Abstract
Caenorhabditis elegans is unusual among animals in having a highly conserved octamer sequence at the 3′ splice site: UUUU CAG/R. This sequence can bind to the essential heterodimeric splicing factor U2AF, with U2AF65 contacting the U tract and U2AF35 contacting the splice site itself (AG/R). Here we demonstrate a strong correspondence between binding to U2AF of RNA oligonucleotides with variant octamer sequences and the frequency with which such variations occur in splice sites. C. elegans U2AF has a strong preference for the octamer sequence and exerts much of the pressure for 3′ splice sites to have the precise UUUUCAG/R sequence. At two positions the splice site has a very strong preference for U even though alternative bases can also bind tightly to U2AF, suggesting that evolution can select against sequences that may have a relatively modest reduction in binding. Although pyrimidines are frequently present at the first base in the exon, U2AF has a very strong bias against them, arguing there is a mechanism to compensate for weakened U2AF binding at this position. Finally, the C in the consensus sequence must remain adjacent to the AG/R rather than to the stretch of U’s, suggesting this C is recognized by U2AF35.
Keywords: mRNA splicing, cross-linking, nematode
INTRODUCTION
How the splicing machinery recognizes the correct splice sites among the many sites that appear to match the consensus sequences is one of the central and most interesting problems in the field of pre-mRNA processing (Reed 2000; Brow 2002; Zhang and Chasin 2004). The 5′ splice site is identified by a base pairing interaction between the U1 snRNP and a sequence on the pre-mRNA. In contrast, the 3′ splice site is a multipart signal comprising a branchpoint consensus, bound by the SF1/BBP protein and subsequently by base pairing to the U2 snRNP (Berglund et al. 1997); the polypyrimidine tract, bound by the large subunit of the essential splicing protein, U2AF; and the AG/R, bound by the U2AF small subunit (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). In mammals these three sequences are generally separated from one another by 10–30 nucleotides, and there is a relatively loose branchpoint consensus but a relatively tight interaction of U2AF65 with a long polypyrimidine tract (Reed 2000). Introns that have short polypyrimidine tracts compensate by having the AG nearby to facilitate interaction with U2AF35. These are called AG-dependent introns (Reed 1989). In budding yeast there is no U2AF35, so all introns are AG independent. Also, most yeast introns lack a strong polypyrimidine tract. Recognition of yeast introns depends instead on the highly conserved branchpoint consensus interaction with BBP/ SF1. In fission yeast, there is a BBP/SF1-U2AF complex that recognizes introns by binding to all three consensus sequences (Huang et al. 2002).
In contrast to vertebrates and yeast, Caenorhabditis elegans appears to have a unique method of identifying 3′ splice sites. There is no consensus sequence for the branch-point, although there is a BBP/SF1 protein (Blumenthal and Steward 1997; Mazroui et al. 1999). Furthermore, most introns do not have a long polypyrimidine tract separated from the splice site itself. Instead, introns have the highly conserved consensus sequence UUUUCAG/R (where / indicates the splice site) at their 3′ ends (Fig. 1A ▶; Blumenthal and Steward 1997; Kent and Zahler 2000). This sequence has been shown to be critical for 3′ splice site recognition (Conrad et al. 1993; Zhang and Blumenthal 1996). The UUUUCAG/R sequence is known to interact with U2AF, the same protein that recognizes the polypyrimidine tract and the AG in mammals (Zorio and Blumenthal 1999). Nonetheless, this sequence is very different from that found in other organisms, even other nematodes, so it is not clear whether it is U2AF or some other component of the splicing machinery that exerts the selective pressure to maintain this sequence. To determine whether U2AF binding is responsible for the high conservation of this octamer, we have performed binding experiments with variants of the consensus sequence. We show here that affinity of U2AF for the octamer corresponds with the frequency of bases in the splice site consensus and thus is sufficient to explain most of its high level conservation.
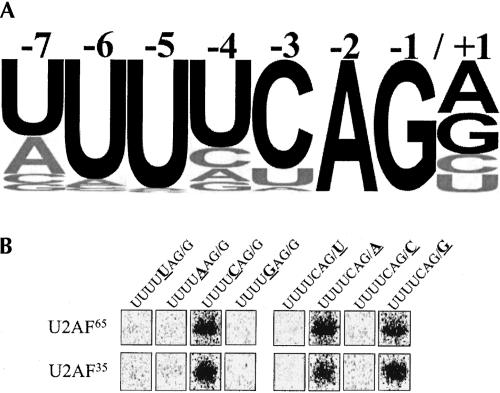
FIGURE 1.
U2AF65 and U2AF35 bind preferentially to the 3′ splice site consensus oligonucleotide. (A) Graphic representation of the C. elegans 3′ splice site consensus sequence from >28,000 confirmed introns. The height of each letter represents the frequency of each base found at that position according to the intron profile (Kent and Zahler 2000). Black letters indicate consensus bases. (B) Oligonucleotides with substitutions at positions −3 and +1 were 32P-labeled, mixed with whole cell extract, ultraviolet-cross-linked, and immunoprecipitated with antibodies to U2AF65.
RESULTS AND DISCUSSION
Cross-linking of consensus sequence and variant oligonucleotides to U2AF65 and U2AF35
Because we have been unable to obtain active recombinant U2AF from C. elegans, we have measured binding in crude embryonic extracts using antibodies specific to U2AF to isolate bound complexes. Our standard procedure was to mix the extract with an end-labeled RNA oligonucleotide containing the splice site, UV cross-link, and immunoprecipitate complexes with anti-U2AF65 (Zorio and Blumenthal 1999). U2AF65 and U2AF35 form a tight complex, so both proteins bound to radiolabeled oligonucleotides were visualized on gels of the immunoprecipitates (Fig. 1B ▶). We also tested in the same assay labeled RNA oligonucleotides containing all single nucleotide substitutions at positions +1 and −3 relative to the splice site. At the −3 position, only the oligonucleotide having the consensus C base showed any binding. Similarly, only oligonucleotides containing a purine at the +1 position bound detectably to U2AF (Fig. 1B ▶).
U2AF binding correlates with the frequency of bases found in the splice site
Because the amount of cross-linking to an RNA oligonucleotide can vary depending on the number of U’s in the sequence, we performed the remainder of the experiments with labeled RNA oligonucleotides containing the consensus octamer along with varying concentrations of competitor RNA oligonucleotides with base substitutions in the octamer. To determine whether alterations in the level of binding result from lack of the consensus base or presence of a nonconsensus base at that position, we tested multiple nonconsensus bases at each position. The hypothesis predicts that oligonucleotides containing mismatches to the consensus will bind in proportion to how conserved that position is in the 3′ splice site (Fig. 1A ▶). An oligonucleotide with a base substitution that occurs at a relatively high frequency in naturally occurring splice sites should be a better competitor than one with a base that occurs rarely.
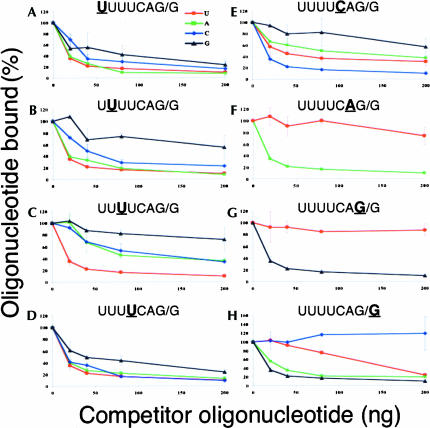
In each case, we observed both U2AF subunits binding with similar profiles, so we show only the quantitation of the U2AF65 results in Figures 2 ▶–4 ▶. The eight panels of Figure 2 ▶ show competition with oligonucleotides containing single nucleotide substitutions at each of the eight positions. The data clearly demonstrate that the alterations in U2AF binding correlate well with the frequency of bases found in the ~28,000 splice sites surveyed (Kent and Zahler 2000). This can be seen in two ways: (1) the most conserved positions tend to be the least tolerant to substitutions with nonconsensus bases, and (2) bases used less frequently in the splice site are more disruptive to U2AF binding.
FIGURE 2.
Competition for U2AF65 binding to the consensus sequence by oligonucleotides with single nucleotide substitutions. Graphic representation of competition experiments with oligonucleotides containing single nucleotide substitutions at the indicated position. Transgenic extract was ultraviolet cross-linked with 32P-labeled U4CAGG-containing oligonucleotides in the presence of variant competitors. Increasing amounts of competitor oligonucleotides were in 2.5-fold, fivefold, 10-fold, and 25-fold molar excess over labeled RNA, which was always present at 8 ng. Bands were quantified and normalized using the PhosphorImager system from Molecular Dynamics. U indicated by red circles; A, green squares; C, blue diamonds; and G, black triangles.
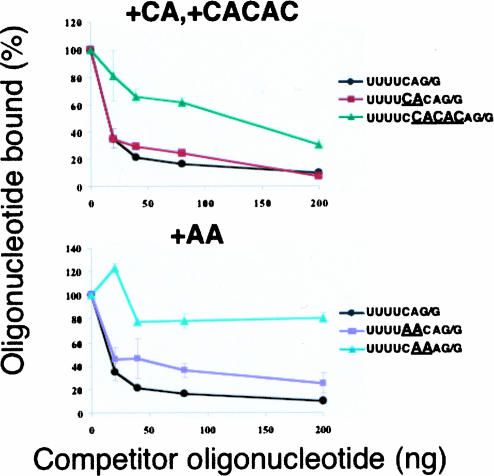
FIGURE 4.
Competition for U2AF65 binding to the consensus sequence by oligonucleotides with multiple base insertions. In the lower panel, an AA insertion between the −4 and −3 positions has a minimal effect on binding, whereas an AA added between the −3 and −2 positions dramatically reduces binding. In the upper panel, a two-base insertion is tolerated far more than a five-base insertion.
At the −7 position, U is favored over the other bases in natural splice sites, but all four bases are found at significant levels (Fig. 1A ▶). Similarly, all four bases are tolerated for U2AF binding, and the strength of binding, as measured by the effectiveness at which they compete for binding to the labeled RNA oligonucleotide containing the consensus sequence, correlates with the frequencies of the four bases in natural splice sites (U > A > C > G) (Fig. 2A ▶).
At the −6 position, U is strongly favored over all four bases in natural splice sites (Fig. 1A ▶), and indeed C- and G-containing oligonucleotides bind substantially less well to U2AF than does the U-containing oligonucleotide. However, an A at this position does not substantially reduce U2AF binding compared with the consensus U (Fig. 2B ▶). Perhaps the requirement for U at this position is exerted by some molecule other than U2AF. Alternatively, natural selection can select against splice sites with A at −6 even though its effect on binding affinity is relatively small. As at most positions in the splice site, the presence of a G at −6 severely compromises U2AF binding and is found very rarely in natural splice sites.
The strongest preference for a single base, other than at the AG itself, is at the −5 position, where 97% of C. elegans splice sites have a U (Fig. 1A ▶). This preference is clearly reflected in the U2AF competition assays, where any base substitution has a severe effect on binding (Fig. 2C ▶). G is used at the lowest frequency and has the strongest negative effect on binding.
The −4 position is much more tolerant of substitutions for the consensus U in natural splice sites (Fig. 1A ▶), and similarly, substitutions at this position have a much smaller effect on U2AF binding (Fig. 2D ▶). Even G is sometimes present in splice sites at the −4 position (8%), and the G-substituted RNA oligonucleotide at this position competes better with the consensus sequence-containing oligonucleotide for U2AF binding than do G-substituted oligonucleotides at the other positions.
The −3 position strongly prefers a C over all other bases in the splice site consensus, and this is also reflected in U2AF binding (Fig. 2E ▶). U is used 15% of the time and binds next most strongly. A is used 2% of the time and binds next most strongly. G is never present at −3, and this oligonucleotide binds least strongly. Again there is a very strong correlation between the consensus sequence and U2AF binding.
Not surprisingly, the perfectly conserved AG at positions −1 and −2, is required for U2AF binding (Fig. 2F,G ▶).
At the +1 position, purines are favored over pyrimidines, but the effect is rather modest: 29% of splice sites have a pyrimidine at the first base in the exon (Fig. 1A ▶). However, quite surprisingly, RNA oligonucleotides with pyrimidines at the +1 position bind to U2AF very weakly or not at all (Fig. 2H ▶). Therefore, something else in the splicing machinery may compensate for the weak U2AF binding to allow for the use of these sites. Alternatively, our binding assay may not perfectly reflect binding of U2AF to a full-length or nascent pre-mRNA, and changes at this position actually have a much more modest effect on U2AF binding in a more natural context. We tested the idea that it is the proximity of this base to the 3′ end of the oligonucleotide that resulted in the strong antipyrimidine bias by testing oligonucleotides in which the three bases from the 5′ were moved to the 3′ end of the sequence, but this did not alter the bias for purines for U2AF binding (data not shown).
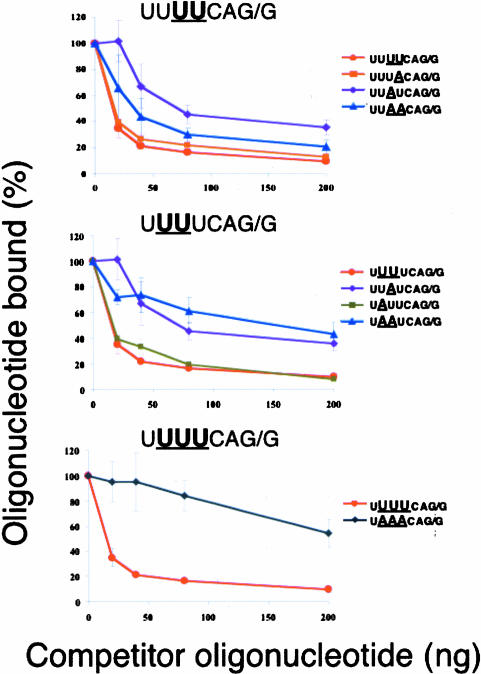
Partial suppression of the U5A phenotype by an additional U4A substitution
We also made a limited number of multiple substitutions and tested these oligonucleotides in the binding competition assay (Fig. 3 ▶). Interestingly, the severe reduction in U2AF binding seen when position −5 U is changed to A is partially rescued when the U at −4 is also changed to A (Fig. 3 ▶, top). The UUAACAG/G-containing RNA oligonucleotide competes more effectively for U2AF binding than does the oligonucleotide containing the UUAU-CAG/G sequence. Nothing about the frequency of this double substitution in the splice site predicted this suppression. Thus, something other than U2AF binding might be selecting against this UUAACAG/R splice site. An oligonucleotide with an A substitution at the −6 position, along with the −5 A substitution, binds weakly to U2AF, much like the oligonucleotide with only the −5 A substitution (Fig. 3 ▶, middle). Clearly, an A substitution at the −4 position improves the binding of an oligonucleotide with a −5 A substitution, whereas an oligonucleotide with a −6 A substitution does not. As expected, the triple substitution of −4, −5, and −6 U’s for A’s eliminated U2AF binding (Fig. 3 ▶, bottom).
FIGURE 3.
Competition for U2AF65 binding to the consensus sequence by oligonucleotides with multiple substitutions. Note the partial suppression of the effect of a −5 A substitution by a concomitant A substitution at −4 but not −6.
Insertion of nucleotides between the U tract and the AG/R can have profound effects on U2AF binding
Is the adjacency of the pyrimidines and the AG/R in the C. elegans 3′ splice site a requirement for U2AF binding? To test that, we inserted two nucleotides, AC, between the pyrimidines and the AG/G to make the sequence UUUUCA CAG/G. As shown in Figure 4 ▶, top panel, this oligonucleotide bound to U2AF nearly as well as the wild-type oligonucleotide, indicating the bases bound to U2AF65 and those bound to U2AF35 do not need to be immediately adjacent for U2AF binding. This suggests that there is an additional selective pressure on the 3′ splice site octamer or that even a change in U2AF affinity too small to be observed in our experimental protocol can have a strong effect at the level of natural selection. Insertion of additional bases, such that the U tract is separated from the AG/R by the sequence CCACAC, instead of the normal C, strongly inhibits binding to U2AF. This indicates that the U tract and the AG/R are normally very close in order to allow formation of a tight complex with U2AF.
We next performed an experiment to determine whether the C at position −3 must be adjacent to the AG/R or the U tract, for tight U2AF binding. That is, is it functionally the 3′ end of the poly(Y) tract or is it associated with the invariant AG at the splice site? The results were unequivocal: Separating the C from the U tract by two bases had no deleterious effect on binding, whereas separating it from the AG/R by the same two bases severely compromised U2AF binding (Fig. 4 ▶, bottom). This result indicates that this C is recognized by U2AF35, the polypeptide that binds to the adjacent nucleotides, AG/R.
Multiple modes of 3′ splice site recognition by the same proteins
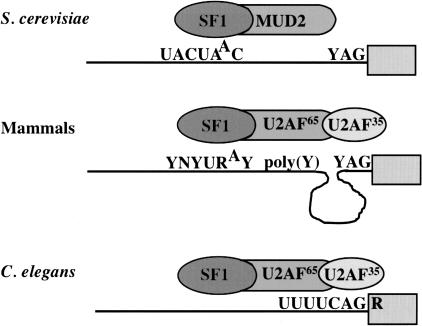
It is surprising that the same three proteins, SF1/BBP, U2AF65, and U2AF35, can be used in such different ways to identify 3′ splice sites. Yeast 3′ splice sites are identified by tight binding of SF1/BBP to the very highly conserved branchpoint sequence, UACUAAC (Berglund et al. 1997). U2AF65, called MUD2, plays an ancillary role, and U2AF35 is not present at all (Fig. 5 ▶). In contrast, 3′ splice site recognition in mammalian cells is much more combinatorial: The weaker branchpoint consensus binds SF1/BBP, the polypyrimidine tract binds U2AF65, and the YAG binds U2AF35, usually with quite a few nucleotides separating the two U2AF subunit binding sites (Fig. 5 ▶). The data reported here make it clear that the C. elegans 3′ splice site is recognized in a third way: by tight binding of the UUUU by U2AF65 and the adjacent CAG/R by U2AF35(Fig. 5 ▶). This recognition is presumably sufficient to position SF1/BBP and the U2 snRNP at an appropriate branchpoint nucleotide, since there is not a long polypyrimidine tract or a branchpoint consensus sequence. Evidently the UUUU is a sufficient polypyrimidine tract, although this is a misnomer since C’s are not acceptable substitutes for U’s in this sequence. It is especially interesting that C’s interrupting the U tract reduce U2AF binding since C’s are acceptable bases for binding mammalian U2AF65(Banerjee et al. 2003). Presumably the UUUU would not be sufficient to attract U2AF65 in the absence of an adjacent U2AF35 binding site, as this is a common tetramer in introns. Thus, it seems that the complete consensus, UUUUCAG/R, is recognized in concert by the U2AF65/35 complex, and that provides enough binding energy to correctly identify 3′ splice sites in C. elegans.
FIGURE 5.
Different modes of recognition of the 3′ splice site in different species. In the yeast, Saccharomyces cerevisiae, introns are AG independent. The splice site is recognized by tight binding of SF1/BBP to the highly conserved branchpoint consensus sequence. In mammals, the site is tripartite; it is identified by weaker binding of SF1/BBP to this site, as well as by binding of the polypyrimidine tract by U2AF65 and the YAG by U2AF35. In C. elegans the octamer at the splice site is bound tightly by the U2AF65/35 complex.
One key question is whether C. elegans utilizes a different mode of 3′ splice site recognition or just an optimal one. Should we be able to identify differences between C. elegans U2AF and U2AF from other animals that are responsible for the differences in the recognition sequences? An alignment of C. elegans U2AF RNA binding region with that of other metazoans shows they are quite similar (Zorio et al. 1997). However, there are 3′ splice sites very much like UUUUCAG/R in other organisms. It may be that these splice sites are recognized in a similar manner, but that other organisms are more likely to use additional modes as well, e.g., binding of U2AF65 to a long poly(Y) tract separated from the AG/R and/or binding of SF1/BBP to the branchpoint consensus. If so, it would be expected that differences between RNA recognition motifs of C. elegans U2AF and other U2AF dimers might be subtle or nonexistent. In any case our data lead us to the conclusion that one mode of 3′ splice site recognition is for the U2AF complex to recognize a short poly(U) tract immediately adjacent to a CAG/R. The fission yeast, Schizosaccharomyces pombe, appears to recognize 3′ splice sites in a very similar way to C. elegans. In this yeast, introns often contain poor matches to a branchpoint sequence and lack a polypyrimidine tract. Furthermore, SF1/BBP forms a stable interaction with the U2AF complex (Huang et al. 2002). This mode may be used for most splice sites in C. elegans, but for fewer splice sites in other organisms. Furthermore, it may not be used for all C. elegans splice sites. Some authentic splice sites match the UUUUCAG/R consensus poorly. These may use a different mode of recognition, or they may represent deliberately weak sites. For example, splice sites with poor matches to the consensus may be involved in alternative splicing.
Other influences on 3′ splice site recognition in C. elegans
The UUUUCAG/R is not the only information that the C. elegans splicing machinery uses to recognize the correct 3′ splice site. This sequence occurs at a boundary between very A/U-rich sequence in the intron or outron and G/C-rich sequence in the exon, and this boundary has previously been shown to be an important feature of 3′ splice site recognition (Conrad et al. 1993). Also, intron length plays a role: Introns with mismatches to the 3′ splice site consensus can still be recognized in vivo, but only when they are quite short (Zhang and Blumenthal 1996). Furthermore, C. elegans has the usual group of SR proteins that can act as splicing enhancers (Longman et al. 2000). Even with an extraordinarily highly conserved U2AF binding sequence, splice-site recognition must be viewed as a highly combinatorial process.
Relevance of the findings to autoregulation of U2AF65 pre-mRNA splicing
C. elegans U2AF65 pre-mRNA is alternatively spliced to yield an RNA that cannot be translated because of a premature stop codon (Zorio et al. 1997). Surprisingly, this RNA is not subject to nonsense-mediated decay. We have shown previously that this is because the RNA is retained in the nucleus, and we have hypothesized that U2AF itself is responsible for causing both the alternative splicing that results in inclusion of the nonsense codon-containing exon and the nuclear retention (MacMorris et al. 1999). We noted that this exon contains 10 copies (18 in the related nematode C. briggsae) of a sequence closely related to the 3′ splice site octamer, and we proposed that binding of U2AF to these sequences resulted in exon inclusion and nuclear retention of the resultant mRNA. The results in this article confirm that UUUUCAGR is the precise sequence recognized by U2AF, and that almost all of the variants of this repeat found in the alternative exon would be expected to bind U2AF tightly. This supports the idea that U2AF could autoregulate the level of its own mRNA by causing inclusion of a poison exon.
MATERIALS AND METHODS
Embryonic crude extract
Embryonic crude extract of transgenic strain BL7014 was prepared as described previously (Zorio and Blumenthal, 1999) with the exception that the heat-shocked embryo homogenate was sedimented at 800 rpm for 2 min, quick frozen, and stored at −80°C.
Cross-linking by ultraviolet light and immunoprecipitation
Cross-linking by ultraviolet light and immunoprecipitation was performed essentially as described previously (Zorio and Blumenthal 1999). Prior to cross-linking, extract was incubated with 100 mM KCl and 0.1% NP40 for 30 min at 4°C followed by centrifugation at 4000 rpm for 2 min. Eight nanograms of 32P-labeled RNA was mixed with buffer (20 mM HEPES-KOH at pH 7.6, 100 mM KCl, 0.2 mM EDTA, 10% glycerol, 0.5 mM DTT, and 50 ng mL−1 BSA) and extract to 25.6 μL. For competition experiments, unlabeled RNA at concentrations of 20 ng, 40 ng, 100 ng, and 200 ng, along with 8 ng of 32P-labeled U4CAGG-containing RNA oligonucleotide, was added to the mixture. After incubation for 30 min at 23°C, samples were irradiated for 10 min at 4°C with ultraviolet light from a Mineralight lamp (Model UVG-54, UVP Inc.; 254-nm bulb) positioned 4 cm away. Following immunoprecipitation with anti-U2AF65 prebound to protein A–Sepharose beads for 1 h at 23°C, samples were washed twice with TBS and resuspended in SDS gel loading buffer. Proteins were separated by electrophoresis on SDS–10% polyacrylamide gels, dried, and exposed on phosphor screens.
Image analysis
Bands were analyzed and quantitated using Image Quant software from Molecular Dynamics. Background values were calculated and subtracted from sample values. For each competition experiment, the relative band intensity was normalized as a percentage value with respect to the average intensity of two lanes of 32P-labeled U4CAGG-containing oligonucleotide without a competitor. The competition results are based on an average of at least two experiments each.
Acknowledgments
This work was supported by Research grant R01 GM59027 from the National Institute of General Medical Sciences.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7221605.
REFERENCES
- Banerjee, H., Rahn, A., Davis, W., and Singh, R. 2003. Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding. RNA 9: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund, J.A., Chua, K., Abovich, N., Reed, R., and Rosbash, M. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T. and Steward, K. 1997. RNA processing and gene structure. In C. elegans II (ed. D.L. Riddle), pp. 117–145. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Conrad, R., Liou, R.F., and Blumenthal, T. 1993. Functional analysis of a C. elegans trans-splice acceptor. Nucleic Acids Res. 21: 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T., Vilardell, J., and Query, C.C. 2002. Pre-spliceosome formation in S. pombe requires a stable complex of SF1-U2AF(59)-U2AF(23). EMBO J. 21: 5516–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J. and Zahler, A.M. 2000. Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res. 10: 1115–1125. [DOI] [PubMed] [Google Scholar]
- Longman, D., Johnstone, I.L., and Caceres, J.F. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19: 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMorris, M.A., Zorio, D.R., and Blumenthal, T. 1999. An exon that prevents transport of a mature mRNA. Proc. Natl. Acad. Sci. 96: 3813–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui, R., Puoti, A., and Kramer, A. 1999. Splicing factor SF1 from Drosophila and Caenorhabditis: Presence of an N-terminal RS domain and requirement for viability. RNA 5: 1615–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino, L., Guth, S., Bilbao, D., Martinez, C., and Valcarcel, J. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Reed, R. 1989. The organization of 3′ splice-site sequences in mammalian introns. Genes & Dev. 3: 2113–2123. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell. Biol. 12: 340–345. [DOI] [PubMed] [Google Scholar]
- Wu, S., Romfo, C.M., Nilsen, T.W., and Green, M.R. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. [DOI] [PubMed] [Google Scholar]
- Zhang, H. and Blumenthal, T. 1996. Functional analysis of an intron 3′ splice site in Caenorhabditis elegans. RNA 2: 380–388. [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.H. and Chasin, L.A. 2004. Computational definition of sequence motifs governing constitutive exon splicing. Genes & Dev. 18: 1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio, D.A. and Blumenthal, T. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838. [DOI] [PubMed] [Google Scholar]
- Zorio, D.A., Lea, K., and Blumenthal, T. 1997. Cloning of Caenorhabditis U2AF65: An alternatively spliced RNA containing a novel exon. Mol. Cell. Biol. 17: 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]