [Google Scholar]
- Lilley, D.M. 2003. The origins of RNA catalysis in ribozymes. Trends Biochem. Sci. 28: 495–501. [DOI] [PubMed] [Google Scholar]
- Maderia, M., Hunsicker, L.M., and DeRose, V.J. 2000. Metal-phosphate interactions in the hammerhead ribozyme observed by P-31 NMR and phosphorothioate substitutions. Biochemistry 39: 12113–12120. [DOI] [PubMed] [Google Scholar]
- Marusic, L., Luzi, E., Barsacchi, G., and Eckstein, F. 1997. Intermolecular cleavage by the newt ribozyme. Eur. J. Biochem. 247: 396–401. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- McKay, D.B. 1996. Structure and function of the hammerhead ribozyme: An unfinished story. RNA 2: 395–403. [PMC free article] [PubMed] [Google Scholar]
- Mitrasinovic, O. and Epstein, L.M. 1997. Differences in the phosphate oxygen requirements for self-cleavage by the extended and prototypical hammerhead forms. Nucleic Acids Res. 25: 2189–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey, S.R., Horton, T.E., Grant, C.V., Hoogstraten, C.G., Britt, R.D., and DeRose, V.J. 1999a. Mn2+-nitrogen interactions in RNA probed by electron spin-echo envelope modulation spectroscopy: Application to the hammerhead ribozyme. J. Am. Chem. Soc. 121: 9215–9218. [Google Scholar]
- Morrissey, S.R., Horton, T.E., Wasowicz, T., and DeRose, V.J. 1999b. Investigation of metal-binding in the hammerhead ribozyme by EPR and cw Q-band ENDOR spectroscopies. Biophys. J. 76: A323. [Google Scholar]
- Morrissey, S.R., Horton, T.E., and DeRose, V.J. 2000. Mn2+ sites in the hammerhead ribozyme investigated by EPR and continuous-wave Q-band ENDOR spectroscopies. J. Am. Chem. Soc. 122: 3473–3481. [Google Scholar]
- Nakano, S., Cerrone, A.L., and Bevilacqua, P.C. 2003. Mechanistic characterization of the HDV genomic ribozyme: Classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry 42: 2982–2994. [DOI] [PubMed] [Google Scholar]
- O’Rear, J.L., Wang, S.L., Feig, A.L., Beigelman, L., Uhlenbeck, O.C., and Herschlag, D. 2001. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA 7: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo, J.C., Wilson, T.J., Jayasena, S.D., Khvorova, A., and Lilley, D.M. 2004. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA 10: 880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley, H.W., Flaherty, K.M., and McKay, D.B. 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74. [DOI] [PubMed] [Google Scholar]
- Prody, G.A., Bakos, J.T., Buzayan, J.M., Schneider, I.R., and Bruening, G. 1986. Autolytic processing of dimeric plant-virus satellite RNA. Science 231: 1577–1580. [DOI] [PubMed] [Google Scholar]
- Rojas, A.A., Vazquez-Tello, A., Ferbeyre, G., Venanzetti, F., Bachmann, L., Paquin, B., Sbordoni, V., and Cedergren, R. 2000. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res. 28: 4037–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Ashtiani, K. and Szostak, J.W. 2001. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414: 82–84. [DOI] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., and Klug, A. 1995. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 81: 991–1002. [DOI] [PubMed] [Google Scholar]
- Slim, G. and Gait, M.J. 1991. Configurationally defined phosphorothioate-containing oligoribonucleotides in the study of the mechanism of cleavage of hammerhead ribozymes. Nucleic Acids Res. 19: 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage-Zimmermann, T.K. and Uhlenbeck, O.C. 2001. A covalent crosslink converts the hammerhead ribozyme from a ribonuclease to an RNA ligase. Nat. Struct. Biol. 8: 863–867. [DOI] [PubMed] [Google Scholar]
- Takagi, Y., Warashina, M., Stec, W.J., Yoshinari, K., and Taira, K. 2001. Recent advances in the elucidation of the mechanisms of action of ribozymes. Nucleic Acids Res. 29: 1815–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, E., Wilson, T.J., Nahas, M.K., Clegg, R.M., Lilley, D.M., and Ha, T. 2003. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl. Acad. Sci. 100: 9308–9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, C.A., Wang, Y., Hoffman, D.W., Krisch, H.M., and Giedroc, D.P. 1998. Non-nearest neighbor effects on the thermodynamics of unfolding of a model mRNA pseudoknot. J. Mol. Biol. 279: 545–564. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck, O.C. 1987. A small catalytic oligoribonucleotide. Nature 328: 596–600. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Less isn’t always more. RNA 9: 1415–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, N.G., Burke, J.M., and Millar, D.P. 1999. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat. Struct. Biol. 6: 544–549. [DOI] [PubMed] [Google Scholar]
- Wang, S., Karbstein, K., Peracchi, A., Beigelman, L., and Herschlag, D. 1999. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry 38: 14363–14378. [DOI] [PubMed] [Google Scholar]
- Zamel, R., Poon, A., Jaikaran, D., Andersen, A., Olive, J., De Abreu, D., and Collins, R.A. 2004. Exceptionally fast self-cleavage by a Neurospora Varkud satellite ribozyme. Proc. Natl. Acad. Sci. 101: 1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. and Epstein, L.M. 1996. Cloning and characterization of extended hammerheads from a diverse set of caudate amphibians. Gene 172: 183–190. [DOI] [PubMed] [Google Scholar]
- Zhao, Z.Y., Wilson, T.J., Maxwell, K., and Lilley, D.M. 2000. The folding of the hairpin ribozyme: Dependence on the loops and the junction. RNA 6: 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]