Abstract
The human 15.5K protein binds to the 5′ stem–loop of U4 snRNA, promotes the assembly of the spliceosomal U4/U6 snRNP, and is required for the recruitment of the 61K protein and the 20/60/90K protein complex to the U4 snRNA. In the crystallographic structure of the 15.5K–U4 snRNA complex, the conformation of the RNA corresponds to the family of kink-turn (K-turn) structural motifs. We simulated the complex and the free RNA, showing how the protein binding and the intrinsic flexibility contribute to the RNA folding process. We found that the RNA is significantly more flexible in the absence of the 15.5K protein. Conformational transitions such as the interconversion between alternative purine stacking schemes, the loss of G-A base pairs, and the opening of the K-turn occur only in the free RNA. Furthermore, the stability of one canonical G-C base pair is influenced both by the binding of the 15.5K protein and the nature of the adjacent structural element in the RNA. We performed chemical RNA modification experiments and observed that the free RNA lacks secondary structure elements, a result in excellent agreement with the simulations. Based on these observations, we propose a protein-assisted RNA folding mechanism in which the RNA intrinsic flexibility functions as a catalyst.
Keywords: splicing, RNA folding, K-turn motif, molecular dynamics, locally enhanced sampling, induced fit
INTRODUCTION
The spliceosome is a large ribonucleoprotein particle that catalytically removes introns from newly transcribed pre-messenger RNAs. Most pre-mRNA introns are removed by the U2-dependent (major) spliceosome, which is composed of the small nuclear ribonucleoprotein particles (snRNPs) U1, U2, U4/U6, and U5 and numerous non-snRNP protein factors (for review, see Burge et al. 1999). Within the U4/U6 snRNP, the U4 and U6 snRNAs form a phylogenetically highly conserved Y-shaped U4/U6 interaction domain, consisting of two intermolecular helices (stems I and II), which are separated by the 5′ stem–loop of U4 (Bringmann et al. 1984; Hashimoto and Steitz 1984; Rinke et al. 1985; Brow and Guthrie 1988).
The 15.5K protein is a specific component of the human U4/U6 snRNP. Its binding to the 5′ stem–loop of U4 sn- RNA is required for the recruitment of the 61K protein and the 20/60/90K protein complex to the U4/U6 snRNA (Nottrott et al. 2002). The 15.5K protein is also a component of box C/D snoRNP and U4/U6atac snRNP in which it is bound to similar RNA motifs (Watkins et al. 2000; Schneider et al. 2002).
In the crystal structure of the complex of the 15.5K protein and the 5′ stem–loop of U4 snRNA, the RNA adopts a conformation typical for the family of kink-turn (K-turn) structural motifs (Vidovic et al. 2000; Klein et al. 2001). This motif has a kink in the phosphodiester backbone that causes a sharp turn in the RNA helix. Two stems are connected by a purine-rich internal asymmetric loop containing one or two flipped out nucleotides and two or three noncanonical base pairs. In the 5′ stem–loop of the U4 snRNA, the canonical stem (C-stem) has three Watson-Crick G-C base pairs and the noncanonical stem (NC-stem) has two Watson-Crick G-C base pairs. Two tandem-sheared G-A base pairs are formed in the internal loop (non-Watson-Crick base pairs in RNA have been described by Leontis and Westhof 2002). The NC-stem is attached to an “external” UUUAU pentaloop, the conformation of which is not revealed in the crystallographic structure. The flipped out uridine (U31), the unpaired adenines (A29, A30), and the guanine (G32) are involved in hydrogen bonds and hydrophobic interactions with the 15.5K protein (Vidovic et al. 2000). The unpaired nucleotides and the tandem-sheared G-A base pairs are crucial for 15.5K protein binding (Nottrott et al. 1999). The 61K protein cross-links with the UUUAU external loop, indicating that the loop is critical to the binding of the 61K protein (Nottrott et al. 2002), although the precise mechanism is not yet understood. Six other K-turns are found in the 23S rRNA of Haloarcula marismortui and two in the 16S rRNA of Thermus thermophilus (Klein et al. 2001). K-turns were also identified in the structure of the box C/D snoRNA bound to archaeal L7Ae protein (Moore et al. 2004) and in the yeast L30e–mRNA complex (Chao and Williamson 2004). Out of 10 K-turns, only one (Kt38) is not associated with proteins (Klein et al. 2001), indicating that the motif could be a candidate for protein-assisted folding. The diversity of RNA structural motifs was reviewed by Leontis and Westhof (2003).
Formation of a wide variety of protein–RNA complexes involves conformational changes in the protein, RNA, or both. In several cases, the folding of the RNA is assisted by protein binding. The terms “induced fit” and “conformational capture” were introduced for designating alternative pathways of conformational change upon complex formation. In the induced fit, the RNA undergoes a transition between two different well-defined conformations, whereas conformational capture refers to the stabilization by the protein of one specific conformation from a pool of conformations reflecting the inherent flexibility of the RNA (Williamson 2000; Leulliot and Varani 2001). One example of protein-assisted RNA folding according to the induced fit mechanism is the binding of the 3′ UTR of U1A pre-mRNA to the U1A protein (Oubridge et al. 1994; Avis et al. 1996; Gubser and Varani 1996; Varani et al. 2000). Previous studies proposed a protein-assisted RNA folding for the K-turn motif (Matsumura et al. 2003; Goody et al. 2004). Goody et al. (2004) characterized two different conformations for the K-turn motif by fluorescence resonance energy transfer (FRET). They showed that the kinked (closed) conformation is in equilibrium with an extended (open) conformation and that the interconversion between the two conformations is a metalion-dependent process. Because of the lack of structural information about the unbound form of the RNA, many questions regarding the folding of the K-turn motif remain unanswered.
In the current study, we used molecular dynamics (MD) simulations to investigate the folding of the K-turn formed by the 5′ stem–loop of U4 snRNA. Computer simulations have become very powerful tools for studying biological processes, largely because of the rapid increase in computer power and improved accuracy. Advances such as the explicit modeling of solvent, further refinement of force fields, and the advent of the particle mesh Ewald (PME) method for treating long-range electrostatic interactions have led to increasingly fruitful simulations of biological systems and processes (Cheatham et al. 1995; Cheatham and Brooks 1998; Cheatham and Kollman 2000; Wang et al. 2001). Molecular dynamics simulations have been applied in studies of RNA structure (Guo et al. 2000; Li et al. 2001; Réblová et al. 2003a,b; Noy et al. 2004), RNA–metalion binding (Auffinger and Westhof 2000; Auffinger et al. 2003, 2004a,b), or RNA–protein interfaces (Reyes and Kollmann 2000; Reyes et al. 2001; Blakaj et al. 2001; Pitici et al. 2002; Réblová et al. 2004). However, limitations remain; for example, MD trajectories are restricted to hundreds of nanoseconds time scales, reducing the range of processes that can be studied to those occurring in this time range. Thus, conformational sampling is still poor in standard MD simulations and large conformational transitions are inaccessible.
Locally enhanced sampling (LES) is a mean field-based theory used to increase the range of conformational sampling during MD trajectories (Elber and Karplus 1990; Simmerling and Elber 1994; Simmerling and Kollman 1996; Simmerling et al. 1998a,b, 2000). Coupled with PME, LES constitutes a powerful tool for locating experimental structures when starting from different conformations (Simmerling et al. 1998a,b). The application of LES leads to a smoother potential energy surface, allowing conformational transitions that are otherwise inaccessible to standard MD simulations (Elber and Karplus 1990).
To elucidate the RNA folding mechanism in the system under study, we performed MD simulations, starting with the 2.9 Å refined crystal structure of the 15.5K protein complexed with the 5′ stem–loop of U4 snRNA (Vidovic et al. 2000). We obtained 10-nsec trajectories for the complex and the unbound RNA, using five different RNA constructs, by varying the external loop attached to the NC-stem (Fig. 1). Since the closed conformation of the unbound RNA was retained and partial opening of the K-turn was observed only briefly during standard MD trajectories, we applied LES to simulate the transition from the closed to the open form of the unbound RNA.
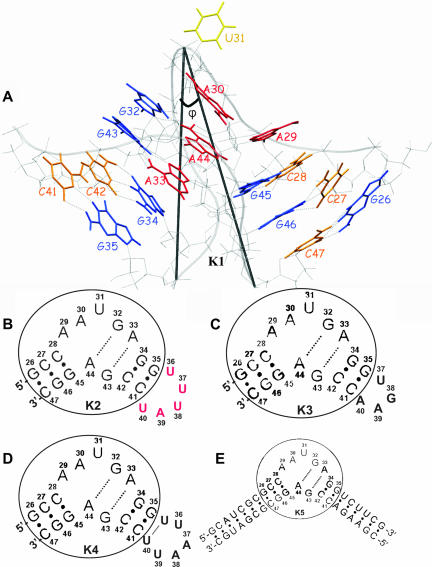
FIGURE 1.
Simulated RNA constructs. (A) The K-turn motif found in the crystal structure of the 5′ stem–loop of U4 snRNA bound to 15.5K protein (K1 RNA); the C-stem has three Watson-Crick base pairs (G26–C47, G46–C27, and G45–C28), the internal loop contains three unpaired nucleotides (A29, A30, U31) and two tandem-sheared G-A base pairs (G32–A44, G43–A33), and the NC-stem consists of two Watson-Crick base pairs (G34–C42 and G35–C41). Guanines are shown in blue, cytosines in orange, adenines in red, and the uracils in yellow. φ is the angle between the P atoms of C47, U31, and G35. (B) K2 RNA (naturally occurring): the pentaloop UUUAU (shown in red) is attached to the NC-stem of K1 RNA. (C) K3 RNA: the tetraloop UGAA was attached to the NC-stem of K1 RNA. (D) K4 RNA: the hexaloop UUAAUU was attached to the NC-stem of K1 RNA. (E) K5 RNA: the C-stem and the NC-stem of K1 RNA were extended with seven and six Watson-Crick base pairs. The core of the K-turn RNA observed in the crystal structure is encircled in B–E.
Based both on the computer simulations and on chemical RNA structure probing experiments, we conclude that the folding of the 5′ stem–loop of U4 snRNA is assisted by the binding of the 15.5K protein and identify different flexible RNA regions that play an important role in the folding process. We propose a model in which the binding of 15.5K protein creates a prefolded structure of the NC-stem and the external loop, thereby creating a suitable structural environment for the subsequent binding of the 61K protein. Nevertheless, the external loop remains flexible after the binding of 15.5K protein, and it is probably folded only after the recruitment of 61K protein to the RNA. In support of this model we present an analogy with the binding of ribosomal proteins L7AE and L15E to another K-turn motif named Kt15. The L7AE protein shows great folding similarities with the 15.5K protein, and the protein–RNA interface is almost identical in both cases.
RESULTS
General analysis of the simulations
Figure 1A shows the crystal structure of the 5′ stem–loop of U4 snRNA bound to 15.5K protein. We characterize the K-turn by the φ angle between the P atoms of residues C47, U31, and G35 (Fig. 1A). The φ angle is independent of instabilities in the RNA structure as opposed to the angle between the helical axes of the two stems, which depends on the stability of the helical regions. In the closed conformation φ has a value of ∼25°, which is slightly smaller than the angle between the two helical axes (∼30°). The naturally occurring RNA (K2) is shown in Figure 1B. We used the crystallographic structure of the RNA (K1), lacking the external pentaloop, as a template to build and simulate five different RNA constructs: (1) K1 (Fig. 1A), (2) K2 (Fig. 1B), (3) K3 (Fig. 1C), (4) K4 (Fig. 1D), and (5) K5 (Fig. 1E). The constructs produced similar trajectories both in the bound and unbound forms, with the differences mainly lying in the stability of the Watson-Crick base pairs of the NC-stem and in the behavior of the external loop. Throughout this study we label the molecular dynamics trajectories as “MD-LES” when LES was not used and “MD+LES” when LES was applied. All the trajectories described were 10 nsec long. In the text we refer to the naturally occurring K2 RNA unless indicated otherwise.
MD-LES simulations
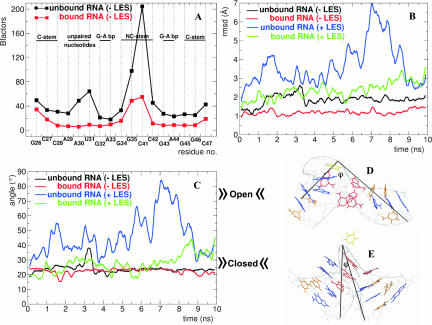
The B-factors per residue were calculated for the bound and unbound RNA during the MD-LES trajectories (Fig. 2A). The plot reveals that the unbound RNA fluctuates more than the bound RNA, indicating that the protein exerts a stabilizing effect on the RNA. Significantly higher fluctuations in the unbound RNA are observed for the unpaired nucleotides A30 and U31 and for the G35–C41 and G34–C42 Watson-Crick base pairs of the NC-stem. Root mean square deviations (RMSD) of the backbone for the core RNA structure (residues G26 to G35 and C41 to C47) were calculated using the initial structure as the reference (Fig. 2B). We observed that the unbound RNA deviates more from the crystal structure than the bound RNA, with peak values for RMSD in the time interval between 1.5 nsec and 3.5 nsec, reflecting a slight opening of the K-turn. After 3.5 nsec the K-turn closes back and stays closed until the end of the trajectory. The φ angle has a peak value of ∼40° after ∼3.2 nsec of the trajectory of the unbound RNA compared to a relatively constant 20° in the bound RNA (Fig. 2C).
FIGURE 2.
General analysis of MD simulations (K2 RNA). (A) B-factors per residue for the RNA structure during the MD-LES trajectories of the bound (red curve) and unbound RNA (black curve). (B) Root mean square deviations (RMSD) of the RNA backbone from the initial structure for the bound and unbound RNA during the MD-LES (red and black curves) and MD+LES trajectories (green and blue curves). (C) φ angle of the bound and unbound RNA during the MD-LES (red and black curves) and MD+LES trajectories (green and blue curves). (D) Closed conformation of the K-turn (φ = 20°–40°). (E) Open conformation of the K-turn (φ = 50°–80°). In C and D the loop attached to the NC-stem is not shown and the φ angle is shown in black in C and D (see Fig. 1A).
MD+LES simulations
When LES is applied, the unbound RNA undergoes a dramatic conformational transition with RMSD values reaching peak values of ∼7 Å (∼4 Å greater than the peak deviation of the bound RNA) after ∼7 nsec (Fig. 2B). Interestingly, the K-turn partially reforms toward the end of the trajectory. The open conformation is characterized by φ values of 50°–90° (Fig. 2C). A snapshot of the open conformation is shown in Figure 2D. Although the bound RNA opens slightly after ∼8.5 nsec of the LES trajectory, the K-turn conformation is not significantly altered (φ is relatively constant). Figure 2E shows a snapshot with the closed conformation of the K-turn. The opening of the K-turn is accompanied by local conformational transitions summarized in Table 1.
TABLE 1.
Conformational transitions in the RNA (K2)
| Conformational transition | Bound RNA (MD − LES) | Unbound RNA (MD − LES) | Bound RNA (MD + LES) | Unbound RNA (MD + LES) |
| Opening of the K-turn Unpaired A30 | No | Partial | No | Yes |
| Syn > high anti/anti C2′-endo > C3′-endo | No | Yes | No | Yes |
| Loss of G-A base pairs | No | No | No | Yes |
| Loss of interstrand contacts | No | Yes | No | Yes |
The purine-rich internal loop is flexible in the absence of the 15.5K protein
The internal loop of the 5′ stem–loop of U4 snRNA consists of two tandem-sheared G-A base pairs (G32–A44, G43–A33) and three unpaired nucleotides (A29, A30, U31). The four adenines establish a “3 + 1” stacking scheme in which A30 stacks on A44, which further stacks on A33. The A29 stacks on the G45–C28 base pair of the C-stem and makes hydrophobic contacts and hydrogen bonds with the Arg97 of the 15.5K protein. The guanines G32 and G43 establish contacts with the 15.5K protein, and U31 is flipped out and trapped in a pocket of the protein. Only A29 and A30 can be mutated to other purines without abolishing protein binding (Nottrott et al. 1999; Vidovic et al. 2000).
MD-LES simulations
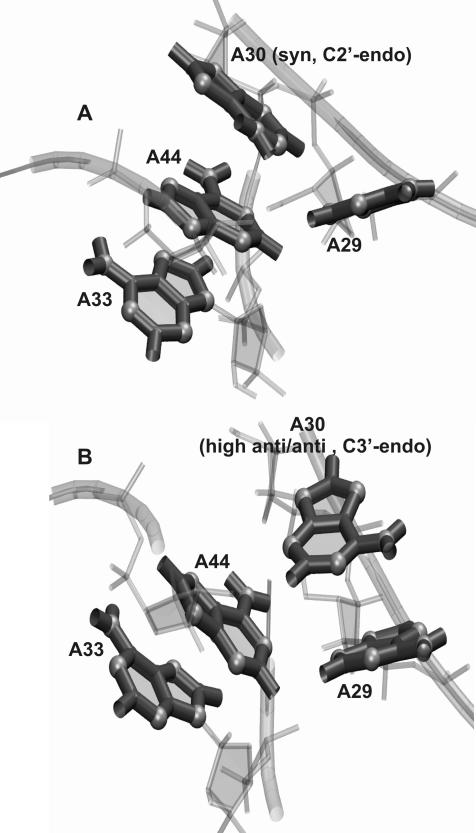
During the MD-LES trajectories of the unbound RNA the “3 + 1” stacking scheme (Fig. 3A) undergoes a conformational transition to a “2 + 2” scheme (Fig. 3B). The adenine A30 changes its conformation from a C2′ endo sugar pucker and a syn base-sugar orientation to a C3′ endo sugar and a high-anti (or anti) base-sugar orientation. The percentages in which A30 is found in either conformation during different trajectories are presented in Table 2. The transition occurs after ∼8 nsec of the MD-LES trajectory of the unbound RNA. In the complex, the A30 is held in its syn/C2′ endo conformation by Lys37, which makes a stable contact network between the N7 atom of A30 and the phosphate backbone of the RNA. The conformational transition of the stacking scheme does not induce the opening of the K-turn; during the last 2 nsec of the MD-LES trajectory of the unbound RNA the K-turn remains closed (φ = ∼20°) while the new stacking scheme forms.
FIGURE 3.
Conformational states of A30 (K2 RNA). (A) “3 + 1” stacking scheme formed by adenines A29, A30, A44, and A33 in the bound RNA, A30 being in a syn/C2′ endo conformation. (B) “2 + 2” stacking scheme formed by adenines A29, A30, A44, and A33 during the trajectories of the unbound RNA, A30 being in a high-anti, anti/C3′ endo conformation.
TABLE 2.
Conformational states of adenine A30
| Sugar pucker (A30) (%) | χ angle (A30) (%) | |||
| Trajectory | C2′- endo | C3′- endo | Syn | High anti/anti |
| Bound K2 RNA (-LES) | 80 | 0 | 93 | 0 |
| Unbound K2 RNA (-LES) | 63 | 9 | 72 | 22 |
| Bound K2 RNA (+LES) | 36 | 0 | 21 | 1 |
| Unbound K2 RNA (+LES) | 16 | 24 | 10 | 84 |
The tandem sheared G-A base pairs are stable during the MD-LES trajectories of the bound and unbound RNA. Nevertheless, several differences are observed when comparing the trajectories of the unbound and bound RNAs. In particular, the displacement of the G32 and G43 is minimized in the unbound RNA, resulting in a better stacking of the two guanines, and triggering a slight increase in the propeller twist of the two G-A base pairs. In the complex, G32 makes contacts with the protein residue Glu40 and G43 establishes hydrogen bonds with Asn40 and Lys44. These contacts result in a displacement between the two guanines that is maintained during the trajectories of the complex.
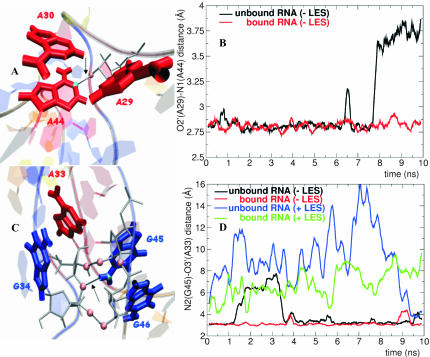
Several interstrand contacts contributing to the stability of the kinked conformation are formed in the region of the internal loop: (1) the 2′ OH group of A30 is hydrogen-bonded with one oxygen atom from the phosphate group of U31; (2) the 2′ OH group of U31 is hydrogen-bonded with one oxygen atom from the phosphate group of A30; and (3) the 2′ OH group of A29 is hydrogen-bonded with the N1 atom of A44 (Fig. 4A). These contacts are well preserved during the MD-LES trajectory of the bound RNA but are lost in the unbound RNA. The hydrogen-bonding distance between the 2′ OH group of A29 and the N1 atom of A44 is plotted in Figure 4B.
FIGURE 4.
Interstrand contacts established in the RNA (K2 RNA). (A) The interstrand contact formed between the O2′ atom of A29 and the N1 atom of A44. (B) Hydrogen-bonding distance between the O2′ atom of A29 and the N1 atom of A44 in the bound (red curve) and unbound RNA (black curve) during the MD-LES trajectories. (C) The interstrand contact formed between the N2 atom of G45 and the O3′ atom of A33. (D) Hydrogen-bonding distance between the N2 atom of G45 and the O3′ atom of A33 in the bound and unbound RNA during the MD-LES (red and black curves) and MD+LES (green and blue curves) trajectories.
MD+LES simulations
When applying LES, we observed the same conformational transition of the stacking scheme in the unbound RNA on a shorter time scale. Owing to the increased conformational sampling, the number of intermediate states between the C2′ endo and C3′ endo conformations increases both for the bound and the unbound RNA. Nevertheless, there is a significant increase in the fractional population of the C3′ endo sugar pucker in the unbound RNA (Table 2).
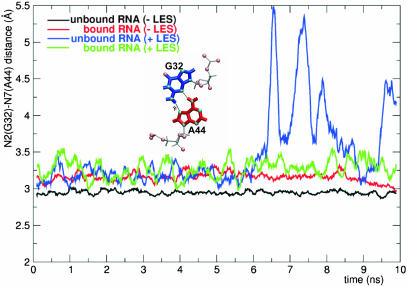
Both G-A base pairs open during the MD+LES trajectories of the unbound RNA. Figure 5 shows the hydrogen-bonding distance between the N2 atom of G32 and the N7 atom of A44. The transition to the open conformation of the K-turn triggers the loss of the G-A base pairs.
FIGURE 5.
The G32–A44 base pair (K2 RNA); hydrogen-bonding distance between the N2 atom of G32 and the N7 atom of A44 in the bound and unbound RNA during the MD-LES (red and black curves) and MD+LES (green and blue curves) trajectories. The hydrogen bond is pointed with a double arrowhead.
The NC-stem is unstable in the absence of the 15.5K protein
To assess the stability of the G34–C42 and G35–C41 base pairs, we calculated the percentage of trajectory frames in which the constituent hydrogen bonds were formed. The criteria chosen for hydrogen-bond formation were (1) maximal donor–acceptor distance of 3.15 Å and (2) maximal donor-hydrogen–acceptor angle of 60°. In Table 3 we show the results for the standard MD trajectories of K1 RNA and the LES trajectories of K2 RNA in the bound and unbound forms.
TABLE 3.
Stability of the Watson-Crick base pairs of the NC-stem
| G34–C42 base pair (%)a | G35–C41 base pair (%)a | |||||
| Trajectory | HB1b | HB2b | HB3b | HB1b | HB2b | HB3b |
| Bound K1 RNA (-LES) | 99 | 98 | 96 | 96 | 95 | 88 |
| Unbound K1 RNA (-LES) | 96 | 83 | 63 | 32 | 24 | 20 |
| Bound K2 RNA (+LES) | 70 | 62 | 53 | 52 | 44 | 44 |
| Unbound K2 RNA (+LES) | 62 | 52 | 36 | 17 | 15 | 14 |
aThe percentage of trajectory frames in which the hydrogen bond is formed
bThe three hydrogen bonds of the G-C base pairs are HB1 = N2(G)–H(G)–O2(C); HB2 = N1(G)–H(G)–N3(C); HB3 = N4(C)–H(C)–O6(G).
MD-LES simulations
In the bound K1 RNA both base pairs are stable, whereas in the unbound K1 RNA the G35–C41 base pair opens and the G34–C42 base pair is less stable. During the MD-LES trajectories of K2, K3, K4, and K5 RNAs the NC-stem is stable (data not shown). The numbers in the first two rows of Table 3 show that the opening of the G35–C41 base pair occurs only in the unbound K1 RNA. Various interstrand contacts are established between the NC-stem and the C-stem: (1) the 2′ OH group of A33 is hydrogen-bonded with the 2′ OH group of G45; (2) the N2 atom of G45 is hydrogen-bonded with the O3′ atom of the sugar of A33; and (3) the 2′ OH group of G46 is hydrogen-bonded with one of the oxygen atoms of the phosphate group of G34. All these contacts are well preserved during all the trajectories of the bound RNA. Figure 4, C and D, shows that the hydrogen-bond distance between the N2 atom of G45 and the O3′ atom of A33 is strictly correlated with the partial opening of the K-turn during the MD-LES trajectories (the loss of the hydrogen bond is observed in the same interval as the increase in the value of φ).
MD+LES simulations
When applying LES, both base pairs remain relatively stable in the bound RNAs. The partial loss in stability compared to the non-LES trajectories reflects the increased sampling density. In both K1 and K2 unbound RNAs, the G35–C41 base pair opens and the G34–C42 base pair is less stable than in the bound RNAs, the opening of G35–C41 base pair occurring much faster in K1 RNA. The numbers in the last two rows of Table 3 show that the stability of the NC-stem is dependent on the protein binding, not only in K1 RNA but also in K2 RNA. Similar results were found for K3 and K4 RNAs, while in K5 RNA the NC-stem is stable regardless of whether the RNA is bound or not to the protein. During the MD+LES trajectories the interstrand hydrogen bond between the N2 atom of G45 and O3′ atom of A33 is lost for both the bound and unbound RNAs. Nevertheless, the distance between the two atoms is significantly larger in the unbound RNA (Fig. 4D).
The external loop has different trajectories in the bound and unbound RNAs
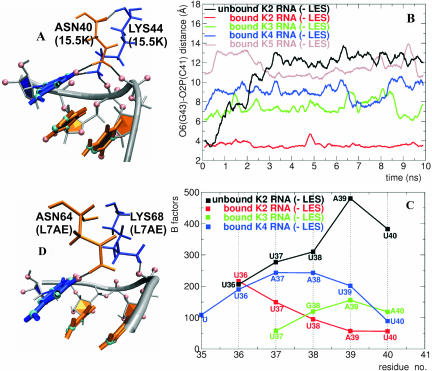
During the non-LES trajectories of the bound K2 RNA, the sugar-phosphate backbone has a turn between the residues U40 and C41 and forms a groove that encloses the G-C base pairs of the NC-stem (Fig. 6A). This structure remains relatively stable during the simulations of the bound RNA but is lost in the unbound RNA after only hundreds of picoseconds. The turn of the backbone is also observed in the crystal structure from the orientation of the phosphate group of C41. Two amino acid residues, Asn40 and Lys44, are important in maintaining this structure, forming a bridge between G43 and the backbone of the external loop. The NH2 group of Asn40 establishes hydrogen bonds with the O6 of G43 and with one oxygen atom of the phosphate group of C41, and the NH3+ group of Lys44 establishes hydrogen bonds with the N7 of G43 and an oxygen atom of the phosphate group of C42 (Fig. 6A). Figure 6B shows that the bridging distance between the O6 atom of G43 and the phosphate group of C41 significantly increases during the trajectories of the unbound RNA when comparing the naturally occurring K2 RNA in its bound and unbound forms, while a comparison of the trajectories of the bound K2 RNA with the bound K3, K4, and K5 RNAs reveals that the bridging distance is dependent on the loop flexibility (Fig. 6B). The UGAA tetraloop has a fold in which the uracil stacks on the guanine and the two adenines stack together (Butcher et al. 1997). The UUAAUU hexaloop has a further U-U base pair lengthening the NC-stem to 3 bp (Zhang et al. 2001). Both structures prevent the backbone from turning over C41 and forming hydrogen bonds with Asn40 and Lys44, as observed in the case of the UUUAU pentaloop. The pentaloop is the only loop in which the nucleotide next to C41 lacks stacking interactions with C41. To analyze the flexibility of the three different loops in the bound and unbound forms of RNA, we calculated the B-factors per residue during the non-LES trajectories. From a comparison of the fluctuations of the UUUAU loop in the bound and unbound RNA, there is a significant decrease in flexibility from U36 to U40 in the bound RNA (Fig. 6C, black and red curves), suggesting that the 15.5K protein stabilizes the orientation of the backbone of the loop in the vicinity of C41. Nevertheless, the absolute values of the B-factors indicate that the pentaloop is still very flexible, suggesting that other factors might contribute to its stability. In the RNAs bound to the 15.5K protein, the flexibility is lower at the end nucleotides and higher at the core nucleotides of the UGAA and UUAAUU loops (Fig. 6C, blue and green curves). This behavior is expected because the end nucleotides are covalently linked to a stable structure (C41 and G35, respectively). Although the UUUAU loop was modeled and the structures of UGAA and UUAAUU loops have been experimentally determined, the flexibility of the nucleotide N40 is lowest in the pentaloop.
FIGURE 6.
Interactions between the 15.5K protein and the external loop. (A) Asn40 and Lys44 of the 15.5K protein establish a bridge between G43 and the sugar-phosphate backbone of the external loop of the 5′ stem–loop of U4snRNA. (B) Bridging distance between the O6 atom of G43 and the O2P atom of C41 for the unbound K2 RNA (black curve) and for the bound K2 (red curve), K3 (green curve), and K4 (blue curve) and K5 (brown curve) RNAs during the MD-LES trajectories; the bridge is shown in A. (C) B-factors per residue for the nucleotides of the external loop for the unbound K2 RNA (black curve) and for the bound K2 (red curve), K3 (green curve), and K4 (blue curve) RNAs during the MD-LES trajectories. (D) Asn64 and Lys68 of the ribosomal L7AE protein establish a similar bridge between G43 and the sugar-phosphate backbone of the Kt15 RNA.
Chemical accessibility of RNA bases: Excellent agreement with the simulations
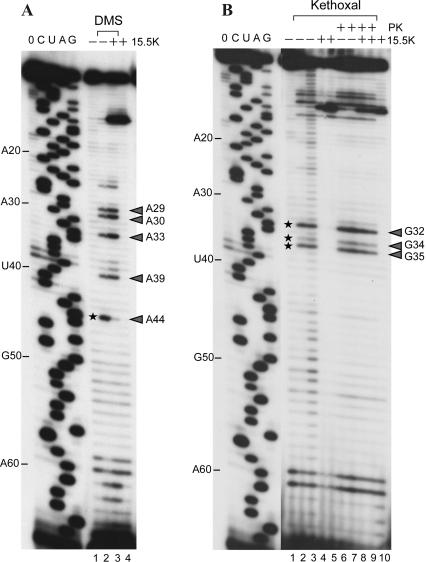
Chemical RNA modification studies (Ehresmann et al. 1987) using dimethylsulfate (DMS) indicate that the N1 position of A44 is clearly accessible, permitting chemical modification in the absence, but not in the presence of 15.5K protein (Fig. 7A, cf. lanes 2 and 3). These data are in excellent agreement with the simulations showing that the interstrand contact between the N1 atom of A44 and the 2′ OH group of A29 is preserved during the trajectories of the bound RNA but is unstable during the trajectories of the unbound RNA.
FIGURE 7.
Chemical RNA structure probing. (A) Primer extension analysis of U4 snRNA after DMS treatment of the RNA either in the absence or presence of recombinant 15.5K protein (lanes 2,3); (lanes 1,4) control lanes, no DMS modification. (B) Primer extension analysis of U4 snRNA after Kethoxal treatment either in the absence (lanes 2,3,6,7) or presence (lanes 4,5,8,9) of 15.5K protein; (lanes 6–9) RNA modification after Proteinase K digestion; (lanes 1,10) control lanes, no Kethoxal treatment; the modification of an RNA base results in a stop of the reverse transcriptase 1 nt before the site of attack (Ehresmann et al. 1987); modified nucleotides are indicated by an arrowhead; nucleotides that are clearly protected from chemical modification in the presence of bound 15.5K protein are marked by asterisks; the presence or absence of the 15.5K protein is indicated by “+” or “-,” respectively. C, U, A, and G refer to dideoxysequencing reactions and correspond to the sequence of human U4 snRNA; 0 indicates a control primer extension with unmodified U4 snRNA where no ddNTPs were added to the reaction; and the position of every tenth nucleotide of the U4 snRNA is indicated on the left in panels A and B, respectively.
RNA structural probing with Kethoxal shows that the nucleotides G32, G34, and G35 are clearly accessible for modification in the absence, but not in the presence of 15.5K protein (Fig. 7B, cf. lanes 2,3 and 6,7 with lanes 4,5). In addition, the nucleotides G32, G34, and G35 are also clearly accessible for modification with Kethoxal after digestion of the bound 15.5K with Proteinase K (Fig. 7B, lanes 8,9). The N2 atom of G32 is involved in the base-pairing interaction of G32 with A44, while the N1 and N2 positions of G34 and G35 form hydrogen bonds with C42 and C41. Therefore, these interactions do not occur in the absence of the 15.5K protein. These findings complement the observations that the tandem-sheared G-A base pairs and the two G-C base pairs of the NC-stem open during the MD+LES trajectories of the unbound RNA. The increased accessibility of G35 compared to G34 reflects the observation from the simulation, that the G34–C42 base pair is more stable than the G35–C41 base pair.
DISCUSSION
Protein-assisted RNA folding
We propose that the folding of the U4 snRNA K-turn is assisted by the protein binding. The folding pathway is largely dependent on the RNA sequence and local flexibility. Our model is based on the susceptibility to chemical modifications of different bases of the unbound and bound RNA and on MD simulations indicating significantly different trajectories for the bound and unbound RNAs. The protein cannot bind to the RNA and promote its folding unless the RNA has the inherent flexibility conferred upon it by the particular nucleotide sequence.
By probing the chemical reactivity of the N1 and N2 positions of guanines (using Kethoxal) and the N1 position of adenines (using DMS) in the presence and in the absence of 15.5K protein we show that certain interactions in the RNA arise only upon protein binding. Conversely, the simulations indicate that conformational transitions with loss of specific secondary structure elements occur only in the unbound RNA. These findings, summarized in Table 4, suggest that the RNA folding is assisted by the binding of 15.5K protein. The K-turn opens and sometimes reforms in the absence of the protein, suggesting that in addition to protein binding, the local flexibility of the RNA plays a crucial role in the folding process. In most of the trajectories the K-turn does not reform from the open conformation within the 10-nsec time limit, only the local conformational transitions being reversible. The transition between the close and open conformations was observed using FRET for the K-turn Kt7, which contains the most of the consensus sequence of K-turn motifs (Goody et al. 2004). The opening–closing process of the K-turn observed by FRET may be quite different and occur on a longer time scale. Nonetheless, the present simulations show a significant degree of opening for the K-turn in the absence of the protein.
TABLE 4.
Interactions in the unbound RNA
| Interaction | Unbound RNA (MD − LES) | Unbound RNA (MD + LES) | Unbound RNA (experiment) |
| G35–C41 base pair | Lost (only K1) | Lost (K1, K2) | Lost |
| G32–A44 base pair | Preserved | Lost | Lost |
| G43–A33 base pair | Preserved | Lost | No data |
| A30–A44 stack | Lost | Lost | No data |
| A30–A29 stack | Formed | Formed | No data |
| O2′ (A29)–H–N1(A44) | Lost | Lost | Lost |
| N2(G45)–H–O3′ (A33) | Preserved | Lost | No data |
The K-turn formed by the 5′ stem–loop of U4 snRNA is of particular interest because the binding of 15.5K protein to this RNA nucleates the assembly of U4/U6 snRNP (Nottrott et al. 2002). Several other studies devoted to the mechanism of K-turn RNA folding suggest that factors such as the association with proteins and the presence of metal ions in solution are important (Matsumura et al. 2003; Goody et al. 2004).
Previously, interactions such as Tat–TAR interaction of HIV-1 or the binding of U1A protein to its RNA substrate have been assigned to the induced fit category (Puglisi et al. 1992, 1995; Oubridge et al. 1994; Avis et al. 1996; Gubser and Varani 1996; Aboulela and Varani 1998; Varani et al. 2000; Reyes et al. 2001). In these cases the conformation of the unbound RNA is stable but significantly differs from that of the bound RNA. The conformational capture (as described by Leulliot and Varani 2001) is more difficult to locate due to the large flexibility of the RNA, which adopts an equilibrium between different conformations in its unbound state such that structure determination is not feasible. The MD simulations do not sample continuous interconversions between conformations since such events occur in the microsecond-to-millisecond time scale. Nevertheless, the FRET study by Goody et al. (2004) and the partial reforming of the kinked conformation during the simulations of the unbound RNA (Fig. 2C) might suggest a “conformational capture” mechanism for the folding of the 5′ stem–loop of the U4 snRNA. In addition, the folding process is influenced by other factors such as ionic strength and magnesium ions, factors that were not considered in the current study. However, localized Mg ions were not found in the crystal structure of the complex, suggesting that they influence the folding of the RNA by a “diffusely bound” rather than “site-bound” mechanism (Misra and Draper 1998; Auffinger et al. 2003, 2004a,b).
From the six K-turns found in the 23S rRNA of H. marismortui (Klein et al. 2001), one (Kt38) is not associated with proteins, suggesting that the folding of the K-turn motif is not always assisted by the protein binding, but it also depends on the structural context in which the K-turn forms. The K-turns from the ribosome are segments of large RNA structures and therefore might have a folding pathway different from the 5′ stem–loop of U4 snRNA, which is an isolated small RNA with a very flexible external loop attached to the NC-stem.
Computer simulations of protein-assisted RNA folding
Structural data are rarely available for both the free and bound RNAs in cases of protein-assisted RNA folding. Computer simulation techniques provide the means for exploring the structure of the RNA in its unbound form and drawing conclusions about flexible regions contributing significantly to the folding process. For MD simulations, the starting structure is essential and dictates the evolution of the system. They are easily trapped in local minima on the potential energy surface. The time scale of conformational transitions important for the RNA folding is often much greater than that available for the simulations. MD simulations have been applied previously to U1A-RNA binding (Pitici et al. 2002). In a recent study, which appeared after the submission of this manuscript, Rázga et al. (2004) reported 35–40-nsec MD simulations of three different K-turns and derived conclusions about the dynamics of the K-turn around the conformation observed in the ribosomal crystal structure. Neither their standard MD simulations nor ours demonstrated the transition between the closed and open conformations of the K-turn, only partial opening being observed. Thus, we opted for LES to increase the conformational sampling during the MD trajectories. A major advantage of this method is that it can be applied in combination with PME and explicit solvent (Simmerling et al. 1998b). Nevertheless, smoothing the potential energy surface might permit transitions that do not occur in reality. Therefore, the choice of the LES regions and the number of copies are crucial.
In this study we presented trajectories with LES systems containing four copies for four different regions. When the entire core structure of the K-turn RNA (K1 RNA) was replaced with three identical copies, the transition between the closed and open conformations was not observed, while with five copies the transition occurred for the unbound RNA on a shorter time scale but also for the bound RNA. The crystal structure of the complex was preserved during all viable simulations, its stability being the criteria for accepting trajectories for further processing. Using different combinations of LES regions, we obtained different trajectories but observed the same local conformational transitions in the unbound RNA. Nevertheless, the transition from a closed to an open K-turn took place only when different LES regions were applied for the internal loop, the NC-stem, and parts of the C-stem, suggesting that the kinked conformation is an intrinsic property of the entire RNA (data not shown).
LES proved very useful for studying large conformational transitions such as the opening of the K-turn, while the standard MD simulations provided a more accurate description of fine contacts such as the interstrand hydrogen bonds.
RNA folding and U4/U6 snRNP assembly
The 15.5K protein is required for the recruitment of other specific proteins to the U4/U6 snRNP such as the 61K or the 20/60/90K protein complex (Nottrott et al. 2002). We propose that the assembly of U4/U6 snRNP is driven by the hierarchical folding of the U4 snRNA 5′ stem–loop. First, the RNA is folded into a K-turn conformation upon binding of the 15.5K protein, while the pyrimidine-rich external loop remains flexible. The simulations of K2 RNA show that the external loop adopts different conformations in the presence and absence of the protein, suggesting that the 15.5K protein creates a pre-folded structure of the external loop. It is very likely that the complete folding of the UUUAU loop is achieved only after recruitment of 61K protein to the U4 snRNA. When replacing the UUUAU loop with the UGAA, the UUAAUU loops, or seven Watson-Crick base pairs, the backbone cannot adopt the orientation required for establishing hydrogen bonds with Asn40 and Lys44, suggesting that this orientation might be the first step in the folding of the external loop.
The naturally occurring RNA (K2) is the only simulated construct in which C41 does not stack on the nucleotide N40 of the loop. Furthermore, electrostatic attractive forces between the protein helix α2 and the RNA backbone trigger structural changes in the external loops of the bound K3 and K4 RNAs: (1) loss of stacking between C41 and A40 in the UGAA loop (K3 RNA) and (2) instability of the U–U40 base pair in the UUAAUU loop (K4 RNA). These findings, together with the experimental observation that the addition of one Watson-Crick G-C base pair to the NC-stem inhibits the binding of the 61K protein in vitro (data not shown), suggest that the lack of a stacking interaction between C41 and N40 may be critical for the recruitment of 61K protein to the U4 snRNA.
During the non-LES simulations, the NC-stem proved less stable in the unbound K1 RNA than in the bound K1 RNA, the external loops (K2, K3, K4 RNAs) or subsequent Watson-Crick base pairs (K5 RNA) stabilizing the stem in the absence of the protein. The LES simulations have shown that protein binding plays a role in the stabilization of the G34–C42 and G35–C41 base pairs when the NC-stem is attached to flexible external loops. Furthermore, the chemical probing experiments have shown that the NC-stem is unstable in the unbound RNA. These findings suggest that a nonrigid external loop is required to keep the NC-stem relatively flexible in the unbound RNA. Therefore, it is likely that the NC-stem folds together with the G-A base pairs and the external loop upon binding of the 15.5K protein.
The K-turn “Kt15” shares many features with the 5′ stem–loop of U4 snRNA
To find whether our model for the external loop shares structural features with other K-turns, we extracted the K-turns and their associated proteins from the big ribosomal subunit of H. marismortui and investigated their 3D structures.
Interestingly, the K-turn “Kt15” shares many features with the 5′ stem–loop of U4 snRNA. The only difference is that instead of the G43–A33 base pair (in U4 snRNA), the Kt15 has an A-U-G triplex. The NC-stem has two Watson-Crick base pairs extended with a loop-like structure different from the UUUAU loop of U4 snRNA. The Kt15 binds to L7AE protein, which has a similar fold to 15.5K protein. The loop-like structure in the Kt15 shows an orientation of the backbone similar to that observed for the UUUAU loop during the trajectories of the bound K2 U4 snRNA. In the L7AE protein two residues (Asn64 and Lys68) establish a bridge between the G nucleotide of the A-U-G triplex and the backbone of the loop-like structure (Fig. 6D) in an almost identical manner as the Asn40 and Lys44 residues from 15.5K protein, suggesting that there might be a common folding path shared by the K-turn of U4 snRNA and Kt15.
MATERIALS AND METHODS
RNA constructs
Five different RNA constructs shown in Figure 1 were used in the MD simulations. K1 RNA (Fig. 1A) is the 5′ stem–loop of U4 snRNA taken from the crystallographic structure of its complex with the 15.5K protein (pdbid 1E7K) (Vidovic et al. 2000). K2 RNA (Fig. 1B) contains the naturally occurring UUUAU external loop attached to the NC-stem. Since a similar pentaloop structure was unavailable in the databases, we applied a restrained simulated annealing (RSA) protocol to obtain an initial structure of the loop. A single-stranded RNA (5′-GGUUUAUCC-3′ ) was constructed using the NAB software (Macke and Case 1998). During the RSA trajectory the two guanines from the 5′ end were forced to form base pairs with the two cytosines from the 3′ end, thus adopting exactly the same conformation as in the crystal structure. The loop was allowed to move freely and explore the conformational space. The RSA protocol consists of 20 psec of vacuum MD simulation with distance-dependent dielectric constant. In the first picosecond the temperature was rapidly increased to 1000 K. For the next 4 psec the temperature was maintained at 1000 K, allowing the loop to sample the conformational space. The annealing step was achieved with a slow cooling phase followed by a fast cooling in the last 2 psec. The two G-C base pairs from the new stem–loop were overlaid on the two G-C base pairs from the NC-stem in the complex. Simulated annealing was then performed using the new complex structure with the loop attached. All the residues from the crystal structure were fixed, so as to allow only loop movement. The resulting structure was then used as input for MD simulations. K3 RNA (Fig. 1C) has a UGAA tetraloop (pdbid 1AFX) in which U37 stacks on G38, C41 stacks on A40, and A40 stacks on A39 (Butcher et al. 1997), while K4 RNA (Fig. 1D) has a UUAAUU loop (pdbid 1HS3) in which the nucleotide U establishes a non-Watson-Crick base pair (N3–H–O2 and N3–H–O4 hydrogen bonds, sugars in cys orientation) with U40 (Zhang et al. 2001). K5 RNA (Fig. 1E) has the C-stem extended with seven Watson-Crick base pairs and the NC-stem with six Watson-Crick base pairs.
Molecular dynamics simulations
For MD simulations, 10-nsec MD trajectories were obtained for the complex, the unbound protein, and the unbound RNA using all the RNA constructs described. The complexes and the unbound RNAs were neutralized with Na+ ions. All the systems were dissolved in a box of TIP3P water with 12 Å distance between the edges of the box and the solute. Periodic boundary conditions were applied. The particle mesh Ewald (PME) (Darden et al. 1993) method was used for the treatment of the electrostatic interactions. The SHAKE algorithm was applied to all the bonds involving hydrogen atoms, thus allowing the use of a 2-fsec integration time step (Ryckaert et al. 1977). All the simulations were run at a constant temperature of 300 K and at a constant pressure of 1 atm using the Berendsen’s coupling algorithm (Berendsen et al. 1984). The calculations were performed using the Cornell et al. force field (Cornell et al. 1996) and the NAMD program (Kale et al. 1999) on a p690 IBM cluster running on 16 CPUs. For the biggest system (the complex of 15.5K protein with K5 RNA) the calculation of one trajectory required ∼500 h of CPU time. For the smallest system (the unbound K1 RNA) the calculation required ∼250 h. For testing the reproducibility of the results we ran multiple trajectories using slightly different initial conditions and different software packages. The visualization and analysis of the trajectories were performed with VMD (Humphrey et al. 1996) and different programs available in the distribution of the AMBER 7 software package (University of California, San Francisco) (Pearlman et al. 1995).
Locally enhanced sampling
In the locally enhanced sampling (LES) method (Elber and Karplus 1990; Simmerling and Elber 1994; Simmerling and Kollman 1996; Simmerling et al. 1998a,b, 2000), the system is divided into separate regions and one or more regions of interest are replaced with multiple copies. The potential energy function of the LES system is constructed in such a way that the potential energy is identical to the corresponding single-copy (non-LES) system in which all the copies occupy the same position. As a result of this feature and the fact that the copies have no direct interaction, the global minimum of this potential energy is the same as for the original system. The copies from one region do not interact with each other but interact in an average way with all the copies of the other regions. In this way the energy potential surface is smoothened and the copies are allowed to sample more of the conformational space. The enhanced sampling is achieved with a small increase in the required CPU time compared to the non-LES system.
We divided K1 RNA into four LES regions: (1) the two G-C base pairs from the NC-stem, (2) the unpaired nucleotides, (3) the two tandem-sheared G-A base pairs, and (4) two of the three G-C base pairs from the C-stem. Each region was replaced by four identical copies. We applied LES coupled with the PME summation method with explicit solvent and periodic boundary conditions. Thus 10-nsec trajectories were obtained for the LES systems of the complex and of the unbound RNA using K1 and K2 RNAs. Shorter test trajectories were run for the unbound K3, K4, and K5 RNAs. The equilibrated non-LES structures have been used as input for the LES simulations.
RNA modification and structural probing
In vitro transcribed U4 snRNA (0.6 pmol) was incubated either in the presence or absence of recombinant 15.5K protein at a final protein concentration of 14 μM as described (Nottrott et al. 1999). Proteinase K digestions were performed for 1 h at 37°C at a final enzyme concentration of 1 μg/μL. DMS modification of the RNA was performed according to Hartmuth et al. (1999). For RNA modification with Kethoxal (Research Organics, Inc., USA), the reaction mixtures were incubated with 10 μL of a 1:10 dilution of the stock reagent in a final volume of 100 μL of 50 mM K+ cacodylate (pH 7.0), 50 mM KCl, and 10 mM MgCl2 for 45 min at 4°C. Reactions were stopped by precipitating the RNA in the presence of ethanol and glycogen, and modification of the RNA was subsequently analyzed by primer extension according to Nottrott et al. (1999). Dideoxy-sequencing reactions on the U4 snRNA transcript were performed in parallel. Primer extension products were separated on a 9.6% (w/v) polyacrylamide/8.3 M urea sequencing gel and visualized by autoradiography. DMS reacts with the N1 position of adenines, while treatment with Kethoxal leads to the formation of a new ring involving the N1 and N2 positions of unpaired guanines and both carbonyl groups of Kethoxal (Ehresmann et al. 1987).
Acknowledgments
We are grateful to Reinhard Lührmann for constant support and helpful discussion. We thank Klaus Hartmuth for advice regarding RNA structural probing experiments, and Pascal Auffinger and Eric Westhof for discussion regarding the simulations. This work is part of the scientific research conducted in the International Ph.D. Program Molecular Biology–International Max Planck Research School at the Georg August University, Göttingen.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7149605.
REFERENCES
- Aboulela, F. and Varani, G. 1998. Recognition of HIV-1 TAR RNA by Tat protein and Tat-derived peptides. J. Mol. Struct. 423: 29–39. [Google Scholar]
- Auffinger, P. and Westhof, E. 2000. Water and ion binding around RNA and DNA (C,G) oligomers. J. Mol. Biol. 300: 1113–1131. [DOI] [PubMed] [Google Scholar]
- Auffinger, P., Bielecki, L., and Westhof, E. 2003. The Mg2+ binding sites of the 5S rRNA loop E motif as investigated by molecular dynamics simulations. Chem. Biol. 10: 551–561. [DOI] [PubMed] [Google Scholar]
- Auffinger, P., Bielecki, L., and Westhof, E. 2004a. Symmetric K+ and Mg2+ ionbinding sites in the 5S rRNA loop E inferred from molecular dynamics simulations. J. Mol. Biol. 335: 555–571. [DOI] [PubMed] [Google Scholar]
- ———. 2004b. Anion binding to nucleic acids. Structure 12: 379–388. [DOI] [PubMed] [Google Scholar]
- Avis, J.M., Allain, F.H.T., Howe, P.W.A., Varani, G., Nagai, K., and Neuhaus, D. 1996. Solution structure of the N-terminal RNP domain of U1A protein: The role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol. 257: 398–411. [DOI] [PubMed] [Google Scholar]
- Berendsen, H.J.C., Postma, J.P.M., Vangunsteren, W.F., Dinola, A., and Haak, J.R. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81: 3684–3690. [Google Scholar]
- Blakaj, D.M., McConnell, K.J., Beveridge, D.L., and Baranger, A.M. 2001. Molecular dynamics and thermodynamics of protein–RNA interactions: Mutation of a conserved aromatic residue modifies stacking interactions and structural adaptation in the U1A-stem loop 2 RNA complex. J. Am. Chem. Soc. 123: 2548–2551. [DOI] [PubMed] [Google Scholar]
- Bringmann, P., Appel, B., Rinke, J., Reuter, R., Theissen, H., and Lührmann, R. 1984. Evidence for the existence of snRNAs U4 and U6 in a single ribonucleoprotein complex and for their association by intermolecular base-pairing. EMBO J. 3: 1357–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow, D.A. and Guthrie, C. 1988. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334: 213–218. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T.H., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosome. In The RNA world (eds. R.F. Gesteland et al.), pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Butcher, S.E., Dieckmann, T., and Feigon, J. 1997. Solution structure of the conserved 16 S-like ribosomal RNA UGAA tetraloop. J. Mol. Biol. 268: 348–358. [DOI] [PubMed] [Google Scholar]
- Chao, J.A. and Williamson, J.R. 2004. Joint X-Ray and NMR refinement of the yeast L30e–mRNA complex. Structure 12: 1165–1176. [DOI] [PubMed] [Google Scholar]
- Cheatham, T.E. and Brooks, B.R. 1998. Recent advances in molecular dynamics simulation towards the realistic representation of bio-molecules in solution. Theor. Chem. Acc. 99: 279–288. [Google Scholar]
- Cheatham, T.E. and Kollman, P.A. 2000. Molecular dynamics simulation of nucleic acids. Annu. Rev. Phys. Chem. 51: 435–471. [DOI] [PubMed] [Google Scholar]
- Cheatham, T.E., Miller, J.L., Fox, T., Darden, T.A., and Kollman, P.A. 1995. Molecular dynamics simulations on solvated biomolecular systems—The particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J. Am. Chem. Soc. 117: 4193–4194. [Google Scholar]
- Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Merz, K.M., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., and Kollman, P.A. 1996. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117: 5179–5197. [Google Scholar]
- Darden, T., York, D., and Pedersen, L. 1993. Particle mesh Ewald—An N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 98: 10089–10092. [Google Scholar]
- Ehresmann, C., Baudin, F., Mougel, M., Romby, P., Ebel, J.P., and Ehresmann, B. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15: 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elber, R. and Karplus, M. 1990. Enhanced sampling in molecular dynamics—Use of the time-dependent Hartree approximation for a simulation of carbon monoxide diffusion through myoglobin. J. Am. Chem. Soc. 112: 9161–9175. [Google Scholar]
- Goody, T.A., Melcher, S.E., Norman, D.G., and Lilley, D.M.J. 2004. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA 10: 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubser, C.C. and Varani, G. 1996. Structure of the polyadenylation regulatory element of the human U1A pre-mRNA 3′-untranslated region and interaction with the U1A protein. Biochemistry 35: 2253–2267. [DOI] [PubMed] [Google Scholar]
- Guo, J., Daizadeh, I., and Gmeiner, W.H. 2000. Structure of Sm binding site from human U4 snRNA derived from a 3 ns PME molecular dynamics simulation. J. Biomol. Struct. Dyn. 18: 335–344. [DOI] [PubMed] [Google Scholar]
- Hartmuth, K., Raker, V.A., Huber, J., Branlant, C., and Lührmann, R. 1999. An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J. Mol. Biol. 285: 133–147. [DOI] [PubMed] [Google Scholar]
- Hashimoto, C. and Steitz, J.A. 1984. U4 and U6 RNAs coexist in a single small nuclear ribonucleoprotein particle. Nucleic Acids Res. 12: 3283–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., and Schulten, K. 1996. VMD: Visual molecular dynamics. J. Mol. Graph. 14: 33–38. [DOI] [PubMed] [Google Scholar]
- Kale, L., Skeel, R., Bhandarkar, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K., and Schulten, K. 1999. NAMD2: Greater scalability for parallel molecular dynamics. J. Comput. Phys. 151: 283–312. [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis, N.B. and Westhof, E. 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. RNA 7: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———.2003. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 13: 300–308. [DOI] [PubMed] [Google Scholar]
- Leulliot, N. and Varani, G. 2001. Current topics in RNA–protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry 40: 7947–7956. [DOI] [PubMed] [Google Scholar]
- Li, W., Ma, B., and Shapiro, B. 2001. Molecular dynamics simulations of the denaturation and refolding of an RNA tetraloop. J. Biomol. Struct. Dyn. 19: 381–396. [DOI] [PubMed] [Google Scholar]
- Macke, T. and Case, D.A. 1998. Modeling unusual nucleic acid structures. In Molecular modeling of nucleic acids (eds. N.B. Leontes and J. Santalucia), pp. 379–393. American Chemical Society, Washington, DC.
- Matsumura, S., Ikawa, Y., and Inoue, T. 2003. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 31: 5544–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, V.K. and Draper, D.E. 1998. On the role of magnesium ions in RNA stability. Biopolymers 48: 113–135. [DOI] [PubMed] [Google Scholar]
- Moore, T., Zhang, Y.M., Fenley, M.O., and Li, H. 2004. Molecular basis of box C/D RNA–protein interactions: Cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure 12: 807–818. [DOI] [PubMed] [Google Scholar]
- Nottrott, S., Hartmuth, K., Fabrizio, P., Urlaub, H., Vidovic, I., Ficner, R., and Lührmann, R. 1999. Functional interaction of a novel 15.5kD [U4/U6 • U5] tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J. 18: 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott, S., Urlaub, H., and Lührmann, R. 2002. Hierarchical, clustered protein interactions with U4/U6 snRNA: A biochemical role for U4/U6 proteins. EMBO J. 21: 5527–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy, A., Perez, A., Lankas, F., Javier Luque, F., and Orozco, M. 2004. Relative flexibility of DNA and RNA: A molecular dynamics study. J. Mol. Biol. 343: 627–638. [DOI] [PubMed] [Google Scholar]
- Oubridge, C., Ito, N., Evans, P.R., Teo, C.H., and Nagai, K. 1994. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372: 432–438. [DOI] [PubMed] [Google Scholar]
- Pearlman, D.A., Case, D.A., Caldwell, J.W., Ross, W.S., Cheatham, T.E., Debolt, S., Ferguson, D., Seibel, G., and Kollman, P. 1995. AMBER, a package of computer programs for applying molecular mechanics, normal-mode analysis, molecular dynamics and free-energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 91: 1–41. [Google Scholar]
- Pitici, F., Beveridge, D.L., and Baranger, A.M. 2002. Molecular dynamics simulation studies of induced fit and conformational capture in U1A-RNA binding: Do molecular substates code for specificity? Biopolymers 65: 424–435. [DOI] [PubMed] [Google Scholar]
- Puglisi, J.D., Tan, R.Y., Calnan, B.J., Frankel, A.D., and Williamson, J.R. 1992. Conformation of the Tar RNA–arginine complex by NMR spectroscopy. Science 257: 76–80. [DOI] [PubMed] [Google Scholar]
- Puglisi, J.D., Chen, L., Blanchard, S., and Frankel, A.D. 1995. Solution structure of a bovine immunodeficiency virus Tat–Tar peptide–RNA complex. Science 270: 1200–1203. [DOI] [PubMed] [Google Scholar]
- Rázga, F., Špačková, N., Réblová, K., Koča, J., Leontis, N.B., and Šponer, J. 2004. Ribosomal RNA kink-turn motif—A flexible molecular hinge. J. Biomol. Str. Dyn. 22: 183–193. [DOI] [PubMed] [Google Scholar]
- Réblová, K., Špačková, N., Štefl, R., Csaszar, K., Koča, J., Leontis, N.B., and Šponer, J. 2003a. Non Watson-Crick basepairing and hydration in RNA motifs: Molecular dynamics of 5S rRNA loop E. Biophys. J. 84: 3564–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová, K., Špačková, N., Šponer, J.E., Koča, J., and Šponer, J. 2003b. Molecular dynamics simulations of RNA kissing-loop motifs reveal structural dynamics and formation of cation-binding pockets. Nucleic Acids Res. 31: 6942–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová, K., Špačková, N., Štefl, R., Csaszar, K., Koča, J., Leontis, N.B., and Šponer, J. 2004. Long-residency hydration, cation binding, and dynamics of Loop E/Helix IV rRNA–L25 protein complex. Biophys. J. 87: 3397–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, C.M. and Kollman, P.A. 2000. Structure and thermodynamics of RNA–protein binding: Using molecular dynamics and free energy analyses to calculate the free energy of binding and conformational change. J. Mol. Biol. 297: 1145–1158. [DOI] [PubMed] [Google Scholar]
- Reyes, C.M., Nifosi, R., Frankel, A.D., and Kollman, P.A. 2001. Molecular dynamics and binding specificity analysis of the bovine immunodeficiency virus BIV Tat–TAR complex. Biophys. J. 80: 2833–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke, J., Appel, B., Digweed, M., and Lührmann, R. 1985. Localization of a base-paired interaction between small nuclear RNAs U4 and U6 in intact U4 U6 ribonucleoprotein-particles by psoralen cross-linking. J. Mol. Biol. 185: 721–731. [DOI] [PubMed] [Google Scholar]
- Ryckaert, J.P., Ciccotti, G., and Berendsen, H.J.C. 1977. Numerical integration of Cartesian equations of motion of a system with constraints—Molecular dynamics of N-alkanes. J. Comput. Phys. 23: 327–341. [Google Scholar]
- Schneider, C., Will, C.L., Makarova, O.V., Makarov, E.M., and Lührmann, R. 2002. Human U4/U6.U5 and U4atac/U6atac.U5 tri-sn-RNPs exhibit similar protein compositions. Mol. Cell. Biol. 22: 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmerling, C. and Elber, R. 1994. Hydrophobic collapse in a cyclic hexapeptide—Computer simulations of chdlfc and caaaac in water. J. Am. Chem. Soc. 116: 2534–2547. [Google Scholar]
- Simmerling, C. and Kollman, P. 1996. Improved simulation of flexible systems through locally enhanced sampling. Abstracts of Papers of the American Chemical Society 212: 153-COMP. [Google Scholar]
- Simmerling, C., Fox, T., and Kollman, P.A. 1998a. Use of locally enhanced sampling in free energy calculations: Testing and application to α→β anomerization of glucose. J. Am. Chem. Soc. 120: 5771–5782. [Google Scholar]
- Simmerling, C., Miller, J.L., and Kollman, P.A. 1998b. Combined locally enhanced sampling and particle mesh Ewald as a strategy to locate the experimental structure of a nonhelical nucleic acid. J. Am. Chem. Soc. 120: 7149–7155. [Google Scholar]
- Simmerling, C., Lee, M.R., Ortiz, A.R., Kolinski, A., Skolnick, J., and Kollman, P.A. 2000. Combining MONSSTER and LES/PME to predict protein structure from amino acid sequence: Application to the small protein CMTI-1. J. Am. Chem. Soc. 122: 8392–8402. [Google Scholar]
- Varani, L., Gunderson, S.I., Mattaj, I.W., Kay, L.E., Neuhaus, D., and Varani, G. 2000. The NMR structure of the 38 kDa U1A protein–PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat. Struct. Biol. 7: 329–335. [DOI] [PubMed] [Google Scholar]
- Vidovic, I., Nottrott, S., Hartmuth, K., Lührmann, R., and Ficner, R. 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell 6: 1331–1342. [DOI] [PubMed] [Google Scholar]
- Wang, W., Donini, O., Reyes, C.M., and Kollman, P.A. 2001. Biomolecular simulations: Recent developments in force fields, simulations of enzyme catalysis, protein–ligand, protein–protein, and protein–nucleic acid noncovalent interactions. Annu. Rev. Biophy. Biom. 30: 211–243. [DOI] [PubMed] [Google Scholar]
- Watkins, N.J., Segault, V., Charpentier, B., Nottrott, S., Fabrizio, P., Bachi, A., Wilm, M., Rosbash, M., Branlant, C., and Lührmann, R. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466. [DOI] [PubMed] [Google Scholar]
- Williamson, J.R. 2000. Induced fit in RNA–protein recognition. Nat. Struct. Biol. 7: 834–837. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Fountain, M.A., and Krugh, T.R. 2001. Structural characterization of a six-nucleotide RNA hairpin loop found in Escherichia coli, r(UUAAGU). Biochemistry 40: 9879–9886. [DOI] [PubMed] [Google Scholar]