Abstract
Plasmids or viral vectors that express short hairpin RNAs (shRNAs) have emerged as important tools for the stable inhibition of specific genes by RNA interference. shRNAs are structural and functional homologs of pre-microRNAs, intermediates in the production of endogenously encoded microRNAs (miRNAs). Therefore, overexpressed shRNAs could inhibit miRNA function by competing for a limiting level of one or more factors involved in miRNA biogenesis or function. Here, we demonstrate that overexpressed shRNAs can saturate the activity of endogenous Exportin 5, a factor required for nuclear export of both shRNAs and pre-miRNAs. While shRNA overexpression can therefore inhibit miRNA function, simultaneous overexpression of Exportin 5 reverses this effect. Moreover, Exportin 5 overexpression can significantly enhance RNA interference mediated by shRNAs. These data have implications for the future clinical utilization of shRNAs and also provide a simple method to enhance RNA interference by shRNAs in culture.
Keywords: microRNAs, nuclear RNA export, RNA interference
INTRODUCTION
RNA interference (RNAi) has emerged as a powerful technique for the specific down-regulation of genes in humans and other animals (Mello and Conte 2004). As a result, RNAi is widely used in research and is also beginning to be used in clinical settings. RNAi is mediated by ~21-nucleotide small interfering RNAs (siRNAs) that guide the RNA-induced silencing complex (RISC) to target mRNA species, leading to mRNA cleavage and degradation. siRNAs can be introduced into vertebrate cells by transfection with synthetic siRNA duplexes, ~21 base-pair (bp) double-stranded RNAs bearing 2-nt 3′ overhangs (Elbashir et al. 2001) or by transfection or transduction with DNA vectors that transcribe short hairpin RNAs (shRNAs) (Brummelkamp et al. 2002; Paddison et al. 2002). shRNAs are ~50-nt-long RNA hairpins, bearing a 2-nt 3′ overhang, that are efficiently processed to siRNAs.
One reason for the efficacy of RNAi is that it utilizes a pre-existing regulatory pathway conserved in all metazoan eukaryotes (Bartel 2004). Humans and other animals encode ~200 endogenous siRNA-like molecules termed microRNAs (miRNAs) that are believed to play a key role in differentiation and development by down-regulating specific target genes. miRNAs are initially transcribed as long (>1000 nt) RNAs termed primary miRNAs (pri-miRNAs) that contain the mature miRNA as one arm of an RNA stem-loop (Lee et al. 2002). This stem-loop is excised by the nuclear RNase III enzyme Drosha to give an ~60-nt RNA hairpin, bearing a 2-nt 3′ overhang, termed a pre-miRNA (Lee et al. 2003a; Zeng and Cullen 2003). Pre-miRNAs are structurally analogous to shRNAs and both are dependent on the karyopherin Exportin 5 (Exp5) for nuclear export (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). Once they reach the cytoplasm, both pre-miRNAs and shRNAs are processed by the RNase III enzyme Dicer to give miRNA or siRNA duplex intermediates, one strand of which is then incorporated into RISC (Hammond et al. 2000; Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001; Paddison et al. 2002; Bartel 2004).
Because shRNAs utilize essentially the same processing pathway as endogenous miRNAs (Bartel 2004), with the exception that Drosha is only required in the latter case, we wondered if shRNA overexpression might competitively inhibit miRNA function. In this paper, we present evidence suggesting that shRNAs can indeed interfere with the inhibition of gene expression mediated by endogenous or over-expressed miRNAs. However, this inhibition could be entirely relieved by simultaneous overexpression of Exp5. Moreover, Exp5 overexpression was shown to significantly enhance the inhibition of target gene expression induced by artificial shRNA expression vectors.
RESULTS
Exp5 overexpression enhances inhibition of gene expression by miRNAs
We have previously described the expression plasmid pCMV-miR-30, which encodes the authentic human miRNA miR-30a (Zeng et al. 2002, 2003; Zeng and Cullen 2003). The initial transcript encoded by pCMV-miR-30 is structurally analogous to a primary miRNA precursor and mature miR-30a expression therefore requires processing by Drosha and Dicer as well as pre-miR-30 nuclear export mediated by Exp5 (Yi et al. 2003; Zeng and Cullen 2003). We have also described the indicator construct pCMV-Luc-miR-30(AP), which encodes the firefly luciferase (F-luc) indicator gene flanked by eight target sites that are perfectly complementary to the miR-30a miRNA (Zeng et al. 2003). As a result, cotransfection of pCMV-Luc-miR-30(AP) with pCMV-miR-30 results in the degradation of the F-luc mRNA encoded by pCMV-Luc-miR-30(AP), with a concomitant drop in F-luc activity (Fig. 1B).
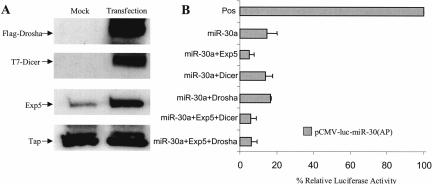
FIGURE 1.
Exp5 overexpression enhances RNAi mediated by overexpressed miR-30a miRNA. 293T cells (24-well plates) were cotransfected with 10 ng of the pCMV-Luc-miR-30(AP) indicator plasmid, 1 ng of the pRL-CMV R-luc internal control, 100 ng of the pCMV-miR-30 effector plasmid, and 50 ng of pKmyc-Exp5 or 100 ng of pCK-Drosha or pK-T7-Dicer. The parental pBC12/CMV or pKmyc plasmids served as negative controls. At 40 h after transfection, the cells were lysed and aliquots used for Western analysis (A) or for assay of F-luc and R-luc activity (B). The endogenous Tap protein served as a loading control in panel A. The data in panel B were compiled from three experiments and were corrected for minor variations in the R-luc internal control. Data are presented relative to the culture transfected with pCMV-Luc-miR-30(AP) in the absence of any miRNA or protein expression plasmid (Pos).
If the biological activity of Drosha, Exp5, or Dicer is rate limiting for the production of mature, biologically active miR-30a miRNA from pCMV-miR-30, then overexpression of the relevant protein would be predicted to increase the observed inhibition of F-luc activity. We therefore transfected 293T cells with the pCMV-Luc-miR-30(AP) indicator and the pCMV-miR-30 effector plasmid together with plasmids expressing Flag-tagged Drosha, T7-tagged Dicer, and/or wild-type Exp5. Approximately 40 h after transfection, the cells were lysed, overexpression of each protein confirmed by Western analysis (Fig. 1A), and F-luc activity determined. As shown in Figure 1B, we did not see any effect of Drosha or Dicer overexpression but did detect a significant reduction in F-luc expression when Exp5 was overexpressed. Coexpression of Exp5 and Dicer, or Exp5 and Drosha, did not further enhance the effect seen with Exp5 alone (Fig. 1B). These experiments were all internally controlled by a cotransfected renilla luciferase (R-luc) expression plasmid, thus arguing that the enhanced inhibition of F-luc expression seen in cells cotransfected with Exp5 and pCMV-miR-30 is not due to toxicity.
To further confirm the specificity of the observed effect of Exp5 on the function of the overexpressed miR-30a miRNA, we asked whether Exp5 overexpression would enhance RNAi caused by transfection of an siRNA duplex specific for the F-luc gene. Exp5 is dispensable for RNAi caused by transfected siRNA duplexes (Yi et al. 2003) and therefore Exp5 overexpression should not enhance inhibition by synthetic siRNAs. As shown in Figure 2A, this is indeed what we observed.
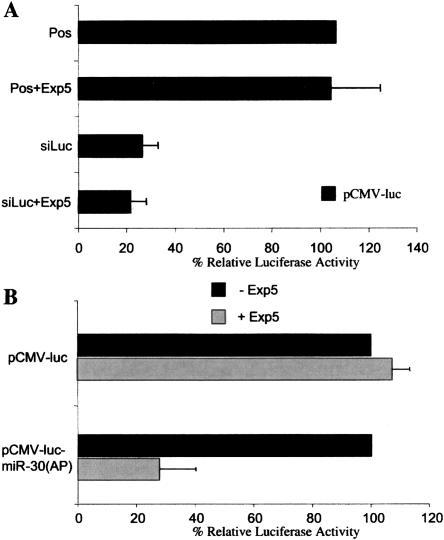
FIGURE 2.
Exp5 overexpression enhances endogenous miRNA but not siRNA function. (A) 293T cells were transfected with pCMV-Luc (10 ng), pRL-CMV (1 ng), 100 nM of a synthetic siRNA specific for F-luc, and 50 ng of pKmyc-Exp5 or the parental pKmyc plasmid. An siRNA specific to the HIV-1 tat gene served as a negative control. At 40 h, cells were harvested and F-luc and R-luc activities determined. Data are presented relative to the culture transfected with pCMV-Luc alone (Pos). The average of three independent transfections with standard deviation is indicated. (B) 293T cells were transfected with pCMV-Luc or with pCMV-Luc-miR-30(AP) (10 ng), with the pRL-CMV internal control (1 ng) and with pKmyc-Exp5 or the parental pKmyc plasmid (50 ng). At 40 h, cells were lysed and F-luc and R-luc activities determined. Data are presented relative to the culture transfected with the pCMV-Luc or pCMV-Luc-miR-30(AP) indicator plasmid in the absence of Exp5 overexpression and were corrected for the R-luc internal control. The average of three experiments with standard deviation is indicated.
Previously, we have reported that 293T cells constitutively express a modest but readily detectable level of the miR-30a miRNA (Zeng et al. 2003) and, presumably as a result, the introduction of eight perfect target sites into the 3′ UTR of the F-luc expression plasmid pCMV-Luc, to give pCMV-Luc-miR-30(AP), reduces F-luc expression by approximately twofold when compared to the R-luc control (Zeng et al. 2003). We therefore asked if overexpression of Exp5 would further attenuate the level of F-luc encoded by pCMV-Luc-miR-30(AP) in cotransfected 293T cells, when compared to the equivalent pCMV-Luc indicator plasmid lacking any miR-30a target sites. As shown in Figure 2B, Exp5 overexpression did not affect luc expression from pCMV-Luc but did inhibit luc expression from pCMV-Luc-miR-30(AP) by approximately threefold. This result suggests that Exp5 overexpression can specifically enhance the selective inhibition of mRNA expression caused by an endogenous miRNA.
If Exp5 activity represents a readily saturable step in miRNA biogenesis and function, then one might predict that overexpression of an artificial shRNA, which is also dependent on Exp5 for nuclear export (Yi et al. 2003), would competitively inhibit miRNA function. To test this hypothesis, we asked whether cotransfection of pSuper-CCR-5, which expresses an shRNA that inhibits expression of the human CCR-5 gene (Lee et al. 2003b), would relieve the inhibition of pCMV-Luc-miR-30(AP) induced by pCMV-miR-30. As shown in Figure 3A, pSuper-CCR5 cotransfection indeed enhanced F-luc enzyme activity by approximately threefold in cotransfected 293T cells, when compared to the R-luc internal control. However, the additional overexpression of Exp5 restored the level of inhibition of F-luc expression seen in the absence of pSuper-CCR5 (Fig. 3A). Similar results were obtained using the previously described indicator construct pCMV-Luc-miR-21(P) (Zeng et al. 2003), which contains eight target sites for the human miR-21 miRNA, and the miR-21 expression plasmid pCMV-miR-21 (Fig. 3B).
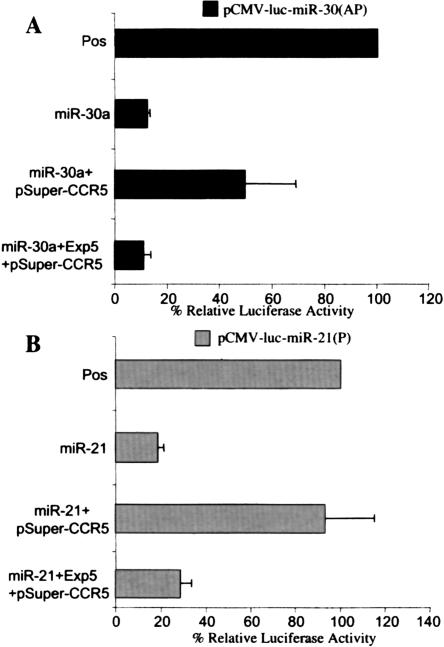
FIGURE 3.
Exp5 overexpression rescues the inhibition of miRNA function caused by an shRNA expression plasmid. (A) Cells were transfected with pCMV-Luc-miR-30(AP) (10 ng), pRL-CMV (1 ng), pCMV-miR-30 or the parental pBC12/CMV plasmid (100 ng), 100 ng of pSuper-CCR-5 or pSuper, and 50 ng of pKmyc-Exp5 or pKmyc. At 40 h, cells were harvested and F-luc and R-luc activities determined. Data are presented as in Figure 2A, where Pos represents the culture transfected with pCMV-Luc-miR-30(AP) together with the pBC12/CMV, pKmyc, and pSuper negative control plasmids. (B) Same as panel A, except that 293T cells were transfected with the pCMV-Luc-miR-21(P) indicator plasmid and the pCMV-miR-21 human miR-21 expression plasmid.
Exp5 overexpression enhances miRNA expression
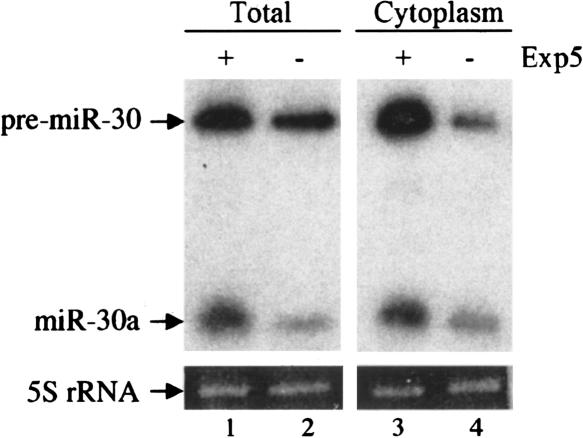
If Exp5-mediated nuclear export of pre-miRNAs represents a rate-limiting step in miRNA biogenesis and function, then one would predict that Exp5 overexpression should enhance the cytoplasmic expression of mature miRNAs and, potentially, also of the pre-miRNA intermediate. To test this hypothesis, we cotransfected 293T cells with the pCMV-miR-30 miRNA expression plasmid in the presence or absence of the Exp5 expression plasmid pKmyc-Exp5 (Brownawell and Macara 2002). At 40 h after transfection, cultures were harvested and either total RNA or the cytoplasmic RNA fraction were isolated and analyzed by Northern blot. As shown in Figure 4, Exp5 had a modest but significant approximately twofold enhancing effect on the level of both mature miR-30 and pre-miR-30 RNA detected at the whole-cell level. Analysis of the cytoplasmic RNA fraction again showed an approximately twofold effect on the level of mature miR-30, as predicted given that mature miRNAs are largely localized to the cytoplasm (Lee et al. 2002). However, these data also revealed a more marked, approximately fivefold increase in the cytoplasmic level of the pre-miR-30 precursor, which is the predicted cargo for nuclear export by Exp5 (Yi et al. 2003; Lund et al. 2004). These observations therefore argue that Exp5 overexpression results in stabilization and enhanced nuclear export of pre-miRNA precursors. This result is consistent with published data demonstrating that binding of endogenous Exp5 by pre-miRNAs can significantly enhance the stability of these RNA intermediates (Yi et al. 2003; Lund et al. 2004; Zeng and Cullen 2004).
FIGURE 4.
Overexpression of Exp5 enhances expression of pre-miR-30 and mature miR-30. 293T cells were transfected as described in Figure 1. At 40 h, wells were pooled into two equal fractions and total cell or cytoplasmic RNA isolated and subjected to Northern analysis. The relative mobility of the 63-nt pre-miR-30 RNA and 23-nt mature miR-30a miRNA are indicated. 5S rRNA served as a loading control.
Exp5 overexpression enhances RNAi induced by an shRNA expression plasmid
Although many transient RNAi experiments continue to utilize artificial siRNA duplexes, other investigators favor shRNA expression constructs, as these can be used to establish stable RNAi and, when introduced into viral vectors, also offer the ability to effectively perform RNAi experiments in primary cells or in vivo (Brummelkamp et al. 2002; Paddison et al. 2002; Lee et al. 2003b). As Exp5 over-expression enhances the biological activity of human miR-NAs in transfected cells (Fig. 1), and as shRNAs also depend on Exp5 for their nuclear export (Yi et al. 2003), one would predict that shRNA-mediated RNAi would also be enhanced by Exp5 overexpression. To test this hypothesis, we knocked down expression of an endogenous gene in 293T cells by transfection of an shRNA expression plasmid, pSuper-Tat-SF1, in the presence or absence of cotransfected pKmyc-Exp5. The target gene chosen, Tat-SF1, is a candidate cofactor for the HIV-1 transcriptional activator Tat (Zhou and Sharp 1996).
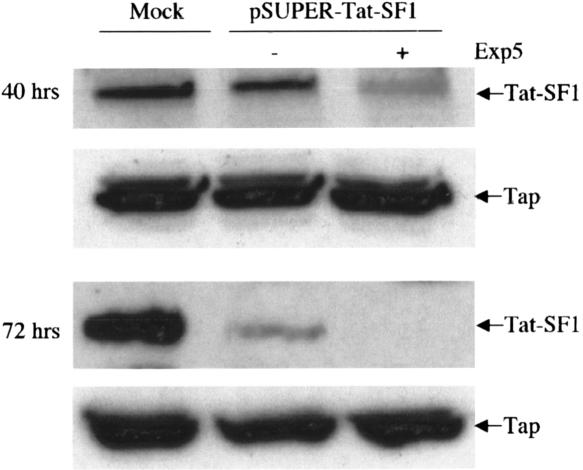
As shown in Figure 5, transfection of 293T cells with pSuper-Tat-SF1 resulted in a modest, two- to threefold drop in Tat-SF1 protein expression at ~40 h after transfection and an ~10-fold drop when measured at 72 h after transfection. In contrast, in the presence of overexpressed Exp5, Tat-SF1 protein expression displayed a more profound approximately eightfold drop at 40 h after transfection and was not detectable at 72 h (Fig. 5). A second endogenously encoded control protein, termed Tap, was unaffected under all conditions analyzed. We have also observed that Exp5 overexpression enhances the selective knockdown of the endogenous human RanBP3 protein in 293T cells transfected with a RanBP3-specific shRNA expression plasmid (data not shown). We therefore conclude that Exp5 overexpression can indeed enhance RNAi mediated by shRNA expression vectors.
FIGURE 5.
Overexpression of Exp5 improves RNAi mediated by an shRNA expression plasmid. 293T cells were mock-transfected or transfected with pSuper-Tat-SF1 (300 ng) and 100 ng of pKmycExp5 or the parental pKmyc plasmid. At 40 h and at 72 h after transfection, individual cell cultures were lysed and subjected to Western blot analysis using antisera specific for the endogenous Tat-SF1 or Tap proteins.
Exp5 is expressed at low levels in many tumor cell lines and in vivo
All of the experiments reported thus far were performed in human 293T cells, a readily transfectable cell line that is widely used for analysis of gene expression in culture. It could be argued that 293T cells are aberrant in that they express unusually low levels of Exp5 and that these data are therefore not predictive of what would be observed in other human or nonhuman cell lines or in vivo.
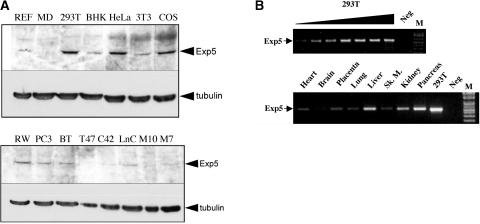
While preliminary experiments also showed enhanced shRNA-mediated RNAi in Exp5 overexpressing HeLa cells (data not shown), we wished to address this concern in a more general way by analyzing the level of Exp5 expression in a range of vertebrate cell lines. As shown in Figure 6A (upper panel), the transformed human cell lines 293T and HeLa, as well as the monkey kidney cell line COS, all express similar, readily detectable levels of Exp5. Somewhat lower, but still detectable, levels were detected in the rodent cell lines BHK and 3T3 and in rat embryo fibroblasts. However, and more importantly, the Exp5 expression levels detected in a series of human cancer cell lines were all substantially lower than seen in 293T cells (Fig. 6A, lower panel). To extend this analysis to human tissues, we performed a semi-quantitative RT-PCR analysis of Exp5 mRNA expression levels using a commercial panel of cDNAs, derived from human tissues, that had been previously normalized based on the expression of four housekeeping genes (β-actin, α-tubulin, GAPDH, and phospholipase A2). A similar level of a cDNA preparation from 293T cells served as a positive control. As may be observed (Fig. 6B), in vivo Exp5 expression levels were somewhat variable, with low levels detected in brain and higher levels in liver, kidney, and pancreas. Most importantly, these RNA data, when combined with the protein data presented in Figure 6A, strongly suggest that 293T cells do not express unusually low levels of Exp5 protein or mRNA and may, in fact, express a relatively high level. Therefore, Exp5 function is predicted to be potentially rate limiting for miRNA and shRNA biogenesis and function in a range of human cell lines and tissues.
FIGURE 6.
Analysis of Exp5 protein or mRNA expression levels. (A) Lysates were prepared from a variety of cell lines, normalized by total protein concentration and 50 μg of each preparation analyzed by Western blot using a polyclonal antibody against full-length human Exp5 or a monoclonal antibody specific for tubulin. Abbreviations and tissue origins are as follows: REF, rat embryo fibroblasts; MD, MDCK canine kidney; 293T, human embryonic kidney; BHK, baby hamster kidney; HeLa, human cervical carcinoma; 3T3, NIH 3T3 murine fibroblasts; COS, African green monkey kidney; RW, RWPE normal human prostate; PC-3, human prostate cancer; BT, BT474 human breast cancer; T47, T-47D human breast cancer; C42, human prostate cancer; LnC, LnCAP human prostate carcinoma; M10, MCF10A human breast epithelial cells; M7, MCF7 human breast cancer. (B) Semiquantitative RT-PCR analysis of Exp5 mRNA expression in human tissues. This experiment utilized a commercially prepared, prenormalized human multiple tissue cDNA panel. A similar level of a 293T cell cDNA sample was used as a positive control and to establish amplification conditions that gave rise to levels of the amplified cDNA fragment that were linearly related to the level of input cDNA. A twofold dilution series, using cDNA prepared from poly(A)+ 293T cell RNA, is shown.
DISCUSSION
In this paper, we provide evidence suggesting that the Exp5-mediated nuclear export of pre-miRNAs and the structurally analogous artificial shRNAs can represent a rate-limiting step in miRNA and shRNA-derived siRNA biogenesis and function. Evidence in support of this hypothesis includes the demonstration that Exp5 overexpression specifically enhances targeted inhibition of gene expression by an authentic, overexpressed miRNA (Fig. 1) or by an shRNA (Fig. 5). This effect was specific, as Exp5 overexpression did not enhance RNAi mediated by an artificial siRNA duplex, which is predicted to gain direct access to the cytoplasm without requiring Exp5-mediated nuclear export (Fig. 2A). Analysis of the expression of the miR-30a miRNA and its precursor pre-miR-30 showed that Exp5 overexpression enhanced miR-30a expression by approximately twofold and enhanced the cytoplasmic expression of pre-miR-30, the target for nuclear export by Exp5, by approximately fivefold (Fig. 4). Together, these data demonstrate that Exp5 function is rate limiting for the biogenesis and function of over-expressed miRNAs and shRNAs in transfected human 293T cells.
While we favor the hypothesis that overexpression of Exp5 primarily enhances miRNA and shRNA function by enhancing their nuclear export, we note that Exp5 also plays an important role in stabilizing pre-miRNAs in the cell (Yi et al. 2003; Lund et al. 2004; Zeng and Cullen 2004). Therefore, it is possible that Exp5 overexpression also increases the pool of pre-miRNAs that are available for nuclear export.
Unlike overexpression of Exp5, overexpression of the RNA processing enzymes Drosha or Dicer did not have any perceptible effect on the biological activity of the miR-30a miRNA (Fig. 1B). If Exp5-mediated nuclear export is indeed rate limiting, then this is perhaps not surprising. Moreover, Drosha is known to participate in the processing of rRNAs, which are expressed at very high levels, and as a result Drosha itself is expressed ubiquitously and at high levels (Abou Elela et al. 1996; Wu et al. 2000). The lack of any effect of Dicer overexpression, including in the presence of excess Exp5 (Fig. 1), was unexpected, especially as over-expression of Exp5 leads to the cytoplasmic accumulation of pre-miRNAs (Fig. 4). We note, however, that work with invertebrates has identified cofactors that bind either to Dicer or Drosha and that may be required for their function in vivo (Liu et al. 2003; Denli et al. 2004). If these cofactors are expressed at a limiting level, then overexpression of Dicer or Drosha would not be expected to have any effect. The lack of any effect of Drosha and Dicer overexpression on miRNA function is therefore not interpretable and simply serves as a negative control for the enhancing effect seen with overexpressed Exp5.
An important issue is whether the 293T cells utilized in these experiments are actually representative of vertebrate cells in general, i.e., do 293T cells express unusually low levels of Exp5? While we were also able to detect saturation of the Exp5 function in HeLa cells (data not shown), we felt this issue would be most effectively addressed by examining the Exp5 protein and mRNA expression levels in a range of cell lines and tissues. As shown in Figure 6A, 293T cells were found to express levels of Exp5 that were comparable to levels expressed in HeLa and in the monkey cell line COS but clearly higher than observed in 12 other cell types, including several human cancer cell lines. As shown in Figure 6B, an RT-PCR analysis of Exp5 mRNA expression in human tissues in vivo also suggests that Exp5 expression is, if anything, relatively high in 293T cells. Therefore, we would argue that our data suggest that Exp5 function may well be close to limiting for shRNA and pre-miRNA function in a range of human cell lines and tissues.
These data have two practical implications. The first of these is that it should be possible to achieve better and/or faster knockdown of specific genes in cultured cells using shRNA expressing plasmids or viral vectors if Exp5 is simultaneously overexpressed. An example of data that confirm this prediction is shown in Figure 5. A second implication of these data is that overexpression of an artificial shRNA could competitively inhibit the function of endogenously transcribed pre-miRNAs, with currently unpredictable consequences for the pattern of cellular gene expression. An example of competitive inhibition of miRNA function is given in Figure 3, which shows that an overexpressed shRNA can inhibit the biological activity of two distinct overexpressed human miRNAs and, moreover, that the biological activity of these miRNAs could be restored by additional overexpression of Exp5. We have previously reported that the level of siRNA expression seen in cells stably transduced with a lentiviral shRNA expression vector is comparable to the level seen in transfected cells (Lee et al. 2003b), so it seems possible that lentiviral shRNA expression vectors could have an inhibitory effect on miRNA function in culture or in vivo. On the other hand, other groups have reported the production of transgenic mice that constitutively express biologically active shRNAs from lentiviral vectors and that are apparently healthy and developmentally normal (Rubinson et al. 2003; Tiscornia et al. 2003), so the shRNA expression levels achieved in these mice are presumably below the threshold level required to significantly impact on miRNA function. Nevertheless, this concern is one that will clearly need to be addressed before shRNA expression vectors can be utilized to treat human disease.
MATERIALS AND METHODS
Plasmids and siRNAs
The parental expression plasmid pBC12/CMV and its derivatives pCMV-miR-30, pCMV-miR-21, pCMV-Luc-miR-30(AP), pCMV-Luc-miR-21(P), and pCMV-Luc have been described (Zeng et al. 2002, 2003). Also described are the expression vector pKmyc and its derivative pKmyc-Exp5 (Brownawell and Macara 2002) and pSuper and its derivative pSuper-CCR5 (Brummelkamp et al. 2002; Lee et al. 2003b) as well as pCK-Drosha (Lee et al. 2003a). The pK-T7-Dicer expression plasmid was derived by insertion of a HindIII/XhoI cDNA fragment, encoding a full-length human Dicer cDNA (a gift from Scott Hammond, Univ. of North Carolina at Chapel Hill) linked to an amino-terminal T7 tag, into pKmyc. This insertion deleted the myc epitope tag present in pKmyc. The pSuper-Tat-SF1 shRNA expression plasmid was derived, as previously described (Lee et al. 2003b), by insertion of a DNA oligonucleotide encoding an shRNA specific for residues 2023–2041 of the 2268-nt human Tat-SF1 open reading frame between the BglII and HindIII sites present in pSuper. The synthetic siRNA duplexes specific for F-Luc and HIV-1 Tat have been described (Elbashir et al. 2001; Coburn and Cullen 2002) and were purchased from Dharmacon.
Transfection assays
All transfections were performed using human 293T cells cultured in 24-well tissue culture plates, as previously described (Yi et al. 2003). Lipofectamine 2000 (Invitrogen) was used for both siRNA and plasmid DNA transfection, according to the manufacturer’s protocols. Dual luciferase assays were performed using DLR kits (Promega).
RNA and protein expression assays
For analysis of miR-30a RNA expression, total and cytoplasmic RNA fractions were isolated from transfected 293T cell cultures, as described in Figure 4, and subjected to Northern analysis as previously described (Yi et al. 2003). Analysis of the tissue expression pattern of Exp5 mRNA was performed using a prenormalized human multiple tissue cDNA panel (BD/Clonetech) or a similar level of cDNA prepared using poly(A)+ 293T cell RNA, essentially as described previously (Wiegand et al. 2004). Briefly, gene-specific primers were used to specifically amplify a fragment of the human Exp5 gene using 30 cycles of PCR performed at the following temperatures and for the indicated lengths of time: 98°C, 45 sec; 42°C, 45 sec; 72°C, 60 sec.
For analysis of Drosha, Dicer, and Exp5 protein expression, 293T cells were transfected as described in Figure 1 and lysates subjected to Western blot analysis using a 1:1000 dilution of an anti-Flag epitope tag mouse monoclonal antibody (Sigma), a 1: 5000 dilution of an anti-T7 epitope tag mouse monoclonal (No-vagen), or a 1:350 dilution of a rabbit polyclonal antiserum raised against recombinant human Exp5 (Brownawell and Macara 2002). This antibody also recognizes the similar Exp5 proteins expressed in other mammalian species. Similarly, levels of endogenous Tap or Tat-SF1 expression were measured using a 1:1000 dilution of rabbit antisera specific for Tap (Coburn et al. 2001) or Tat-SF1 (a gift of Mariano Garcia-Blanco, Duke Univ. Medical Center), while levels of Exp5 and tubulin-α expression in cell lines (Fig. 6A) were assayed by Western blot analysis using a 1:200 dilution of the rabbit polyclonal anti-Exp5 antiserum and a 1:1000 dilution of a monoclonal specific for tubulin-α (Sigma). Horseradish peroxidase-conjugated secondary antibodies (Amersham) and enhanced chemiluminescence followed by autoradiography were used to detect reactive proteins.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and by National Institutes of Health grant R01 GM071408. The authors thank Mariano Garcia-Blanco, Narry Kim, Michael Lee, and Scott Hammond for reagents used in this research.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7233305.
REFERENCES
- Abou Elela, S., Igel, H., and Ares Jr., M. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85: 115–124. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bohnsack, M.T., Czaplinski, K., and Görlich, D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownawell, A.M. and Macara, I.G. 2002. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol. 156: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- Coburn, G.A. and Cullen, B.R. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76: 9225–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn, G.A., Wiegand, H.L., Kang, Y., Ho, D.N., Georgiadis, M.M., and Cullen, B.R. 2001. Using viral species specificity to define a critical protein/RNA interaction surface. Genes & Dev. 15: 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli, A.M., Tops, B., Plasterk, R.H.A., Ketting, R.F., and Hannon, G.J. 2004. Processing of pri-microRNAs by the microprocessor complex. Nature 432: 231–235. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–295. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., McLachlan, J., Pasquinelli, A.E., Bálint, É., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.-T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21: 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, H., Yim, J., Lee, J., Provost, P., Rådmark, O., Kim, S., et al. 2003a. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Lee, M.-T.M., Coburn, G.A., McClure, M.O., and Cullen, B.R. 2003b. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J. Virol. 77: 11964–11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.-E., Smith, D.P., and Wang, X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Lund, E., Güttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. 2004. Nuclear export of microRNA precursors. Science 303: 95–98. [DOI] [PubMed] [Google Scholar]
- Mello, C.C. and Conte Jr., D. 2004. Revealing the world of RNA interference. Nature 431: 338–342. [DOI] [PubMed] [Google Scholar]
- Paddison, P.J., Caudy, A.A., Bernstein, E., Hannon, G.J., and Conklin, D.S. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 16: 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson, D.A., Dillon, C.P., Kwiatkowski, A.V., Sievers, C., Yang, L., Kopinja, J., Zhang, M., McManus, M.T., Gertler, F.B., Scott, M.L., et al. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33: 401–406. [DOI] [PubMed] [Google Scholar]
- Tiscornia, G., Singer, O., Ikawa, M., and Verma, I.M. 2003. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. 100: 1844–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand, H.L., Doehle, B.P., Bogerd, H.P., and Cullen, B.R. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23: 2451–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Xu, H., Miraglia, L.J., and Crooke, S.T. 2000. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 275: 36957–36965. [DOI] [PubMed] [Google Scholar]
- Yi, R., Qin, Y., Macara, I.G., and Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y. and Cullen, B.R. 2003. Sequence requirements for micro RNA processing and function in human cells. RNA 9: 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2004. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 32: 4776–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed microRNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Yi, R., and Cullen, B.R. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. 100: 9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q. and Sharp, P.A. 1996. Tat-SF1: Cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274: 605–609. [DOI] [PubMed] [Google Scholar]