Abstract
RNA-guided nucleotide modification complexes direct the post-transcriptional nucleotide modification of both archaeal and eukaryotic RNAs. We have previously demonstrated that efficient 2′-O-methylation activity guided by an in vitro reconstituted archaeal box C/D sRNP requires juxtaposed box C/D and C′/D′ RNP complexes. In these experiments, we investigate the importance of spatially positioning the box C/D and C′/D′ RNPs within the sRNP complex for nucleotide modification. Initial sequence analysis of 245 archaeal box C/D sRNAs from both Eukyarchaeota and Crenarchaeota kingdoms revealed highly conserved spacing between the box C/D and C′/D′ RNA motifs. Distances between boxes C to D′ and C′ to D (D′ and D spacers, respectively) exhibit highly constrained lengths of 12 nucleotides (nt). Methanocaldococcus jannaschii sR8 sRNA, a model box C/D sRNA with D and D′ spacers of 12 nt, was mutated to alter the distance between the two RNA motifs. sRNAs with longer or shorter spacer regions could still form sRNPs by associating with box C/D core proteins, L7, Nop56/58, and fibrillarin, comparable to wild-type sR8. However, these reconstituted box C/D sRNP complexes were severely deficient in methylation activity. Alteration of the D and D′ spacer lengths disrupted the guided methylation activity of both the box C/D and C′/D′ RNP complexes. When only one spacer region was altered, methylation activity of the corresponding RNP was lost. Collectively, these results demonstrate the importance of box C/D and C′/D′ RNP positioning for preservation of critical inter-RNP interactions required for efficient box C/D sRNP-guided nucleotide methylation.
Keywords: archaeal sRNA, box C/D RNA, box C/D RNP, snoRNA, nucleotide modification
INTRODUCTION
The RNA-guided nucleotide modification complexes utilize guide RNAs possessing complementary sequences, which base pair with target RNA(s) to direct the modification of specific nucleotides (Kiss 2001; Bachellerie et al. 2002; Terns and Terns 2002; Decatur and Fournier 2003; Omer et al. 2003). Based upon conserved sequence elements or “boxes”, the guide RNAs have been divided into two major families and designated the box C/D and the box H/ACA RNAs (Balakin et al. 1996). The primary function of the box C/D RNAs is to direct the 2′-O-methylation of target nucleotides, whereas the box H/ACA RNAs guide the conversion of specific uridine residues to pseudouridine. The presence of box C/D and H/ACA guides RNAs in both Eukarya and Archaea has indicated that these nucleotide modification RNPs are evolutionarily ancient RNA:protein enzymes (Omer et al. 2000; Dennis et al. 2001; Terns and Terns 2002).
Guide RNAs directing nucleotide modification were first described and characterized in eukaryotes (Kiss-Laszlo et al. 1996; Ganot et al. 1997; Ni et al. 1997). Scores of small nucleolar RNAs (snoRNAs) have now been identified and their important roles in ribosome biogenesis have been revealed. In addition to guiding the site-specific modification of ~200 ribosomal RNA (rRNA) nucleotides, a select few also play roles in pre-rRNA folding and the processing of the rRNA precursor transcript (Tycowski et al. 1994; Liang and Fournier 1995; Peculis 1997; Borovjagin and Gerbi 1999). For each of these roles, the snoRNA guides sequence base pairs with the pre-rRNA to select the site of snoRNA function. Box C/D and H/ACA guide RNAs are also found in Archaea. These “snoRNA-like” or sRNAs also use guide sequences for target RNA recognition and nucleotide modification. The strong conservation of box sequence elements and RNA folded structures between the eukaryotic and archaeal guide RNAs suggests a common evolutionary origin (Tran et al. 2004). This suggests that the RNA-guided nucleotide modification complexes arose before the divergence of the Eukarya and Archaea >2 billion yr ago. Recent investigations of both eukaryotic and archaeal guide RNAs have also extended the target RNAs beyond the rRNAs. These targets include the eukaryotic splicing snRNAs, archaeal pre-tRNAs, and even some eukaryotic mRNAs (Tycowski et al. 1998; Cavaille et al. 2000; Gaspin et al. 2000; d’Orval et al. 2001; Darzacq et al. 2002; Tang et al. 2002).
The box C/D RNAs of both eukaryotes and Archaea possess a highly conserved RNA structure located at the terminus of the folded RNA designated the terminal box C/D core motif (Caffarelli et al. 1996; Cavaille and Bachellerie 1996; Watkins et al. 1996; Xia et al. 1997). In eukaryotes, the box C/D core motif has been shown to be essential for snoRNA biosynthesis and nucleolar transport as well as for directing nucleotide 2′-O-methylation (Caffarelli et al. 1996; Cavaille and Bachellerie 1996; Watkins et al. 1996; Lange et al. 1998; Samarsky et al. 1998). Sequence analysis has revealed a second box C/D-related element designated the C′/D′ motif, which is located internally within the RNA molecule (Kiss-Laszlo et al. 1998; Gaspin et al. 2000; Omer et al. 2000). Experiments have demonstrated that the C′/D′ motif with its respective guide sequence can also guide methylation of targeted nucleotides. Both motifs direct 2′-O-methylation of the target nucleotide positioned within the guide RNA:target RNA duplex and located 5 nucleotides (nt) upstream from the D or D′ box. Despite both motifs possessing box C and D sequences, the phylogenetically conserved nucleotides found in the terminal box C/D core motif are frequently degenerate in the C′ and D′ boxes. This is particularly true for the eukaryotic snoRNAs where many snoRNA species do not appear to possess an internal C′/D′ motif. For some eukaryotic snoRNAs such as U14, the C′/D′ motif does not guide nucleotide methylation but alternatively functions in pre-rRNA processing and 18S rRNA production (Liang and Fournier 1995).
Investigations have defined both snoRNP and sRNP core proteins for the box C/D RNAs. In eukaryotes, biochemical and genetic experiments have defined four box C/D snoRNP core proteins; the 15.5kD protein (Snu13p in yeast), nucleolar proteins Nop56 and Nop58, and the methylase fibrillarin (Gauthier et al. 1997; Lafontaine and Tollervey 1999, 2000; Newman et al. 2000; Watkins et al. 2000). In vivo crosslinking of the core proteins has indicated a differential distribution on the terminal box C/D core and internal C′/D′ motif (Cahill et al. 2002; Szewczak et al. 2002). The 15.5kD protein, which initiates snoRNP assembly, binds only the terminal core motif, whereas the Nop56 and Nop58 proteins crosslink to the C′/D′ and core C/D motifs, respectively. Only the methylase fibrillarin is bound to both motifs. Although Archaea contain homologs to the eukaryotic snoRNA core proteins, analysis of the archaeal sRNP complex has revealed a distinctly different RNP structure (Omer et al. 2002; Tran et al. 2003). The three archaeal sRNP core proteins include ribosomal protein L7 (the archaeal homolog of the 15.5kD protein), a single Nop56/58 protein homolog, and an archaeal fibrillarin homolog. In contrast to the eukaryotic snoRNP complex, all three core proteins are bound to both the box C/D core and C′/D′ motifs, forming identical RNPs at both the C/D and C′/D′ motifs.
In vitro assembly systems for the archaeal box C/D sRNP have recently been established (Omer et al. 2002; Tran et al. 2003). These complexes assembled with bacterially expressed recombinant core proteins are enzymatically active and direct 2′ O-methylation from both the terminal box C/D and internal C′/D′ RNPs. During our recent studies characterizing the methylation activity of the reconstituted Methanocaldococcus jannaschii sR8 box C/D sRNP, we demonstrated that efficient methylation activity required juxtaposed box C/D and C′/D′ RNPs (Tran et al. 2003). These observations indicated that inter-RNP interactions played an important role in the RNP-guided methylation reactions. We therefore decided to explore the nature of these crosstalk interactions by determining the importance of spacing between the two RNA motifs for methylation activity. Here we demonstrate that the spacing between the box C/D and C′/D′ motifs is highly constrained in archaeal box C/D sRNAs and that the spatial positioning of the two constituent RNPs within the sRNP complex is critical for nucleotide modification activity.
RESULTS
Archaeal box C/D sRNAs exhibit conserved spacing between the box C/D and C′/D′ motifs
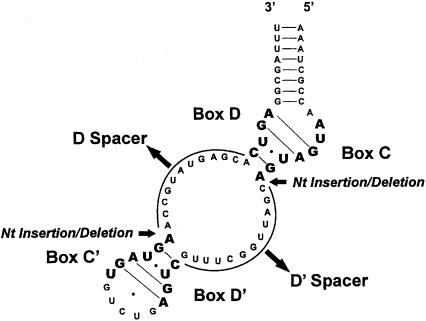
Illustrated in Figure 1 ▶ is the folded structure of M. jannaschii sR8 sRNA, a model box C/D RNA. The terminal box C/D core motif includes boxes C and D, whereas internally located C′ and D′ sequences fold to form the C′/D′ motif. Both motifs form a unique RNA structure called the kink-turn (K-turn) motif, which is typified by two tandem-sheared G:A pairs hydrogen-bonding across the asymmetric bulge (Watkins et al. 2000; Klein et al. 2001; Kuhn et al. 2002; Goody et al. 2004). Guide regions, which base pair through complementarity to the selected target sequence and designate a nucleotide for modification, are found immediately upstream of boxes D and D′. In vitro reconstitution of the sR8 sRNP has demonstrated that each RNA motif binds ribosomal protein L7, Nop56/58, and fibrillarin to assemble identical RNPs within the sRNP complex. Both complexes are functional in guiding the 2′-O-methylation of target RNAs that base pair to the respective D and D′ guide sequences (Tran et al. 2003). For our analyses, we have designated the M. jannaschii sR8 spacing distances between the folded box C/D and C′/D′ motifs as the D and D′ spacers. These two spacers include the D and D′ guide regions, respectively (see Fig. 1 ▶). For sR8 sRNA, the D and D′ guide sequences are 12 nt in length and constitute the entire spacer regions. However, for other archaeal box C/D sRNAs, these spacer regions include additional nucleotides that do not base pair to the target RNAs and are, therefore, not part of the guide sequence.
FIGURE 1.
Folded secondary structure of M. jannaschii sR8 box C/D RNA, a model archaeal box C/D sRNA. The spacing between the box C/D and C′/D′ motifs in archaeal box C/D sRNAs is conserved. Consensus sequence elements (boxes C and D of the terminal core motif and internal boxes C′ and D′) are indicated in bold lettering. The spacer regions between boxes C and D′ (D′ spacer) and boxes C′ and D (D spacer) are designated. These spacer regions include, but are not limited to, the sRNA guide sequence that base pairs with the respective target RNAs. Bold arrows indicate the sites of nucleotide insertions and deletions for the constructed sR8 sRNA mutants.
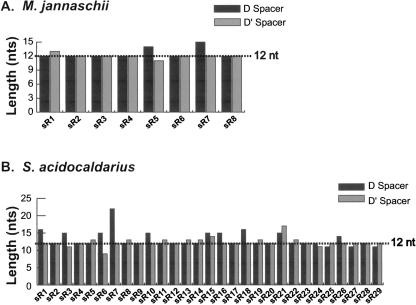
Previous analysis from the Bachellerie laboratory has noted a constrained or conserved spacing between the box C/D core motif and the internal C′/D′ motif of the archaeal box C/D sRNAs (Gaspin et al. 2000). This conservation was attributed to the idea that Archaea are simple organisms and therefore possess “minimalist” box C/D sRNAs. Consistent with this observation, inspection of M. jannaschii box C/D sRNAs revealed that most RNA species possess D and D′ spacers 12 nt in length (Fig. 2A ▶). Analysis of Sulfolobus acidocaldarius box C/D sRNAs also revealed similar box C/D and C′/D′ spacing distances. Interestingly, specific box C/D sRNAs deviating from the 12-nt average frequently possess one spacer region that exhibits the typical inter-motif distance of 12 nt (i.e., S. acidocaldarius sR7 sRNA). Finally, a more complete examination of available box C/D sRNAs from both Euryarchaeota and Crenarchaeota organisms revealed that the spatial constraints between the two RNA motifs is widespread in Archaea (Table 1 ▶). Of the 245 box C/D sRNAs analyzed, ~40% had both D and D′ spacer distances of 12 nt and >80% had at least one 12-nt spacer region.
FIGURE 2.
The spacing between the box C/D and C′/D′ motifs in archaeal box C/D sRNAs is conserved. The length (nucleotides) of the D (black) and D′ (gray) spacer regions is plotted versus specific sRNA species of M. jannaschii (A) and S. acidocaldarius (B) box C/D sRNAs. The average spacer length of 12 nt is indicated by the dashed lines. Box C/D sRNA sequences were obtained from the Eddy snoRNA database (http://rna.wustl.edu/snoRNAdb/).
TABLE 1.
Euryarchaeota and Crenarchaeota box C/D sRNA spacing between the box C/D and C′/D′ RNA motifs
| Archaeal species | Box C/D sRNAs | Average D spacer length (nt) | Average D′ spacer length (nt) |
| Euryarchaeota | |||
| P. horikoshii | 54 | 12.6 ± 1.4 | 12.3 ± 0.7 |
| P. furiosis | 55 | 12.7 ± 1.3 | 12.3 ± 0.9 |
| P. abyssi | 59 | 12.7 ± 1.7 | 12.3 ± 0.9 |
| A. fulgidus | 4 | 11.5 ± 1.0 | 11.5 ± 1.0 |
| M. jannaschii | 8 | 12.6 ± 1.2 | 12.0 ± 0.5 |
| Crenarchaeota | |||
| S. solfactaricus | 13 | 13.3 ± 2.0 | 11.5 ± 0.7 |
| S. acidocaldarius | 29 | 13.2 ± 2.3 | 12.3 ± 1.3 |
| A. pernix | 23 | 13.1 ± 2.2 | 13.0 ± 3.0 |
| Total | 245 | 12.8 ± 1.6 | 12.3 ± 1.1 |
Box C/D sRNA sequences were obtained from the Eddy snoRNA database (http://rna.wustl.edu/snoRNAdb/). Spacer lengths are reported as the average plus or minus the standard deviation for the D or D′ spacer of each archael species analyzed.
Alteration of D and D′ spacer distances does not affect box C/D and C′/D′ RNP assembly
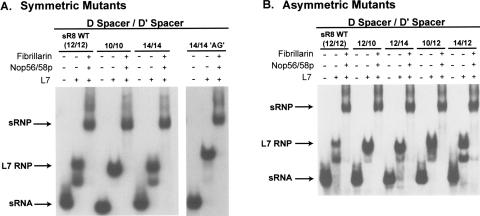
To assess the importance of box C/D and C′/D′ motif spacing for sRNP assembly, various M. jannaschii sR8 mutants were constructed with altered D and/or D′ spacer lengths. sR8 RNA was selected as a model archaeal box C/D RNA because it possesses 12-nt D and D′ spacer regions, the most commonly observed spacer lengths in Archaea. Nucleotide deletions and insertions were made at the 3′ end of each spacer (see Fig. 1 ▶) so as to not alter the guide region immediately downstream of the D and D′ box sequences and thus affect guide sequence:target RNA base pairing. Corresponding deletions/insertions were made in both spacer regions to construct “symmetric” mutants, while nucleotide deletion/insertion in only one spacer created “asymmetric” mutants. Constructed mutants included deletion mutants with final spacer lengths of 10 nt as well as insertion mutants increasing spacer distances by increments of 2 nt (uridine insertions) up to 22 nt in length.
sRNP assembly was first assessed by binding of L7, the core protein that first binds both motifs and initiates box C/D and C′/D′ RNP assembly (Tran et al. 2003). The appearance of two RNPs when the sRNAs were incubated with only L7 demonstrated that this core protein was binding both the box C/D and C′/D′ motifs (Fig. 3A ▶). For all sR8 mutants, L7 bound to both box C/D and C′/D′ RNA motifs. Note that for the 10/10 sRNP mutant, two L7 RNPs are not readily visible. This is most likely due to a higher L7 binding affinity for the box C/D and C′/D′ motifs for this particular sRNA. Subsequent incubation of Nop56/58 and fibrillarin with the L7 RNP formed with wild-type sR8 resulted in the formation of a fully assembled sRNP complex. Similarly, the addition of all three core proteins to both deletion and insertion symmetric as well as asymmetric RNA mutants mimicked wild-type sR8 sRNP assembly, thus demonstrating complete sRNP reconstitution (Fig. 3A,B ▶). For simplicity, a subset of mutant RNP assemblies is shown, but all deletion and insertion mutants, both symmetric and asymmetric, assembled full sRNP complexes (data not shown). Finally, sRNP assembly was independent of the identity of the nucleotides inserted into the spacer regions. Symmetric spacing mutant 14/14 AG (14-nt spacer length for both the D and D′ spacers), which inserted the dinucleotide AG in place of the typical UU dinucleotide, was also able to bind all three core proteins and assemble the complete sRNP complex (Fig. 3A ▶).
FIGURE 3.
Alteration of the spacing distance between the sR8 box C/D and C′/D′ motifs does not affect sRNP assembly. sR8 sRNA D and D′ spacer regions were shortened or lengthened in 2-nt increments by deleting or inserting nucleotides upstream and downstream of boxes C′ and C, respectively, as indicated in Figure 1 ▶. Spacing mutants lengthening the D and D′ spacers were typically created by inserting two uridine nucleotides unless otherwise indicated (spacing mutant 14/14 AG inserted the dinucleotide AG in place of UU). sR8 spacing mutants were either symmetric (A), where both the D and D′ spacers were shortened/lengthened, or asymmetric (B), where only one spacer region was altered. Altered spacing distances are indicated at the top (D spacer/D′ spacer). sR8 sRNPs were assembled in vitro and analyzed by electrophoretic mobility-shift analysis. The sequential addition of the box C/D sRNP core proteins to radiolabeled wild-type and mutant spacer sR8 RNAs is indicated at the top and the migration positions of the resulting RNP complexes are designated at the side. The slightly altered migration of the 14/14 (AG) RNPs is a result of slightly different electrophoretic conditions.
Altered D and D′ spacer distances severely affect box C/D and C′/D′ RNP-guided methylation of target RNAs
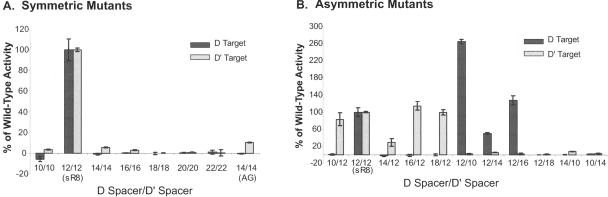
Alteration of the conserved spacing between the box C/D and C′/D′ motifs clearly did not affect sRNP assembly. We then reasoned that the observed constraint in motif spacing might be required for box C/D and C′/D′ RNP-guided methylation activity. To assess this possibility, the methylation capabilities of the box C/D and C′/D′ RNPs assembled on the mutant sR8 sRNAs were determined and compared with wild-type sR8 sRNP activity. Strikingly, deletion or insertion of nucleotides in both D and D′ spacer regions (symmetric mutants) resulted in the loss of guided methylation from both box C/D and C′/D′ RNP complexes as compared with wild-type sR8 (Fig. 4A ▶). Insertion of an AG dinucleotide in place of the typically inserted uridines also resulted in the loss of methylation activity (mutant 14/14 AG). The possibility that the sum of the spacer distances was critical for methylation activity was assessed with two mutants that deleted one spacer while extending the other. sR8 mutants 14/10 and 10/14 maintained a spacer length sum of 24 nt. Both of these mutant sRNAs were incapable of guiding methylation from either RNP. In contrast to the symmetric sR8 mutants, alteration of the length of only one spacer region (asymmetric mutants) had severe effects upon guided methylation only for the RNP complex corresponding to the shortened or lengthened spacer (Fig. 4B ▶). Guided methylation activity from the RNP complex corresponding to the nonmutated or wild-type spacer of 12 nt was either unaffected or even enhanced. Indeed, for asymmetric sR8 mutant 12/10, methylation activity was increased to ~250% of wild-type activity. Collectively, these results demonstrated the importance of conserved spacing between the box C/D and C′/D′ motifs for efficient 2′-O-methylation activity of the corresponding RNP complexes.
FIGURE 4.
Alteration of the distance between the sR8 box C/D and C′/D′ motifs disrupts 2′-O-methylation guided by the box C/D and C′/D RNP complexes. sR8 sRNAs possessing either shortened or lengthened D and D′ spacer regions were assembled in vitro with the three sRNP core proteins and incubated in the presence of 3H-S-adenosyl-methionine and target RNA oligonucleotides complementary to the D or D′ guide regions. 2′-O-methylation guided by the box C/D and C′/D′ RNPs was determined by measuring 3H-methyl incorporation into the respective D and D′ target RNAs as detailed in Materials and Methods. Target RNA oligonucleotides synthetically methylated at the target nucleotide prior to incubation served as controls to establish background levels of TCA-precipitable 3H counts as well as demonstrate site-specific nucleotide modification. Box C/D and C′/D′ RNP-guided methylation activities of symmetric (A) and asymmetric (B) mutant sR8 sRNAs are presented as the percentage of wild-type sR8 sRNP (12/12)-guided methylation activity minus the background of the synthetically methylated target RNA controls. The reported activities are the averages of three determinations. 14/14 (AG) is the sR8 mutant where the dinucleotide AG was inserted in place of UU.
DISCUSSION
Previous investigation of the M. jannaschii sR8 sRNP assembled in vitro demonstrated that juxtaposed box C/D and C′/D′ RNPs are required for efficient sRNP-guided nucleotide methylation (Tran et al. 2003). In the present studies, we have examined the importance of the spatial positioning of the two RNPs within the complete sRNP complex for enzyme activity. The coordinate shortening or lengthening of both D and D′ spacers by as few as 2 nt had a detrimental impact upon the methylation capabilities of this RNA:protein enzyme. Alteration of a single spacer region specifically affected the catalytic activity of its corresponding RNP complex while most often leaving the other RNP’s methylation capabilities largely functional. Collectively, these results demonstrate the importance of spatial positioning of the box C/D and C′/D′ RNPs within the sRNP complex for efficient nucleotide modification.
Sequence analysis of 245 archaeal box C/D sRNAs revealed the highly constrained spatial positioning of the box C/D and C′/D motifs. However, a few sRNA species clearly exhibit spacer regions that deviate significantly from the 12-to 13-nt average, thus calling into question the general requirement of constricted inter-motif spacing for nucleotide methylation. From these exceptions, it may be tempting to speculate that motif spacing is not important for all archaeal double-guide box C/D sRNAs. However, at present, it is not known how many archaeal box C/D species actually function as double-guide sRNAs. Like some eukaryotic box C/D snoRNAs, select archaeal sRNAs may carry out nonnucleotide modification functions such as chaperoning pre-rRNA folding or facilitating cleavage of the pre-rRNA (Tycowski et al. 1994; Liang and Fournier 1995; Peculis 1997; Borovjagin and Gerbi 1999). Thus, constrained inter-motif spacing for these particular sRNAs may not be critical. The sR7 sRNA of S. acidocaldarius may be such a candidate for alternative sRNA function. This sRNA species deviates markedly from the typical box C/D sRNA with a D spacer of >20 nt (Fig. 2 ▶). sR7 does possess two strong guide sequences of 9 and 10 nt that are perfectly complementary to target sequences in 23S rRNA. However, when 23S rRNA methylation was assayed via standard primer extension analysis, only the target nucleotide for the D′ guide sequence was methylated. The lack of target nucleotide methylation for the D guide sequence suggests a nonnucleotide modification function for the box C/D motif, thus potentially explaining sR7’s lack of conserved inter-motif spacing.
It is clear that crosstalk interactions between the box C/D and C′/D′ RNPs play an important role in their methylation capabilities, but it is unclear at the present time the nature of the inter-RNP interactions. Box C/D and C′/D′ RNP crosstalk could result from the physical interactions such as protein:protein contacts between core proteins of the two complexes. Alternatively, optimization of methylation activities could result from induced structural changes in either the RNA motifs or the assembled RNPs caused by juxtaposed complexes. L7 core protein binding to the box C/D and C′/D′ motifs is co-operative, but neither physical interaction of the two proteins nor induced structural changes has yet been demonstrated (Tran et al. 2003). From analysis of the crystal structure of the Nop56/58-fibrillarin dimer, Aittaleb and coworkers have suggested that the Nop56/58 proteins of each RNP could interact via an extended coiled-coil region of this core protein (Aittaleb et al. 2003). This model suggests that crosstalk interactions between the box C/D and C′/D′ RNPs could be due to protein:protein contact of the two Nop56/58p proteins bound to each motif mediated by the extended coiled-coil arm. However, molecular modeling of the self-dimerized Nop56/58 proteins positioned upon the box C/D and C′/D′ RNA motifs has revealed that the spatial distances required for their coiled-coil interaction precludes the correct positioning of the Nop56/58-fibrillarin dimers upon the two RNA motifs that is needed to carry out the corresponding methylation reactions (B. Brown, E. Tran, and S. Maxwell, unpubl.).
The conserved inter-RNP spacing of archaeal box C/D sRNPs leads to the question of whether a similar, constrained spacing of the box C/D and C′/D′ motifs is observed for the eukaryotic box C/D snoRNAs. We have carried out a preliminary examination of motif spacing for a limited number of box C/D snoRNAs from human and yeast (Fig. 5 ▶). While the analyzed snoRNAs do not represent the full complement of box C/D species for these two organisms, it is clear from a cursory examination that the spacing between the two motifs is not highly constrained. For the human box C/D snoRNAs presented in Figure 5A ▶, D and D′ spacer lengths vary from as little as 3 (U41) to as many as 75 nt (U15). Similarly, those yeast box C/D snoRNAs with identified C′ and D′ boxes vary from <20 to ~130 nt (Fig. 5B ▶). Variation in spacer lengths is consistent with the fact that eukaryotic snoRNAs are typically larger than the archaeal sRNAs. From these preliminary analyses, one can conclude that the eukaryotic snoRNA box C/D and C′/D′ motifs are not spatially constrained.
FIGURE 5.
D and D′ spacer lengths of human and yeast box C/D snoRNAs. The length (nucleotides) of the D (black) and D′ (gray) spacer regions of selected box C/D snoRNAs of human and the yeast Saccharomyces cerevisiae is presented in A and B, respectively. These selected species from Kiss-Laszlo et al. (1998) possess known or putative C′ and D′ sequences. Asterisks indicate those box C/D snoRNAs known to guide 2′-O-methylation from both the terminal box C/D and internal C′/D′ RNPs.
Additionally, our limited survey of eukaryotic box C/D snoRNAs has revealed that analysis of motif spacing is not straightforward. In contrast to Archaea where the C′ and D′ boxes closely reflect the box C and D consensus sequences, the eukaryotic C′ and D′ sequences are often quite variable. Identification of C′ sequences is particularly demanding as this box element seems less conserved than the D′ box. D′ sequences have often been predicted as they are located immediately downstream of a second snoRNA guide sequence. However, even for numerous yeast snoRNAs possessing designated D′ boxes delineated by associated guide sequences, identification of the corresponding C′ sequences has been problematic and they are often left undesignated. The inherent difficulty in defining such variable sequences elements calls into question many of the C′ and D′ boxes designated in the absence of functional analysis and/or identification of target RNAs for their putative guide sequences.
A first glance, it is tempting to conclude that since the spatial distancing of the box C/D and C′/D′ motifs within eukaryotic snoRNAs is not conserved, inter-RNP crosstalk between the box C/D and C′/D′ complexes plays no role in guided methylation. However, such a conclusion may be premature. Many box C/D snoRNAs with identified C′ and D′ boxes do not utilize both box C/D and C′/D′ motifs for nucleotide modification. Thus, the spatial positioning of juxtaposed RNA motifs may or may not be critical for their alternative functions. For those snoRNAs that do guide 2′-O-methylation from both the box C/D and C′/D′ RNPs (snoRNAs of Fig. 5 ▶, A and B designated by asterisks), these species still appear to lack constrained and conserved D and D′ spacer lengths. However, it is possible that these double-guide snoRNAs possess box C/D and C′/D′ motifs that are spatially and functionally linked using alternative schemes. It is reasonable to suggest that within the fully assembled snoRNP, the folding of spacer regions possessing extra sequence into novel secondary structures could spatially link the box C/D and C′/D′ RNPs. Such may be the case for the double-guide snoRNA U15, where extra sequence in the D spacer region (~65 nt) has been proposed to fold and juxtapose box C/D and C′/D′ motifs (Tycowski et al. 1993; Pellizzoni et al. 1994). Alternatively, species-specific snoRNP proteins could function to spatially position the box C/D and C′/D′ RNPs by binding novel sequences in the expanded spacer regions. These species-specific proteins could alter motif positioning by folding expanded spacer regions or even bridge the two RNP complexes via protein:protein interactions.
The functional importance of RNP subcomplex spatial positioning within the overall architecture of the RNA-guided nucleotide modification complexes requires further investigation. The observation that the pseudouridylation of nucleotides targeted by the eukaryotic H/ACA snoRNAs is affected by the second hairpin of their characteristic bipartite snoRNA structure implies inter-RNP interactions for this guide RNA family as well (Bortolin et al. 1999). It is interesting to consider that the spatial positioning of the RNP subcomplexes may ultimately be required for the precise positioning of each RNP at their respective sites of action on the target RNA(s). For example, dual guide snoRNAs might direct the activities of both constituent RNPs simultaneously upon a single pre-rRNA transcript being assembled into ribosomal subunits within the nucleolus. In this context, the dual actions of a given box C/D or H/ACA RNP may also be temporally controlled. Such is the case for archaeal pre-tRNAtrp methylation, where the two nucleotide methylation events guided by the pre-tRNA’s two intron-encoded box C/D RNPs are accomplished sequentially (Singh et al. 2004). Ultimately, such temporal and/or spatial constraints may be critical for coordinating specific, post-transcriptional processing steps in the biosynthetic pathway of the target RNA.
MATERIALS AND METHODS
DNA template construction and RNA synthesis
Target RNA substrates were purchased from Dharmacon Research, Inc. Their sequences are reported here in the 5′ to 3′ direction with methylated nucleotides preceded by an “m.” D target (D): CUGAUGCUCAUACGGUCUGCU; methylated D target (D-CH3): CUGAUGCUmCAUACGGUCUGCU; D′ target (D′): GCUCAAAGCCAAUCGC; methylated D′ target (D′-CH3): GCUCAAAmGCCAAUCGC. The DNA templates used for RNA transcription of wild-type and mutant sR8 RNAs were produced by PCR amplification of template M. jannaschii genomic DNA as previously described (Tran et al. 2003). In vitro transcription of the DNA templates was carried out using AmpliScribe T7-Flash Transcription Kit (Epicentre Technologies) following the manufacturer’s protocol. RNA transcripts were purified from denaturing polyacrylamide gels, treated with calf intestine phosphatase (New England Biolaboratories), and then 5′-labeled with γ-32P-ATP using T4 polynucleotide kinase (New England Biolaboratories) following the manufacturer’s protocol. DNA primer pairs (in parentheses) used for DNA template synthesis are indicated below followed by the primary sequences of the individual primers. Wild-type sR8 DNA template (a + b); Symmetric mutants: 10/10 (c + d), 14/14 (e + f), 16/16 (g + h), 18/18 (i + j), 20/20 (k + l), 22/22 (m + n), 14/14 AG (o + p); Asymmetric mutants: 12/10 (a + d), 12/14 (a + f), 12/16 (a + h), 12/18 (a + j), 10/12 (c + b), 14/12 (e + b), 16/12 (g + b), 18/12 (i + b).
C TAAT ACGACTCACTATAGGCCAAATCGCCAATGATGA CGATTG
AAATCGCCTCAGTGCTCATACGG
C TAATACGAC TCACTATAGGCCAAATCGCCAATGATGA ATTGG
AAATCGCCTCAGTGCTCATACGTCATCACAGAC
C TAATACGACTCAC TATAGGCCAAATCGCCAATGATGA TTCGATTGGC
AAATCGCCTCAGTGCTCATACGGTAATCATCACAG
C TAATACGACTCACTATAGGCCAAATCGCCAATGATGA TTTTCGATTGGC
AAATCGCCTCAGTGCTCATACGGTAAAATCATCACAG
C TAATACGACTCAC TATAGGCCAAATCGCCAATGATGA TTTTTTCGATTGGC
AAATCGCCTCAGTGCTCATACGGTAAAAAATCATCACAG
C TAATACGACTCAC TATAGGCCAAATCGCCAATGATGA TTTTTTTTCGATTGGC
AAATCGCC TCAGTGC TCATACGGTAAAAAAAATCATCA CAG
C TAATACGACTCAC TATAGGCCAAATCGCCAATGATGA TTTTTTTTTTCGATTGGC
AAATCGCC TCAGTGC TCATACGG TAAAAAAAAAATCAT CACAG
C TAATACGACTCACTATAGGCCAAATCGCCAATGATGA AGCGATTGGC
AAATCGCCTCAGTGCTCATACGGTCTTCATCACAG
RNP analysis and in vitro methylation of target RNAs
Wild-type and mutant sR8 sRNAs were incubated with purified, recombinant M. jannaschii core proteins (L7, Nop56/58p, and fibrillarin) and in vitro assembled sRNP complexes analyzed by native gel electrophoresis as previously described (Tran et al. 2003). The in vitro methylation activities of assembled sRNP complexes were determined as previously described (Tran et al. 2003). Briefly, sRNP complexes were assembled at 70°C for 10 min in 40-μL volumes containing 0.65 μM guide RNA, 0.5 μM L7, 1.5 μM Nop56/58p, and 1.5 μM fibrillarin. Assembled RNPs were then mixed with 6.5 μM target RNA, 3.3 μM SAM (S-adenosyl-L-methionine dihydrogen sulfate; Calbiochem) and 0.8 μCi of [3H] SAM (55 Ci/mmol; MP Biomedicals) to a final volume of 55 μL. After incubation at 68°C for 1 h, 20-μL aliquots were removed and TCA precipitated onto filter paper. Methyl incorporation into the target RNAs was measured by scintillation counting. Methylation assays were performed in triplicate and are reported as the average of three experiments.
Acknowledgments
This work was supported by NSF Grant MCB-0215545 to E.S.M. and an NSF REU to L.L.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7223405.
REFERENCES
- Aittaleb, M., Rashid, R., Chen, Q., Palmer, J., Daniels, C., and Li, H. 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 10: 256–263. [DOI] [PubMed] [Google Scholar]
- Bachellerie, J.P., Cavaille, J., and Huttenhofer, A. 2002. The expanding snoRNA world. Biochimie 84: 775–790. [DOI] [PubMed] [Google Scholar]
- Balakin, A., Smith, L., and Fournier, M.J. 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834. [DOI] [PubMed] [Google Scholar]
- Borovjagin, A. and Gerbi, S. 1999. U3 small nucleolar RNA is essential for cleavage at sites 1, 2, and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol. 286: 1347–1363. [DOI] [PubMed] [Google Scholar]
- Bortolin, M., Ganot, P., and Kiss, T. 1999. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 18: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli, E., Fatica, A., Prislei, S., DeGregorio, E., Fragapane, P., and Bozzoni, I. 1996. Processing of intron-encoded U16 and U18 snoRNAs: The conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 15: 1121–1131. [PMC free article] [PubMed] [Google Scholar]
- Cahill, N., Friend, K., Speckman, W., Li, Z., Terns, R., Terns, M., and Steitz, J. 2002. Site-specific cross-linking analyses reveal an asymmetric distribution for a box C/D snoRNP. EMBO J. 21: 3816–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille, J. and Bachellerie, J.P. 1996. Processing of fibrillarin-associated snoRNAs from pre-rRNA introns: An exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie 78: 443–456. [DOI] [PubMed] [Google Scholar]
- Cavaille, J., Buiting, K., Kiefmann, M., Lalande, M., Brannan, C., Horsthemke, B., Bachellerie, J.P., Brosius, J., and Huttenhofer, A. 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. 97: 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq, X., Jady, B., Verheggen, C., Kiss, A., Bertrand, E., and Kiss, T. 2002. Cajal body-specific small nuclear RNAs: A novel class of 2 ′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21: 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur, W. and Fournier, M.J. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278: 695–698. [DOI] [PubMed] [Google Scholar]
- Dennis, P., Omer, A., and Lowe, T. 2001. A guided tour: Small RNA function in Archaea. Mol. Microbiol. 40: 509–519. [DOI] [PubMed] [Google Scholar]
- d’Orval, B., Bortolin, M., Gaspin, C., and Bachellerie, J.P. 2001. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 29: 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot, P., Bortolin, M., and Kiss. T. 1997. Site-specific pseudouridine formation in pre-ribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809. [DOI] [PubMed] [Google Scholar]
- Gaspin, C., Cavaille, J., Erauso, G., and Bachellerie, J.P. 2000. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: Lessons from the Pyrococcus genomes. J. Mol. Biol. 297: 895–906. [DOI] [PubMed] [Google Scholar]
- Gauthier, T., Berges, T., Tollervey, D., and Hurt, E. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17: 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody, T., Melcher, S., Norman, D., and Lilley, D. 2004. The kink-turn motif in RNA is dimorphic, and metal ion dependent. RNA 10: 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20: 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M., and Kiss, T. 1996. Site-specific ribose methylation of pre-ribosomal RNA: A novel function for small nucleolar RNAs. Cell 85: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., and Kiss, T. 1998. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 17: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, D., Schmeing, T., Moore, P., and Steitz, T. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, J., Tran, E., and Maxwell, E.S. 2002. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 30: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, D. and Tollervey, D. 1999. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 5: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Synthesis and assembly of the box C + D small nucleolar RNPs. Mol. Cell. Biol. 20: 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, T., Borovjagin, A., Maxwell, E.S., and Gerbi, S. 1998. Conserved nucleotide boxes C and D are essential nucleolar localization elements of U8 and U14 snoRNAs. EMBO J. 17: 3176–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, W. and Fournier, M.J. 1995. U14 base-pairs with 18S rRNA: A novel snoRNA interaction required for rRNA processing. Genes & Dev. 9: 2433–2443. [DOI] [PubMed] [Google Scholar]
- Newman, D., Kuhn, J., Shanab, G., and Maxwell, E.S. 2000. Box C/D snoRNA-associated proteins: Two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 6: 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, J., Tien, A., and Fournier, M.J. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573. [DOI] [PubMed] [Google Scholar]
- Omer, A., Lowe, T., Russell, A., Ebhardt, H., Eddy, S., and Dennis, P. 2000. Homologs of small nucleolar RNAs in Archaea. Science 288: 517–522. [DOI] [PubMed] [Google Scholar]
- Omer, A., Ziesche, S., Ebhardt, H., and Dennis, P. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribo-nucleoprotein complex. Proc. Natl. Acad. Sci. 99: 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer, A., Zeische, S., Decatur, W., Fournier, M.J., and Dennis, P. 2003. RNA-modifying machines in Archaea. Mol. Microbiol. 48: 617–629. [DOI] [PubMed] [Google Scholar]
- Peculis, B. 1997. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol. Cell. Biol. 17: 3702–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni, L., Crosio, C., Campioni, N., Loreni, F., and Pierandrei-Amaldi, P. 1994. Different forms of U15 snoRNA are encoded in the introns of the ribosomal protein S1 gene of Xenopus laevis. Nucleic Acids Res. 22: 4607–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky, D., Fournier, M.J., Singer, R., and Bertrand, E. 1998. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17: 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S., Gurha, P., Tran, E., Maxwell, E.S., and Gupta, R. 2004. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonu-cleoprotein of pre-tRNA. J. Biol. Chem. 279: 47661–47671. [DOI] [PubMed] [Google Scholar]
- Szewczak, L.W., DeGregorio, S., Strobel, S., and Steitz, J. 2002. Exclusive interaction of the 15.5kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9: 1095–1107. [DOI] [PubMed] [Google Scholar]
- Tang, T., Bachellerie, J.P., Rozhdestvenshy, T., Bortolin, M., Huber, H., Drungowski, M., Elge, T., Brosius, J., and Huttenhofer, A. 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. 99: 7536–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns, M. and Terns, R. 2002. Small nucleolar RNAs: Versatile transacting molecules of ancient evolutionary origin. Gene Expr. 10: 17–39. [PMC free article] [PubMed] [Google Scholar]
- Tran, E., Zhang, X., and Maxwell, E.S. 2003. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 22: 3930–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, E., Brown, J., and Maxwell, E.S. 2004. Evolutionary origins of the RNA-guided nucleotide modification complexes: from the primitive translation apparatus? Trends Biochem. Sci. 29: 343–350. [DOI] [PubMed] [Google Scholar]
- Tycowski, K., Shu, M., and Steitz, J. 1993. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S13. Genes & Dev. 7: 1176–1190. [DOI] [PubMed] [Google Scholar]
- ———. 1994. Requirement for intron-encoded U22 small nucleolar RNA for 18S ribosomal RNA maturation. Science 266: 1558–1561. [DOI] [PubMed] [Google Scholar]
- Tycowski, K., You, Z., Graham, P., and Steitz, J. 1998. Modification of U6 spliceosomal RNA is guided by other small RNAs. Cell 92: 629–638. [DOI] [PubMed] [Google Scholar]
- Watkins, N., Leverette, R., Xia, L., Andrews, M., and Maxwell, E.S. 1996. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA 2: 118–133. [PMC free article] [PubMed] [Google Scholar]
- Watkins, N., Segault, V., Charpentier, B., Nottrott, S., Fabrizio, P., Bachi, A., Wilm, M., Roshbash, M., Branlant, C., and Luhrmann, R. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466. [DOI] [PubMed] [Google Scholar]
- Xia, L., Watkins, N., and Maxwell, E.S. 1997. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA 3: 17–26. [PMC free article] [PubMed] [Google Scholar]