Abstract
Based on the contents of the database S/MARt DB, the most comprehensive data collection of scaffold/matrix-attached regions (S/MARs) publicly available thus far, we initiated a systematic evaluation of the stored data. By analyzing the 245 S/MAR sequences presently described in this database, we found that the S/MARs contained in this collection are generally AT-rich, with certain significant exceptions. Comparative analyses showed that most of the AT-rich motifs which were found to be enriched in S/MARs are also enriched in randomized S/MAR sequences of the same AT content. Some sequence patterns previously suggested to be characteristic for S/MARs were also investigated, among them potential binding sites for homeodomain transcription factors. Even though hexanucleotides containing the core motif of homeodomain factors were frequently observed in S/MARs, only a few potential binding sites for these factors were found enriched when compared with regulatory regions or exon sequences. All our analyses indicated that, on average, the observed frequency of motifs in S/MAR elements is largely influenced by the AT content. Our results can serve as a guideline for further improvements in the definition of S/MARs, which are now believed to constitute the functional coordinate system for genomic regulatory regions.
INTRODUCTION
Within the past 20 years the model of the interphase nucleus changed from a ‘bag of chromatin immersed in homogeneous nucleoplasm’ to an anchored loop domain model in which chromatin is organized into loops fastened by a nuclear scaffold or matrix consisting of non-histone proteins (1,2). Attachment of these loops to the nuclear scaffold or matrix occurs at discrete regions, the scaffold or matrix-attached regions (S/MARs). It has been estimated that the human genome contains approximately 100 000 nuclear matrix attachment sites (3).
S/MARs reside at the bases of the DNA loops that become visible as a halo around extracted nuclei and are retained in nuclear scaffold/matrix preparations from interphase nuclei. A mostly overlapping group of elements is shown to exhibit a high affinity when interacting with preparations of the nuclear matrix (4,5).
S/MARs have been implicated in a variety of important functions, such as genome organization and gene expression. In metaphase chromosomes S/MARs appear to be juxtaposed in the center of each chromatid (6). Thereby they are in a position to contribute to chromosome condensation during nuclear division (6). The similarity between metaphase chromosomes and lampbrush chromosomes suggests the same principal organization for both transcriptionally inactive and active chromosomes (7).
S/MARs have also been assigned to function in gene expression, where they are regarded as a distinct class of cis-acting elements affecting transcription regulation (8,9). Along these lines, several reports have demonstrated that, after stable integration of a template, S/MARs may provide a significant enhancement of transcription (10–13). Other reports have suggested S/MARs to function as boundary elements or insulators (14–18). S/MARs may also play a fundamental role in carcinogenesis (19).
As soon as the first S/MAR sequences became available they were analyzed for common sequence characteristics (see for example 1,20–22). The features which emerged in these analyses were considered characteristic of this group of elements. In order to assist S/MAR prediction and to enhance our knowledge of their function we have set up a database, S/MARt DB, which, at present, contains 245 S/MAR sequences (23,24). Based on this database, we attempted to verify some sequence characteristics of S/MARs by comparison with sequences from regulatory (extended promoter sequences) and non-regulatory (exon sequences) regions.
MATERIALS AND METHODS
S/MAR sequences were obtained from the S/MAR transaction database [http://transfac.gbf.de/SMARtDB/], release 2.0 (24).
For comparison the following additional data sets were compiled: four sets of randomized S/MAR sequences were generated by Perl script. To shuffle the bases in the individual sequences we used a Roulette Wheel selection algorithm with adaptive weights for the probabilities of getting one of the four base types. The Roulette Wheel selection is a stochastic selection algorithm where an individual is selected with a probability directly proportional to its fitness (here: frequency of occurrence).
Extended promoter sequences between –499 and +101 relative to the transcription start site were retrieved from the Eukaryotic Promoter Database (EPD), release 59 (25). Some extended promoter sequences may be shorter than 600 bp because the available genomic sequences did not cover the whole range.
Exon sequences of vertebrates were extracted from the EMBL data library, release 56 (26). Only sequences from exon 3 and following exons were considered. Sequences of exons 1 and 2, which were shown earlier to contain a certain number of proven transcription factor-binding sites, were omitted to ensure a minimal concentration of functional regulatory elements in the final sequence set (27).
The sequence sets were analyzed using the program MatInspector professional 4.2, locally installed under Linux (28). Using an integrated matrix library mainly derived from the TRANSFAC database (29), we screened the S/MAR sequences for the presence of transcription factor cognate motifs. To analyze the S/MAR sequences for hexanucleotide composition the analysis was carried out using the user defined IUPAC string option of the MatInspector program. This option was also used to check the sequence sets for A-box and T-box motifs, for consensus sequences for Drosophila and vertebrate topoisomerase II and for the S/MAR-specific sequence defined by van Drunen et al. (22). To enable comparison the analysis of these motifs was also performed with matrices. For this purpose, we transformed the IUPAC consensus strings into matrices of comparable format according to the meaning of the degenerate codes, e.g. C gives mC = 4 and mA = mG = mT = 0, S is transformed to mC = mG = 2 and mA = mT = 0 and N is represented by mA = mC = mG = mT = 1. The search for insect and vertebrate transcription factor-binding sites was performed using TRANSFAC-derived weight matrices. The analyses were carried out with Matrix Family Library v.1.7. For searches employing matrices the core similarity was set to 75 or 90%, respectively, and the matrix similarity was set to ‘calculated optimized’. To calculate S/MAR enrichment factors over distinct negative training sets, we determined the ‘concentration’ of individual patterns a in the distinct sequence sets. Thus, the concentration csm(a) of pattern a in the S/MAR set of ssm sequences comprising nsm nucleotides is
csm(a) = [hsm(a)]/{nsm – [ssm·w(a) – 1]}
with hsm(a) being the number of hits of pattern a in this sequence set and w(a) the width (length) of pattern a. The denominator represents the number of positions that are capable of matching; csm(a) thus ranges between 0 and 1. The concentrations of pattern a in exon sequences [cex(a)] and extended promoters [cep(a)] were determined accordingly. Thus, the enrichment factor rsm/ex of pattern a in S/MAR over exon sequences is defined as
rsm/ex(a) = [csm(a)]/[cex(a) + csm(a)]
Enrichment factors for comparison with extended promoters (rsm/ep) were calculated correspondingly. As is evident from this equation, a rsm/ex(a) value of 0.5 indicates an equal concentration of motif a in both sequence sets, higher values (up to 1) over-, and lower values (down to 0) under-representation of a in S/MARs compared to exons.
The AT content of the individual search patterns represented by positional weight matrices was calculated as
rAT(a) = (∑w(a)i = 1{[mA(i) + mT(i)]/
[ ∑N A,C,G,TmN(i)]})/w(a)
with mN(i) being the weight (occurrence or frequency, depending on the type of matrix used) of nucleotide N in position i. IUPAC consensus strings were transformed into matrices as stated above.
The output of the MatInspector searches for the motifs mentioned above and putative transcription factor-binding sites was further analyzed by making use of MS Excel statistic functions (i.e. calculation of linear regression line, standard error of regression, coefficient of correlation, coefficient of determination).
Deviation from regression was calculated as follows:
dsm/ex(a) = [rsm/ex(observed) – rsm/ex(expected)]/σ(expected)
where rsm/ex(expected) is the y value of a point on the respective regression line.
In a different approach the matrices were grouped by their AT content in such a way that each group consists of at least three matrices (<0.240, 0.241–0.260, 0.261–0.280, …, 0.781–0.800, >0.801). For each group the mean enrichment factor and the standard deviation were calculated and used to assess the deviations from the means.
RESULTS
Nucleotide composition of S/MARs
Until recently, most researchers in the field have agreed that a common feature of all S/MARs is their elevated AT content (6,30,31) and it was only Boulikas who has also considered a group of S/MARs rich in GA or CT motifs (30). Since S/MARs are supposed to be involved in gene regulation and to reside in non-coding regions (9,32), we have compared the AT content of S/MAR sequences in our database with promoter and exon sequences. Promoter sequences have been included in the analysis as a distinct class of regulatory sequences whereas exon sequences were taken as representatives of sequences which are supposed to be involved neither in gene regulation nor in matrix attachment (Table 1; 25,26). Both comparisons revealed that the mean AT content of the S/MARs covered here was significantly higher (62%) than that of the respective reference sequences, which did not differ considerably (exons 47%, extended promoters 51%). The difference becomes particularly evident when comparing the portion of sequences with an AT content of >70% in S/MARs (12% of all sequences) with those in exons (0.4%) and in extended promoter regions (3%). Conversely, the portion of sequences that exhibit a low AT content (≤50%) is extremely low in the S/MAR set (6.5%) compared to the other sequence sets (67 and 47% in exons and extended promoters, respectively). Thus, S/MAR sequences contained in S/MARt DB tend to support the idea that prototype S/MARs are AT-rich.
Table 1. AT content in different data sets.
| S/MARs | Exons | Extended promoters | |
|---|---|---|---|
| No. of nucleotides | 521 366 | 3 686 246 | 813 600 |
| No. of sequences | 245 | 19 238 | 1356 |
| Mean AT ratio | 61.68% | 46.85% | 50.63% |
| Proportion of sequences with AT ratio ≤50% | 6.53% | 66.87% | 47.49% |
| Proportion of sequences with AT ratio >70% | 12.24% | 0.41% | 2.95% |
S/MARs, S/MAR sequences in the S/MARt DB database; Exons, exon sequences of vertebrates (EMBL release 56, exon 3 and greater sequences were considered); Extended promoter regions, 600 bp promoter region (–499 to +101) according to EPD release 59.
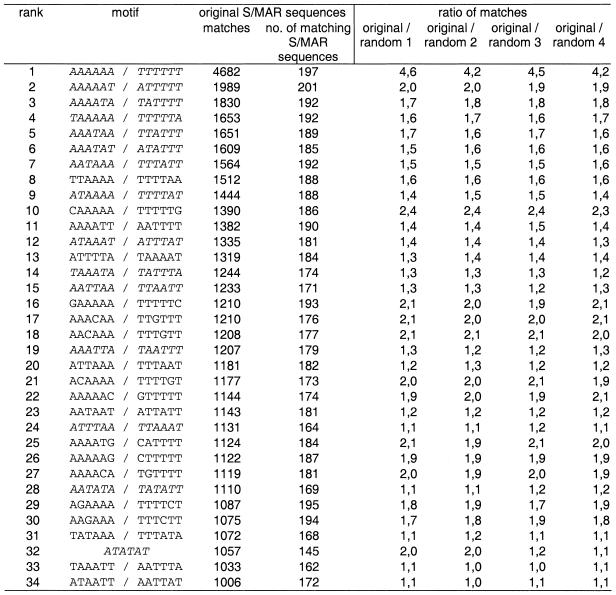
The collection of S/MAR sequences compiled in the S/MARt DB was scanned with all possible hexanucleotides (4096) to identify those that are significantly over-represented. Results were cleared of redundant patterns yielding a non-redundant list of 2080 hexamer patterns, each of them expected to identify about 250 hexameric sites in the S/MAR sequence set. This analysis was repeated with four randomized S/MAR sequence sets. Even though the number of matches obtained ranged between 4682 and 17 per hexanucleotide for the original S/MAR sequences, none of the hexanucleotides is present in all S/MARs. Table 2 lists the hexanucleotides that have been found most frequently in the S/MAR sequence set and exhibit a match number of >4-fold of the expected frequency, i.e. >1000. Out of these 34 sequence elements, about two-thirds consist exclusively of A and T; none of them contains more than one C or G residue. These non-A/T bases are (with only one exception) interspersed in pure T stretches, as might be expected since by far the most frequent motif consists of a stretch of six T residues (or A in the case of the complementary strand). The second most frequent pattern, which has been found in even slightly more S/MARs than the T6 motif, is ATTTTT (or TAAAAA). All five other A5T hexamers are in the list as well, as are 12 of the 15 possible A4T2 hexanucleotides, but only 4 of 14 non-redundant A3T3 hexamers have been found in the list of the most over-represented hexamer patterns, and these are in the lower third. Among those over-represented hexamers that contain a C or G are all six A5C and four of the six possible A5G hexamers, and only one hexamer in this list contains a G that is not embedded in or adjacent to a A or T run (AAAATG). These findings agree with previous observations that S/MARs often contain AT-rich motifs such as oligo(A) runs (10,33). Furthermore, the compilation of frequent motifs contains two hexanucleotides (ATATTT and ATATAT) which, in the context of a certain sequence (ATC) environment were correlated with the unwinding propensity of S/MARs, i.e. constituting ‘core unpairing motifs’ (CUEs) (5,34,35). Table 2 also shows that hexanucleotides containing the core motif of homeodomain transcription factors (ATTA or TAAT) belong to the frequently found motifs in S/MARs.
Table 2. Most frequent 34 hexanucleotides in S/MARt DB.
A total of 521 366 nt in 245 sequences, giving a maximum of 520 141 positions that are capable of matching, was scanned with all hexanucleotide patterns. The analysis was done with both strands, therefore the complementary hexanucleotides (giving the same number of matches) are omitted from the list. Theoretically, each hexanucleotide can give rise to about 250 matches. Italic indicates motifs that were also found to be over-represented in randomized S/MAR sequences.
Analyzing the different sets of randomized S/MAR sequences, between six and nine motifs matched the criteria defined above. All these motifs are composed exclusively of A and/or T residues and are also found over-represented in the original S/MAR sequence set (Table 2, motifs in italic). Of note, the two motifs most frequently observed in the original S/MAR sequence set showed up in all sets of shuffled S/MARs. On the other hand, of the hexanucleotides listed in Table 2 the T6 (or A6) and T5G motifs were consistently found at least twice as often in the original S/MAR sequences than in the shuffled S/MARs. Beyond this finding there were no other marked deviations in the numbers of hits between the original and randomized sequence sets. In particular, we did not observe a significant enrichment of the hexameric CUEs (see above; 5,34) and hexamers containing the core motif of homeodomain transcription factors in S/MARs compared to random sequences of the same AT content.
Potential transcription factor-binding sites in S/MARs
A number of previously published reports implicated transcription factors in S/MAR function (21,30,36–38). One of them proposed homeodomain protein-binding sites to be characteristic for S/MARs (21,30). Therefore, the MatInspector program (28) was used to search with weight matrices for potential binding sites for transcription factors of insects and vertebrates in the sequence sets comprising S/MARs, shuffled S/MARs, exons and extended promoters. For systematic comparison we calculated the S/MAR enrichment factor r for each matrix as explained in Materials and Methods.
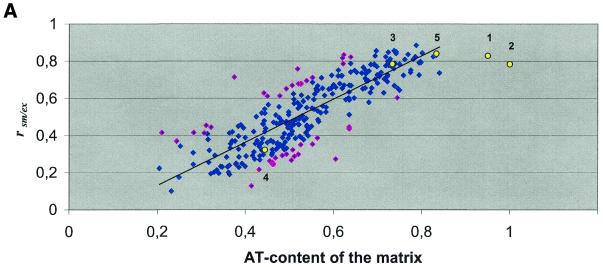
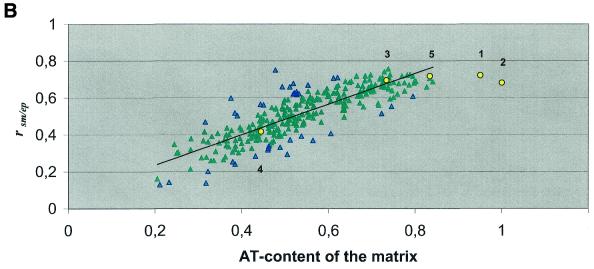
Since prototype S/MARs exhibit an elevated AT content (Table 1), we suspected that the AT content of a matrix might have an influence on whether or not a certain DNA transcription factor-binding motif is over-represented in the S/MAR data set. Therefore, we computed the AT content of the matrices used (see Materials and Methods). When plotting the S/MAR enrichment factors determined for all matrices against their AT content, we noticed a positive correlation. This holds true for comparison of the ‘wild-type’ S/MAR sequences with either exons or extended promoters (Fig. 1) and for distinct MatInspector search stringencies (data not shown). Essentially the same picture emerged when the original S/MARs were replaced by randomized S/MAR sequences. In contrast, no such correlation was found when S/MARs were compared to either set of randomized S/MAR sequences and plotted against the AT content of the matrices used (data not shown).
Figure 1.
Correlation of S/MAR enrichment factors with AT content of the search patterns applied. A matrix search for putative binding sites of insect and vertebrate transcription factors or sequence elements previously reported to be S/MAR-specific was performed. The MatInspector program was adjusted to a core similarity of 0.75 and the matrix similarity was set to ‘calculated optimized’. (A) Enrichment factors (rsm/ex) indicated by blue diamonds refer to over-representation of putative transcription factor-binding sites in S/MARs when compared to exon sequences. The enrichment factors in pink indicate results that exceed the confidence interval of 1.5σ. The correlation coefficient is 0.83 for this comparison. (B) Enrichment factors (rsm/ep) indicated by green triangles refer to over-representation of putative transcription factor-binding sites in S/MARs when compared to extended promoter sequences. The enrichment factors in blue indicate results that exceed the confidence interval of 1.5σ. Here the correlation coefficient is 0.84. The obtained correlations are significant (significance level P < 0.001). Yellow circles with numbers attached refer to matrices for previously reported S/MAR-specific sequence elements re-evaluated here. The numbering refers to the footnotes to Table 4. Regression lines are shown as solid black lines.
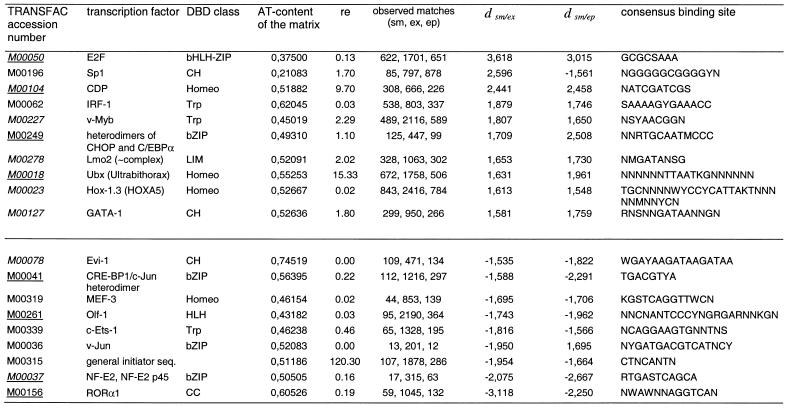
In order to determine matrices which describe motifs that may be significantly over- or under-represented in S/MARs, we calculated, as a first approximation, linear regression lines and their standard errors. Those patterns a were considered to be significantly over- or under-represented for which the corresponding r(a) value exceeded the ±1.5-fold standard error value (see dsm/ex and dsm/ep values). This table also indicates the ‘re values’ given by MatInspector, indicating the number of hits per kb the corresponding matrix produces in random sequences and, hence, its stringency. Routine analyses normally do not use matrices with a ‘re value’ greater than 7. Applying the criterion |dsm/ex| or |dsm/ep| > 1.5, the matrices listed in Table 3 describe motifs that are over- or under-represented in S/MARt DB. Comparable numbers of matrix patterns were found to be over- and under-represented. Among these patterns, there is no clear-cut relation between their AT content and the degree of over- or under-representation. For instance, the GC-rich Sp1 pattern is significantly over-represented in S/MARs when taking into account that it has an extremely low a priori probability of occurring in these AT-rich genome regions. On the other hand, the (relatively AT-rich) recognition sites for Evi-1 and RORα1 are clearly among the under-represented patterns. With few exceptions, the observed trends are generally the same for S/MAR enrichment factors that refer to exon or extended promoter sequences. The proteins that are supposed to bind to these over- and under-represented sequence patterns belong to nearly all known classes of DNA-binding domains (39). Among the matrices listed in Table 3 only M00104 describes the DNA-binding profile for a homeodomain factor, CDP/Cux, which has been mentioned in relation to S/MARs before (38,40). Motifs described by two other matrices for homeodomain factors (M00018, Ubx; M00023, Hox-1.3) were also found to be enriched in S/MARs. However, other matrices which also describe the DNA-binding properties of CDP/Cux did not match with S/MARs at a significantly higher rate beyond that expected from the correlation shown in Figure 1, and DNA-binding profiles of other homeodomain proteins also did not exhibit significant matching frequencies. Likewise, potential binding sites for other transcription factors that have been mentioned in connection with S/MARs did not show up markedly, as far as positional weight matrices are available for them (YY1 and NF-1) (36,37).
Table 3. Transcription factor-binding motifs exhibiting enhanced or lowered levels in S/MARs.
The MatInspector program was adjusted to core sim 0.75. TRANSFAC matrix accession numbers given in italics indicate that the respective matrix also showed up in analyses of randomized S/MAR sequences. An underlined matrix accession number indicates that this matrix was also over- or under-represented when non-parametric statistics were applied. re, ‘random expectation’ values of MatInspector indicating how many hits per kb the corresponding matrix may produce in one megabase of random sequences; dsm/ex and dsm/ep are the differences of the respective enrichment factors from the linear regression shown in Figure 1. The consensus binding sites are given as shown by MatInspector professional.
When randomized S/MAR sequence sets were compared to the exon or extended promoter sequences, again about half of the matrices listed in Table 3 were found to be over- or under-represented, respectively. The relevant TRANSFAC accession numbers of matrices are given in italic in Table 3. A different approach in which the outliers were calculated from the mean enrichment factors and standard deviations of distinct classes yielded a similar picture, i.e. 8 of 14 over- or under-represented motifs described by weight matrices were shared with the former analysis. The TRANSFAC accession numbers of these matrices are underlined in Table 3.
Scanning for previously reported S/MAR-specific sequence elements
Topoisomerase II sites have been reported to be indicative of S/MAR sequences (30,32,41). Other sequence motifs that have often been recognized within nuclear matrix-attached regions are A- and T-box motifs (1,14,42,43). Still another motif characteristic of S/MARs (TAWAWWWNNAWW RTAANNWWG) has been proposed by van Drunen et al. (22). Therefore we searched S/MARt DB sequences and the sets of shuffled S/MAR sequences as well as the other data sets of regulatory and non-regulatory sequences for these sequence motifs. To facilitate overall comparison the IUPAC consensus strings were transformed into matrices (see Materials and Methods for details); thus, the search was performed under the same conditions as for transcription factor-binding sites.
We observed an enrichment of sites predicted by the matrix derived from the Drosophila topoisomerase II consensus in S/MARs and randomized S/MARs when compared to exon sequences or to the extended promoter sequence data set (44; Table 4A). A converse picture emerged when we scanned the data sets with the matrix derived from the published consensus site for vertebrate topoisomerase II (45): the vertebrate consensus appeared to be under-represented in the S/MAR data set as well as in the shuffled sequences when compared to other sequence sets (Table 4A).
Table 4. Enrichment factors for previously reported S/MAR consensus patterns.
| rsm/ex | rsm/ep | rsm/non6 | r̄sm/rand | r̄rand/ex | r̄rand/ep | ||
|---|---|---|---|---|---|---|---|
| A | A-box1 | 0.83 | 0.72 | 0.75 | 0.62 | 0.47 | |
| T-box2 | 0.78 | 0.68 | 0.66 | 0.65 | 0.53 | ||
| Topo II (Drosophila)3 | 0.78 | 0.70 | 0.55 | 0.75 | 0.65 | ||
| Topo II (vertebrates)4 | 0.32 | 0.42 | 0.50 | 0.33 | 0.42 | ||
| S/MAR motif5 | 0.84 | 0.72 | 0.55 | 0.81 | 0.67 | ||
| B | A-box1 | 0.84 | 0.74 | 0.82 | 0.72 | 0.66 | 0.53 |
| T-box2 | 0.86 | 0.74 | 0.90 | 0.81 | 0.58 | 0.40 | |
| Topo II (Drosophila)3 | 0.82 | 0.74 | nd | 0.57 | 0.78 | 0.68 | |
| Topo II (vertebrates)4 | 0.42 | 0.45 | nd | 0.52 | 0.40 | 0.43 | |
| S/MAR motif5 | 0.90 | 0.65 | nd | 0.70 | 0.79 | 0.59 |
(A) A matrix search was performed with matrices derived from the respective IUPAC consensi (see Materials and Methods for details). MatInspector was adjusted to core sim 0.75. (B) Search with the IUPAC consensus using the user defined IUPAC string option of the MatInspector program allowing at maximum 1 mismatch. Enrichment factors r are defined as explained in Materials and Methods and refer to over-representation of the respective element in S/MARs or randomized S/MAR sequences when compared with exon sequences (rsm/ex, rrand/ex) or extended promoter sequences (rsm/ep, rrand/ep); r ranges between 0 and 1, equal distribution between S/MARs, and the reference set is indicated by 0.5. When the means of the enrichment factors determined for the comparisons of all randomized S/MAR sequence sets with a certain reference set are shown this is indicated by r̄rand/ex, r̄rand/ep and r̄sm/rand. nd, not determined.
1A-box sequence (5′-AATAAAYAAA-3′) (32).
2T-box sequence (5′-TTWTWTTWTT-3′) (32).
3Topoisomerase II consensus string for Drosophila (5′-GTNWAYATTNATNNR-3′) (44).
4Topoisomerase II consensus string for vertebrates (5′-RNYNNCNNGYNGKTNYNY-3′) (45).
5S/MAR-specific sequence (5′-TAWAWWWNNAWWRTAANNWWG-3′) (22).
6Recalculated from Amati and Gasser (46). Here rsm/non refers to over-representation of A- and T-box motifs in S/MARs when compared to non-S/MARs.
Analyses for A- and T-box motifs revealed that S/MARs are enriched in these motifs whereas an enrichment in randomized sequences is observed only when compared to the exon sequence set. The ‘S/MAR-specific sequence’ proposed by van Drunen et al. (22) was enriched in the original and shuffled S/MAR sequences when compared to either exon or promoter sequences (Table 4A). As described above, we analyzed whether the enrichment is influenced by the AT content of the consensus-derived matrices. Therefore their AT contents were determined as described in Materials and Methods and plotted against the r(a) values. For comparison this plot has been included in Figure 1. The points for both topoisomerase II matrices as well as that for the ‘S/MAR-specific sequence’ appear in close vicinity to the regression line, indicating that these motifs are neither over- nor under-represented in S/MARs when the AT content is taken into account. Confirmation also comes from yet another approach in which the matrices were grouped by their AT content and r(a) values of individual matices were compared to the mean r(a) of the group it belongs to. This approach allows the tentative estimation of the enrichment of all motifs. It suggests that all previously reported S/MAR-specific sequence elements re-evaluated in our analyses are neither over- nor under-represented in S/MARs when the AT content is taken into account.
Calculating the enrichment factors from analyses with IUPAC consensus sequences as search strings (Table 4B) and plotting them against the AT content of the motif yielded essentially the same picture, thus confirming the results obtained by employing the consensus-derived matrices.
DISCUSSION
Previous attempts to analyze S/MARs for any kind of pattern were hampered by the low number of S/MAR sequences available. Thus definition of motifs or rules was previously performed on the basis of 5, 7, 15 or 31 S/MAR sequences (references 1,22,30,46, respectively). With the help of our data collection, S/MARt DB, which presently contains 245 S/MAR sequences from a variety of species (24), we analyzed certain sequence characteristics of S/MARs. It has been shown in re-association assays that S/MAR elements from one species bind to matrices prepared from tissues of another species (13,22,46), although these species may be as apart as animals, yeast and plants. Therefore, taking advantage of an improved basis for statistical analyses, we decided to use the whole set of S/MARs collected in S/MARt DB. This decision is supported by the observation that the enrichment of selected motifs in S/MARs did not reveal significant species specificities (see below). Even so, the coverage of experimentally verified S/MARs in the analyses is still low compared to the estimated number of 100 000 nuclear matrix-attached sites alone for the human genome, a number which is essentially supported by Frisch et al. (47). Analyses were carried out by comparing S/MAR sequences with sequences from regulatory (promoter sequences) and non-regulatory regions (exon sequences) and random sequences of the same AT content (shuffled S/MAR sequences). Such a comparison should aid in identifying common features as well as those that separate S/MARs from genomic regions of distinct function.
These attempts confirmed that a certain type of S/MAR, which forms the bulk of the available data material, exhibits an enhanced AT content as described earlier by others (3,33,48). The S/MARs contained in our collection have been obtained by a range of methods and according to varying activity criteria. While similar results seem to originate from various (re-)association protocols (4,5), binding thresholds which qualify an element as a S/MAR have rarely been specified. Moreover, a few S/MARs contained in our collection exhibit a low AT ratio while others have been mapped to rather large restriction fragments that may contain unspecified portions of non-S/MAR sequence. Among others, these reasons may affect the reported mean AT ratio. Nevertheless, as a whole, our collection of S/MAR sequences exhibits a markedly higher AT content than any of the reference sequence sets, even though it is lower than outlined in previous reports (31,33).
In order to study the nucleotide composition of S/MARs in more detail, the occurrence frequency of all possible non-redundant hexanucleotides was determined. By this means our results support previous observations that S/MARs often contain AT-rich motifs such as oligo(A) runs (10,30,33). Comparative analyses of shuffled S/MAR sequences showed that most of the AT-rich motifs are shared with the original S/MARs. The A6 motif especially was again found most often in the shuffled sequence set, although at a >4-fold lower frequency. While this seems to support the idea that homopolymeric stretches of A are a characteristic of S/MARs, the following consideration should also be taken into account: the absolute number of matches obtained for the original S/MARs might be biased by the fact that our search detected and independently counted overlapping A6 motifs in homo(A) runs (thus the high number of hits). On the other hand, there is a high chance that sequence shuffling inserts another nucleotide into a homo(A) run causing each insertion to erase up to six A6 occurrences in the randomized sequences (thus resulting in fewer hits in the randomized sequence sets). Still, they are enriched in this motif when compared to sequences with a more equal nucleotide distribution. The hexanucleotide motifs that were demonstrated to represent CUEs within an appropriate sequence context (ATATTT and ATATAT) (5,34) are also contained in S/MAR and shuffled S/MAR sequences. Similarly, we could confirm the observation that hexanucleotides containing the core motif of homeodomain transcription factors (ATTA or TAAT) occur frequently in S/MARs. In the latter case even the numbers of hits were similar in the original S/MARs and randomized sequences.
In summary, the observed hexanucleotide composition of S/MARs is clearly influenced by their AT-richness. This conclusion is supported at least in part by an analysis of the hexanucleotide distribution of 3′-UTRs taken from UTRdb (49), which exhibit a similar AT content (60.12% compared to 61.68% for S/MARs; see Table 1). About one-third of the hexanucleotides frequently met in S/MARs occurred often in 3′-UTRs as well (data not shown). Support also comes from a survey by Ganapathy and Singh (50) in which 122 S/MARs were analyzed for 39 S/MAR-related sequence motifs. Their motif collection ranged from a dinucleotide to motifs comprising >20 nt. In that study four of the five most frequently observed motifs (ATTA, AAA, AAAA, WWWWWW and YR) were entirely composed of A and/or T nucleotides (50). All of the motifs recognized by Ganapathy and Singh as occurring frequently in S/MARs appear in Table 2 as well. On the other hand, some recent studies emphasize that, in particular, (A)n·(T)n motifs are important for matrix attachment when they exceed a critical length or are spaced according to certain distance criteria which are referred to as either AT-patches or 90% AT-boxes (51,52).
Following the observations made for the hexanucleotides containing the core motif of homeodomain transcription factors only relatively few weight matrices for homeodomain factors showed up in our systematic analyses for potential transcription factor-binding sites. In particular, this search revealed an elevated frequency of CDP/Cux-binding sites in original and randomized S/MARs when compared to other sequences. In addition, these sites were identified with just one of about six matrices for this transcription factor. Similar observations were made for E2F, GATA-1 and Sp1. We also observed an enrichment for three matrices which represent putative binding sites for homeodomain factors (CPP/Cux, Hox 1.3 and Ubx), but our collection of matrices includes many more matrices for homeodomain factors, most of which are rather inconspicuous. On the other hand, motifs described by another matrix for a homeodomain factor were found to be under-represented in S/MARs. Taking these observations into account it is hard to generalize that binding sites for homeodomain factors as a whole are over-represented in S/MARs. For the time being we therefore suggest that potential binding sites be treated as singular sequence motifs, leaving a potential role of the respective homeodomain factors in S/MAR function to further experimental analyses.
To the best of our knowledge only Amati and Gasser also investigated the distribution of A- and T-boxes between S/MAR and non-S/MAR sequences (46). The data sets they used in their analysis were considerably smaller (15 S/MARs and seven non-S/MARs) and different in base composition. Their ‘ratio of occurrence’, which corresponds to the enrichment factors r(a) of our study, was computed in a different way. Therefore, we recalculated the r(a) values according to our formula from the frequency values given in their publication to allow for comparison (Table 4B). Even though Amati and Gasser observed a slightly stronger enrichment for the T-box motif, their results do not contradict ours, especially if the differences in the sizes of the data sets are taken into account (46; Table 4B). We have noticed a higher enrichment of topoisomerase II sites when scanning against a matrix derived from the consensus for the insect enzyme, but not for vertebrate topoisomerase II (Table 4). Although the potential sites for the insect enzyme appear to be much more enriched in S/MARs and randomized S/MARs, both these sites occur in S/MARs and randomized S/MAR sequences at the levels expected from their AT content. The same seems to be true for the remaining S/MAR-specific sequence elements considered in our analyses (Fig. 1). Equivalent observations concerning these five selected motifs were made in analyses confined to either vertebrate, insect or plant S/MARs (data not shown), thus justifying the use of the undivided S/MAR sequence set in other analyses.
In the light of our analyses, at least some of the previously described S/MAR consensus patterns appear debatable. For the investigated group of conventional S/MARs, the elevated AT content seems to be most indicative for their presence while the enrichment of a number of motifs may just be due to this general feature. In fact, an attractive hypothesis would be that prototype S/MARs are AT-rich just to facilitate a higher concentration of (certain) AT-motifs to occur. This implies the possible occurrence of sequences with a balanced AT/GC ratio in which these sequence features may have evolved by selection pressure. As there are definitely S/MARs that exhibit a low AT content (see for example 53,54), we have to consider that there must exist characteristics beyond mere AT-richness that confer scaffold/matrix attachment potential. In this regard it might be useful to elucidate the relevance of purine-rich S/MAR segments. Systematic wet lab investigation of the ‘ATC sequence’ context should also generate valuable results. The ATC sequence context has been shown to be important for the S/MAR-binding proteins SATB 1 and BRIGHT; it might be relevant for other factors as well (55,56).
Previously, development of new effective algorithms for the recognition of scaffold/matrix-attached regions or the improvement of existing ones was impeded by the small number of sequences available and the non-availability of a ‘classical consensus’ sequence shared by all S/MARs. Our analyses and that by Ganapathy and Singh (50) put emphasis on the latter point. Having said that, this does not necessarily mean that it might be impossible to find such motifs, at least for certain subsets, the definition of which will be possible by S/MARt DB.
We would have tried to establish subsets of S/MARs on the basis of alternative biological or biophysical features that are connected to S/MAR function, e.g. their matrix-binding specificity, but these characteristics have not generally been investigated. So far the use of binding strength is critical as it calls for a rigorous standardization (compare results in 57 and 58,59).
Despite these shortcomings, progress in defining common motifs on the basis of certain contigs with reported binding potential was made by Frisch et al. (47), who defined matrices for a novel tool, SMARTest. Since it is based on our database, so far SMARTest has also concentrated on S/MARs with a high AT content. The MAR-Finder program uses rules for combinations of a number of structural motifs that have been reported to occur in the vicinity of S/MARs (60,61). Yet another approach makes use of the established unwinding propensity of S/MAR elements (62).
Having complete eukaryotic genomes at hand, it will be an exciting task to systematically identify their S/MARs by appropriate in silico methods which, however, will have to be accompanied by rigorous in vitro and in vivo experiments. If the results of these analyses are capable of complementing the accumulating experimental knowledge about the locations and properties of S/MARs, the possibility arises of using complete genomic maps for S/MARs as a functional coordinate system for regulatory genomic units.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Thomas Werner and Matthias Frisch for providing free access to their tools, communicating results prior to publication and helpful discussions. We thank Maik Christensen for his help in establishing the shuffling algorithm. This work was supported by grants from the Federal Ministry of Education, Science and Research (project nos 0311640 and 01SF9988/4).
REFERENCES
- 1.Gasser S.M. and Laemmli,U.K. (1986) Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell, 46, 521–530. [DOI] [PubMed] [Google Scholar]
- 2.Berezney R. (1991) The nuclear matrix: a heuristic model for investigating genomic organization and function in the cell nucleus. J. Cell. Biochem., 47, 109–123. [DOI] [PubMed] [Google Scholar]
- 3.Bode J., Stengert-Iber,M., Kay,V., Schlacke,T. and Dietz-Pfeilstetter,A. (1996) Scaffold/matrix-attached regions: topological switches with multiple regulatory functions. Crit. Rev. Eukaryot. Gene Expr., 6, 115–138. [DOI] [PubMed] [Google Scholar]
- 4.Bode J. and Maass,K. (1988) Chromatin domain surrounding the human interferon-beta gene as defined by scaffold-attached regions. Biochemistry, 27, 4706–4711. [DOI] [PubMed] [Google Scholar]
- 5.Mielke C., Kohwi,Y., Kohwi-Shigematsu,T. and Bode,J. (1990) Hierarchical binding of DNA fragments derived from scaffold-attached regions: correlation of properties in vitro and function in vivo. Biochemistry, 29, 7475–7485. [DOI] [PubMed] [Google Scholar]
- 6.Hart C.M. and Laemmli,U.K. (1998) Facilitation of chromatin dynamics by SARs. Curr. Opin. Genet. Dev., 8, 519–525. [DOI] [PubMed] [Google Scholar]
- 7.Pikaard C.S. (1998) Chromosome topology—organizing genes by loops and bounds. Plant Cell, 10, 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laemmli U.K., Käs,E., Poljak,L. and Adachi,Y. (1992) Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev., 2, 275–285. [DOI] [PubMed] [Google Scholar]
- 9.Schübeler D., Mielke,C., Maass,K. and Bode,J. (1996) Scaffold/matrix-attached regions act upon transcription in a context-dependent manner. Biochemistry, 35, 11160–11169. [DOI] [PubMed] [Google Scholar]
- 10.Bode J., Kohwi,Y., Dickinson,L., Joh,T., Klehr,D., Mielke,C. and Kohwi-Shigematsu,T. (1992) Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science, 255, 195–197. [DOI] [PubMed] [Google Scholar]
- 11.Allen G.C., Hall,G.E.,Jr, Michalowski,S, Newman,W., Spiker,S., Weissinger,A.K. and Thompson,W.F. (1996) High-level transgene expression in plant cells: effects of a strong scaffold attachment region from tobacco. Plant Cell, 8, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blasquez V.C., Xu,M., Moses,S.C. and Garrard,W.T. (1989) Immunoglobulin kappa gene expression after stable integration. I. Role of the intronic MAR and enhancer in plasmacytoma cells. J. Biol. Chem., 264, 21183–21189. [PubMed] [Google Scholar]
- 13.Dietz A., Kay,V., Schlacke,T., Landsmann,J. and Bode,J. (1994) A plant scaffold attached region detected close to a T-DNA integration site is active in mammalian cells. Nucleic Acids Res., 22, 2744–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy-Wilson B. and Fortier,C. (1989) The limits of the DNaseI-sensitve domain of the human apolipoprotein B gene coincide with the locations of chromosomal anchorage loops and define the 5′ and 3′ boundaries of the gene. J. Biol. Chem., 264, 21196–21204. [PubMed] [Google Scholar]
- 15.van der Geest A.H.M., Hall,G.E.,Jr, Spiker,S. and Hall,T.C. (1994) The β-phaseolin gene is flanked by matrix attachment regions. Plant J., 6, 413–423. [Google Scholar]
- 16.Kalos M. and Fournier,R.E. (1995) Position-independent transgene expression mediated by boundary elements from the apolipoprotein B chromatin domain. Mol. Cell. Biol., 15, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Namciu S.J., Blochinger,K.B. and Fournier,R.E.K. (1998) Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol. Cell. Biol., 18, 2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antes T.J., Namicu,S.J., Fournier,R.E.K. and Levy-Wilson,B. (2001) The 5′ boundary of the human apolipoprotein B chromatin domain in intestinal cells. Biochemistry, 40, 6731–6742. [DOI] [PubMed] [Google Scholar]
- 19.Will K., Warnecke,G., Albrechtsen,N., Boulikas,T. and Deppert,W. (1998) High affinity MAR-DNA binding is a common property of murine and human mutant p53. J. Cell. Biochem., 69, 260–270. [DOI] [PubMed] [Google Scholar]
- 20.Cockerill P.N. and Garrard,W.T. (1986) Chromosomal loop anchorage of the kappa immunoglobin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell, 44, 273–282. [DOI] [PubMed] [Google Scholar]
- 21.Boulikas T. (1993) Homeodomain protein binding sites, inverted repeats and nuclear matrix attachment regions along the human beta-globin gene complex. J. Cell. Biochem., 52, 23–36. [DOI] [PubMed] [Google Scholar]
- 22.van Drunen C.M., Oosterling,R.W., Keultjes,G.M., Weisbeek,P.J., van Driel,R. and Smeekens,S.C. (1997) Analysis of the chromatin domain organisation around the plastocyanin gene reveals a MAR-specific sequence element in Arabidopsis thaliana. Nucleic Acids Res., 25, 3904–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Prüß,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liebich I., Bode,J., Frisch,M. and Wingender,E. (2002) S/MARt DB: a database on scaffold/matrix attached regions. Nucleic Acids Res., 30, 372–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Périer R.C., Junier,T., Bonnard,C. and Bucher,P. (1999) The eukaryotic promoter database (EPD): recent developments. Nucleic Acids Res., 27, 307–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoesser G., Tuli,M.A., Lopez,R. and Sterk,P. (1999) The EMBL nucleotide sequence database. Nucleic Acids Res., 27, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickert L., Reuter,I., Klawonn,F. and Wingender,E. (1998) Transcription regulatory region analysis using signal detection and fuzzy clustering. Bioinformatics, 14, 244–251. [DOI] [PubMed] [Google Scholar]
- 28.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wingender E., Chen,X., Fricke,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhäuser,R., Prüß,M., Schacherer,F., Thiele,S. and Urbach,S. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulikas T. (1993) Nature of DNA sequences at the attachment regions of genes to the nuclear matrix. J. Cell. Biochem., 52, 14–22. [DOI] [PubMed] [Google Scholar]
- 31.Fukuda Y. (1999) Characterization of matrix attachment sites in the upstream region of a tobacco chitinase gene. Plant Mol. Biol., 39, 1051–1062. [DOI] [PubMed] [Google Scholar]
- 32.Gasser S.M. and Laemmli,U.K. (1987) A glimpse of a chromosomal order. Trends Genet., 3, 16–22. [Google Scholar]
- 33.Käs E., Izaurralde,E. and Laemmli,U.K. (1989) Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA)·oligo(dT) tracts. J. Mol. Biol., 210, 587–599. [DOI] [PubMed] [Google Scholar]
- 34.Dickinson L.A. and Kohwi-Shigematsu,T. (1995) Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol., 15, 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bode J., Fetzer,C.P., Nehlsen,K., Scinteie,M., Hinrichs,B.-H., Baiker,A., Piechaczek,C., Benham,C. and Lipps,H.J. (2001) The hitchhiking principle: optimizing episomal vectors for the use in gene therapy and biotechnology. Gene Ther. Mol. Biol., 6, 33–46. [Google Scholar]
- 36.Farache G., Razin,S.V., Targa,F.R. and Scherrer,K. (1990) Organization of the 3′-boundary of the chicken alpha globin gene domain and characterization of a CR 1-specific protein binding site. Nucleic Acids Res., 18, 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dworetzky S.I., Wright,K.L., Fey,E.G., Penman,S., Lian,J.B., Stein,J.L. and Stein,G.S. (1992) Sequence-specific DNA-binding proteins are components of a nuclear matrix-attachment site. Proc. Natl Acad. Sci. USA, 89, 4178–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banan M., Rojas,I.C., Lee,W.H., King,H.L., Harriss,J.V., Kobayashi,R., Web,C.F. and Gottlieb,P.D. (1997) Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J. Biol. Chem., 272, 18440–18452. [DOI] [PubMed] [Google Scholar]
- 39.Wingender E. (1997) Classification scheme of eukaryotic transcription factors. Mol. Biol. (Mosk.), 31, 483–497. [PubMed] [Google Scholar]
- 40.Wang Z., Goldstein,A., Zong,R.-T., Lin,D., Neufeld,E.J., Scheuermann,R.H. and Tucker,P.W. (1999) Cux/CDP homeoprotein is a component of NF-µNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the Bright transcription activator. Mol. Cell. Biol., 19, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bode J., Bartsch,J., Boulikas,T., Iber,M., Mielke,C., Schübeler,D., Seibler,J. and Benham,C. (1998) Transcription-promoting genomic sites in mammalia: their elucidation and architectural principles. Gene Ther. Mol. Biol., 1, 551–580. [Google Scholar]
- 42.Slatter R.E., Dupree,P. and Gray,J.C. (1991) A scaffold-associated DNA region is located downstream of the pea plastocyanin gene. Plant Cell, 3, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanson R.D. and Ley,T.J. (1992) A-T-rich scaffold attachment regions flank the haematopoietic serine protease genes clustered on chromosome 14q11.2. Blood, 79, 610–618. [PubMed] [Google Scholar]
- 44.Sander M. and Hsieh,T. (1983) Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem., 258, 8421–8428. [PubMed] [Google Scholar]
- 45.Spitzner J.R. and Muller,M.T. (1988) A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res., 16, 5533–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amati B. and Gasser,S.M. (1990) Drosophila scaffold-attached regions bind nuclear scaffolds and can function as ARS elements in both budding and fission yeast. Mol. Cell. Biol., 10, 5442–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frisch M., Frech,K., Klingenhoff,A., Cartharius,K., Liebich,I. and Werner T. (2002) In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome Res., 12, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romig H., Fackelmayer,F.O., Renz,A., Ramsperger,U. and Richter,A. (1992) Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J., 11, 3431–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pesole G., Liuni,S., Grillo,G., Licciulli,F., Mignone,F., Gissi,C. and Saccone,C. (2002) UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res., 30, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ganapathy S. and Singh,G. (2001) Statistical mining of S/MARt Database. In Proceedings of the Atlantic Symposium on Computational Biology and Genome Information System Technology (CBGIST), Duke University, NC, 15–17 March. pp. 235–239.
- 51.Tsutsui K. (1998) Synthetic concatemers as artificial MAR: importance of a particular configuration of short AT-tracts for protein recognition. Gene Ther. Mol. Biol., 1, 581–590. [Google Scholar]
- 52.Michalowski S.M., Allen,G.C., Hall,G.E.,Jr, Thompson,W.F. and Spiker,S. (1999) Characterization of randomly-obtained matrix attachment regions (MARs) from higher plants. Biochemistry, 28, 12795–12804. [DOI] [PubMed] [Google Scholar]
- 53.Christova R., Bach,I. and Galcheva-Gargova,Z. (1992) Sequences of DNA fragments contacting the nuclear lamina in vivo. DNA Cell Biol., 11, 627–636. [DOI] [PubMed] [Google Scholar]
- 54.Mielke C., Maass,K., Tümmler,M. and Bode,J. (1996) Anatomy of highly expressing chromosomal sites targeted by retroviral vectors. Biochemistry, 35, 2239–2252. [DOI] [PubMed] [Google Scholar]
- 55.Dickinson L.A., Joh,T., Kohwi,Y. and Kohwi-Shigematsu,T. (1992) A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell, 70, 631–645. [DOI] [PubMed] [Google Scholar]
- 56.Herrscher R.F., Kaplan,M.H., Lelsz,D.L., Das,C., Scheuermann,R.H. and Tucker,P.W. (1995) The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev., 9, 3067–3082. [DOI] [PubMed] [Google Scholar]
- 57.Jarman A.P. and Higgs,D.R. (1988) Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J., 7, 3337–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartjeliotou A.J. and Dimitriadis,G.J. (1992) The association of the human epsilon-globin gene with the nuclear matrix: a reconsideration. Mol. Cell. Biochem., 115, 105–115. [DOI] [PubMed] [Google Scholar]
- 59.Neri L.M., Fackelmayer,F.O., Zweyer,M., Kohwi-Shigematsu,T. and Martelli,A.M. (1997) Subnuclear localization of S/MAR-binding proteins is differently affected by in vitro stabilization with heat or Cu2+. Chromosoma, 106, 81–93. [DOI] [PubMed] [Google Scholar]
- 60.Kramer J.A., Singh,G.B. and Krawetz,S.A. (1996) Computer assisted search for sites of nuclear matrix attachment. Genomics, 33, 302–308. [DOI] [PubMed] [Google Scholar]
- 61.Singh G.B., Kramer,J.A. and Krawetz,S.A. (1997) Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res., 25, 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benham C., Kohwi-Shigematsu,T. and Bode,J. (1997) Stress-induced duplex DNA destabilization in scaffold/matrix attachment regions. J. Mol. Biol., 274, 181–196. [DOI] [PubMed] [Google Scholar]