Abstract
The internal ribosome site RNA of the cricket paralysis-like viruses (CrPV-like) binds directly to the ribosome, assembling the translation machinery without initiation factors. This mechanism does not require initiator tRNA, and translation starts from a non-AUG codon. A wealth of biochemical data has yielded a working model for this process, but the three-dimensional structure and biophysical characteristics of the unbound CrPV-like IRES RNAs are largely unexplored. Here, we demonstrate that the CrPV-like IRESes prefold into a two-part structure in the presence of magnesium ions. The largest part is a prefolded compact RNA domain that shares folding and structural characteristics with other compactly folded RNAs such as group I intron RNAs and RNase P RNA. Chemical probing reveals that the CrPV-like IRES’ compact domain contains RNA helices that are packed tightly enough to exclude solvent, and analytical ultracentrifugation indicates a large change in the shape of the IRES upon folding. Formation of this compact domain is necessary for binding of the 40S subunit, and the structural organization of the unbound IRES RNA is consistent with the hypothesis that the IRES is functionally and structurally preorganized before ribosome binding.
Keywords: internal ribosome entry site (IRES), translation, CrPV-like, RNA folding, RNA structure
INTRODUCTION
Translation of the vast majority of eukaryotic mRNAs is driven by the presence of a 7-methyl guanosine cap on their 5′ end. This cap is bound by eukaryotic translation factor (eIF) 4E. Binding of this protein factor leads to the recruitment of the 43S complex (containing the 40S subunit, eIF3, and the initiator tRNA•eIF2•GTP ternary complex) through the action of eIF4G and associated factors. This 40S subunit-containing complex scans the message to the start codon, at which time the codon–anticodon interaction forms. This interaction induces GTP hydrolysis, factor release, and recruitment of the large subunit to form the 80S ribosome (Hershey and Merrick 2000; Pestova et al. 2001). In contrast to the canonical cap-dependent mechanism, internal initiation of translation is a cap-independent process driven by structured RNA sequences called internal ribosome entry sites (IRESes) (Hellen and Sarnow 2001; Vagner et al. 2001; Sarnow 2003). Some IRESes can bind directly to the ribosome without the use of the canonical initiation factors, while others require a subset of the canonical initiation factors or additional trans-activating factors. In all cases, IRESes direct internal entry of the translation machinery onto the message. IRESes were first discovered in viruses, but since have been identified in cellular mRNAs (Stoneley and Willis 2004). In IRES-using viruses, proper function of the IRES is critical for the success of the virus, and eukaryotic cellular IRESes may serve as important regulators of gene expression at the translational level (Stoneley and Willis 2004).
IRES RNAs have diverse sequences, structures, functions, and requirements for cofactors (Bonnal et al. 2003). Two groups of IRES RNAs have been shown to bind directly to the 40S subunit in vitro in the absence of any additional protein factors (Sarnow 2003). The first is the hepatitis C virus (HCV) IRES and structurally related IRESes from classical swine fever virus (CSFV) and bovine viral diarrhea virus (BVDV) (Pestova et al. 1998; Kolupaeva et al. 2000b). Folding of the HCV IRES in the presence of divalent cations results in regions of local compact structure, but there is considerable conformational flexibility in the free IRES RNA, and globally the IRES RNA remains extended in both the free and ribosome-bound forms (Kieft et al. 1999; Spahn et al. 2001). The HCV IRES recognizes both the 40S ribosomal subunit and eIF3, using this extended architecture to contact several parts of the ribosome (Pestova et al. 1998; Sizova et al. 1998; Kieft et al. 1999, 2001; Kolupaeva et al. 2000a; Spahn et al. 2001). The HCV IRES RNA contains three independently folded regions that are organized around structured RNA junctions (Kieft et al. 1999). Hence, the strategy employed by the free HCV IRES RNA is to organize into an extended scaffold-like conformation where emerging stem–loops are placed correctly in space to recognize the translational machinery.
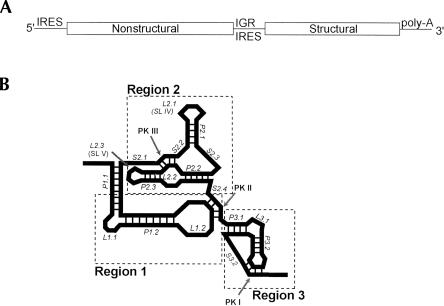
The second group of IRES RNAs that has been shown to bind directly to the ribosome is the intergenic region (IGR) of the Dicistroviridae, also referred to as the cricket paralysis virus-like (CrPV-like) IRESes (Fig. 1A ▶; Christian and Scotti 1998; Sasaki and Nakashima 1999; Wilson et al. 2000b; Mayo 2002). These viruses are single-stranded positive-sense RNA viruses that infect primarily insects. The family includes 11 clearly identified members; despite sequence differences, the IGR IRESes of all members are proposed to form similar secondary structures, with most of the variation in region 3 (Fig. 1B ▶; Kanamori and Nakashima 2001; Hatakeyama et al. 2004). These IRESes initiate protein synthesis from a non-AUG codon and without methionine initiator tRNA (Sasaki and Nakashima 1999, 2000; Wilson et al. 2000a,b). Furthermore, translation initiates from the A-site of the ribosome, and the first translocation event occurs before a peptide bond is synthesized (Wilson et al. 2000a; Pestova and Hellen 2003). Assembly of the ribosome on these IRESes occurs without initiation factors, and they also can bind directly to preformed 80S ribosomes (Pestova et al. 2004). Recent cryo-EM reconstructions and directed hydroxyl-radical cleavage experiments reveal the location of the bound CrPV IRES on the 40S subunit and lend insight into the interaction of the IRES RNA with the ribosome decoding groove (Pestova et al. 2004; Spahn et al. 2004). Well-supported mechanistic models of CrPV-like IRES-driven translation initiation now exist based on numerous biochemical studies (Sasaki and Nakashima 2000; Jan et al. 2001, 2003; Thompson et al. 2001; Jan and Sarnow 2002; Nishiyama et al. 2003; Shibuya et al. 2003; Pestova et al. 2004). These models support the hypothesis that the streamlined mode of translation initiation used by the CrPV-like IRESes depends on the specific three-dimensional structure of the IRES RNA.
FIGURE 1.
Location and secondary structure of the Dicistroviridae IGR IRESes. (A) The genome of the Dicistroviridae consists of two open reading frames. The second open reading frame contains the structural proteins and is under control of an IRES in the intergenic region (IGR). (B) Cartoon depicting the consensus secondary structure for the Dicistroviridae IGR IRESes. To provide a more detailed nomenclature for discussing the structural characteristics of the CrPV-like IRES RNAs, we have adopted the convention shown. This convention allows each paired helical segment (P), internal and hairpin loop (L), and single-stranded region (S) to be unambiguously identified and also maintains the divisions between regions 1, 2, and 3 originally named by Kanamori and Nakashima (2001). Throughout this text we use this nomenclature, but parenthetically include the nomenclature used in previous publications. Regions 1 + 2 and region 3 have been shown to be functionally independent. Region 3 contains an additional stem–loop in three members of the IGR IRES group (not shown).
Despite the emerging understanding regarding the function of the CrPV-like IRESes, there still is a gap in our understanding of the three-dimensional conformation of the unbound IRES RNA, and hence the mechanism by which the RNA organizes itself for ribosome binding. Visual inspection of the proposed secondary structure suggests a convoluted fold, and the cryo-EM structure shows that the CrPV IRES is compact when docked on the ribosome (Kanamori and Nakashima 2001; Spahn et al. 2004). Biochemical analyses identify two separate elements in the CrPV-like IRESes. The first (region 1 + 2) contains the majority of the affinity for the 40S subunit, and the second (region 3) is a pseudoknot element that occupies the ribosome’s P-site and is critical for the function of the IRES (Fig. 1B ▶; Kanamori and Nakashima 2001; Jan and Sarnow 2002; Jan et al. 2003; Nishiyama et al. 2003).
What is the nature of the unbound CrPV-like IRES RNA fold? Do these RNAs prefold into a compact structure, or is the RNA extended prior to binding? How does the IRES RNA organize itself prior to interacting with the translational machinery? Do conserved secondary structure motif and long-range pseudoknot interactions affect the structure of free, folded RNA? Using biochemical and biophysical approaches designed to investigate the three-dimensional architecture of a representative member of the CrPV-like IRESes, we show that the unbound IRES contains a compact fold that contains a substantial amount of tightly packed RNA and comprises a preorganized 40S subunit binding domain. Consistent with previous results, the IRES folds into two separate domains. The larger of these two domains contains the compact fold. Our data suggest that much more of the CrPV-like IRES RNAs are devoted to forming the fold than is the case with the HCV IRES, although both preorganize before binding to the ribosome. This illustrates that different viral IRES RNAs have evolved different global architectures to achieve a similar biological function.
RESULTS
The folded IGR IRES has a solvent-inaccessible core
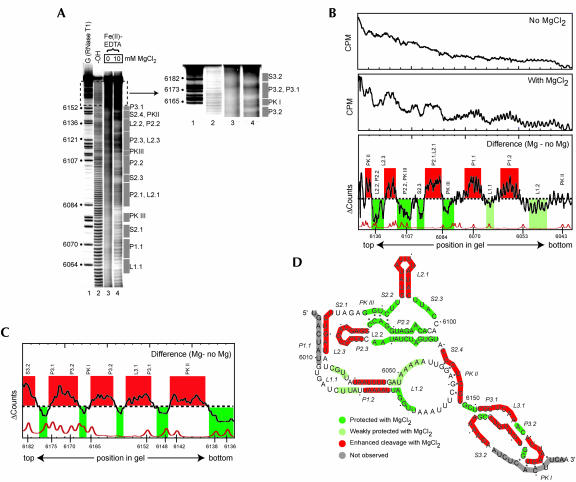
To probe the tertiary structure of unbound CrPV-like IRES RNA in solution, we employed hydroxyl radical probing on a representative member of the family, the plautia stali intestinal virus (PSIV) IRES RNA (Fig. 2A ▶). Hydroxyl radicals cleave the RNA backbone in a non-sequence-specific manner; only RNA backbones packed close enough to be protected from solvent are protected from cleavage (Latham and Cech 1989; Celander and Cech 1991). Because folding of complex RNA molecules into their three-dimensional conformations depends on divalent cations, we conducted the hydroxyl radical cleavage in the presence and absence of magnesium chloride (Fig. 2A ▶). The gel of Figure 2A ▶ shows changes in the cleavage pattern upon addition of the cation (cf. lanes 3 and 4), indicative of a folding transition that involves changes in the solvent accessibility of the RNA backbone. This particular gel resolves nucleotides 6058–6182; we used longer or shorter gel run times and both 5′ and 3′ end-labeled RNA to visualize almost the entire backbone (data not shown).
FIGURE 2.
Hydroxyl radical probing of the IGR IRES. (A) Example of a hydroxyl radical probing experiment performed on 5′ end-labeled PSIV IRES RNA. Lanes 3 and 4 contain the probed RNA in the absence and presence of MgCl2. The top portion of the gel is expanded and the contrast adjusted to show the cleavage pattern near the 3′ end of the RNA. The location of various secondary structural domains are noted to the right of the gel. Lane 1 contains an RNase T1 ladder that is annotated on the left and lane 2 is a partial hydrolysis ladder. (B) Quantitative analysis of the hydroxyl radical probing profile of a portion of PSIV IRES RNA. Gel lanes were quantitated as a function of position in the gel, normalized, and compared as described in the text. The top is a lane with no added magnesium, the center is a lane with 10 mM added magnesium, and the bottom is the calculated difference. A trace of the RNase T1 sequencing lane (bottom) allows precise identification of the locations of the protections on the RNA backbone. Regions that we assigned as protected upon folding are shown in green, with weaker protections in light green. Regions that show enhanced cleavage are shown with red. The locations of corresponding secondary structures features are shown above the trace. (C) Results of an analysis identical to part B on a gel that resolved the portion of the RNA backbone corresponding to region 3 at the 3′ end of the PSIV IRES RNA. (D) Map of hydroxyl radical cleavage profile on the PSIV IRES secondary structure. Color scheme is identical to that in B and C. Gray areas denote parts of the backbone not observed on the gel.
To map the observed hydroxyl radical protection onto the RNA sequence we quantitatively analyzed the cleavage pattern. The intensity of cleavage was measured as a function of location in the gel and hence location on the RNA backbone, yielding two traces (Fig. 2B ▶, top and center). The difference between these two traces reveals the location of regions of enhanced or reduced cleavage in response to magnesium (Fig. 2B ▶, bottom). Regions were assigned as protected if the trace dropped below the zero (no difference) line for two or more consecutive nucleotides. Weaker, but reproducible, protections were assigned to regions where the degree of protection was only slightly below the zero line. Similarly, enhanced regions were assigned based on the trace rising above the line. Parts of the backbone where the trace oscillated around the zero line are due to the tightening of each cleavage band in the lane with magnesium and were considered to be areas of no change. An adjacent RNase T1 ladder aided in precise mapping of these effects onto the sequence. Application of this analysis to several repetitions of the experiment with different gel run times and both 3′ and 5′ end-labeled RNA allowed us to map folding-dependent cleavage changes onto nearly the entire backbone, including region 3 (Fig. 2C ▶).
The map of protections and enhancements superimposed on the PSIV IRES secondary structure reveals several features of the fold (Fig. 2D ▶). First, most of the RNA becomes either more protected or enhanced upon addition of magnesium, and these different regions often lie directly adjacent to one another. This suggests that the ion-induced structural change is one that involves most of the RNA. Second, region 1 + 2 contains approximately equal areas of protection and enhancement, while region 3 is mostly enhanced. This suggests that region 1 + 2 contains a substantial amount of tightly packed RNA while region 3 does not. Third, the parts of the RNA that are involved in tight RNA packing include several conserved secondary structure elements. Pseudoknot III (PK III) is protected from solvent, as is the adjacent helix (P2.2) and single-stranded regions (S2.2 and S2.3) immediately upstream and downstream of L2.1 and P2.1 (stem–loop IV). Nucleotides 6062–6065 of L1.1 are protected; they are highly conserved among the various members of the IGR IRES family (Kanamori and Nakashima 2001). In general, region 2 is the most protected part of the IRES, suggesting it makes up much of the packed core. Correspondingly, RNase T1 probing experiments also indicated strong protection in region 2 as a result of RNA folding (data not shown). This agrees with an earlier prediction (Kanamori and Nakashima 2001).
To assess if this probing pattern (and hence folding pattern) is shared by other Dicistroviridae IGR IRESes, we conducted identical hydroxyl radical probing and RNAse T1 analyses on both the CrPV and Himetobi P virus (HiPV) IGR IRESes. The results of these experiments show that the overall features of the fold are retained by all three IRES RNAs, although there are some local variations (D. Costantino and J. Kieft, in prep.).
Parts of the IRES RNA are more solvent-exposed in the folded form
We observed several regions of the PSIV IRES backbone with enhanced susceptibility to hydroxyl radicals in the folded molecule. In the case of other RNAs with tightly folded structures, these enhanced regions correspond to parts of the RNA that are extended into solution. The explanation for this phenomenon is that when unstructured, the RNA is moving between protected and exposed states, but as the molecule folds these regions are maintained in an exposed state, leading to enhanced cleavage (Takamoto et al. 2002). In both P2.1 and L2.1 (stem–loop IV) and in the apical portion of L2.3 (stem–loop V) we observed a folding-dependent enhancement in the cleavage by hydroxyl radicals, suggesting that the folding of the RNA forces these stem–loops to extend into solution. Consistent with this, RNase T1 probing of all three IRES RNAs showed enhanced cleavage of nucleotides in these apical loops when the RNA folded (data not shown). These stem–loops previously have been implicated as likely 40S subunit binding surfaces, and their extension into solution places them in an ideal conformation for this role (Jan and Sarnow 2002; Nishiyama et al. 2003). The backbone within the PK II interaction also shows enhancement, but on only one strand. At present we cannot explain this result, but we speculate that the previously reported dynamic nature of the PK II interaction may play a role (Jan and Sarnow 2002). Much of region 3 also became more susceptible to cleavage in the folded form, consistent with it extending away from the folded region 1 + 2. This is consistent with region 3 and region 1 + 2 existing as independently prefolded entities in the unbound form.
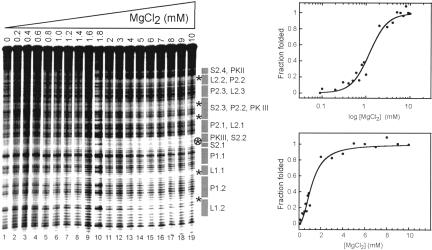
Magnesium ion dependence of the fold
We monitored the hydroxyl radical cleavage pattern as a function of magnesium ions to determine the concentration of magnesium needed to fold the PSIV IRES RNA (Fig. 3 ▶). The degree of protection was measured at several different locations along the backbone and each set of data was fit to a Hill equation. The folding transition occurred at approximately the same magnesium concentration for all monitored locations, suggesting the molecule folds all at once. The folding transition occurred at ~ 1.9 mM MgCl2 with a fitted Hill coefficient of 1.9 ± 0.2. Thus, full-length unbound PSIV IRES RNA forms a stable higher-order fold in the presence of physiological concentrations of MgCl2, and uptake of magnesium upon folding is cooperative. Similar behavior is exhibited by the HCV IRES RNA (Kieft et al. 1999).
FIGURE 3.
Hydroxyl radical probing of the PSIV IRES as a function of [MgCl2]. The amount of magnesium added to each lane is shown above the gel. The location of secondary structure elements is annotated to the right of the gel. Regions where the degree of protection was quantitated as a function of [MgCl2] are delineated with stars. The circled star indicates the region of protection that was used to construct the graph on the right. For each graph, a fitted Hill equation is shown. The top graph is the data plotted on a logarithmic scale; the bottom graph is the same data on a linear scale.
CrPV-like IRES RNA folding involves a substantial change in shape
Our hydroxyl radical probing experiments reveal that the unbound PSIV IRES RNA folds in the absence of the ribosome, and that the fold contains packed RNA helices. Transition from an unfolded state to a compact fold should be accompanied by a substantial change in the overall size and shape of the RNA molecule. To test this, we conducted velocity sedimentation experiments by analytical ultracentrifugation (AUC), measuring hydrodynamic properties of the RNA that are dependent on the shape of the molecule. AUC measurements with the compactly folded group I intron revealed the global collapse induced by the addition of cations and the measured parameters correspond well with the known structure (Takamoto et al. 2002). This illustrates the robustness of the technique as an assay for monitoring folding of RNA into compact structures.
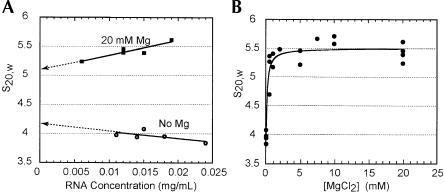
We measured the sedimentation coefficient of the PSIV IRES RNA at various RNA concentrations in both 0 and 20 mM MgCl2. In the absence of magnesium, the measured sedimentation coefficient shows a slight negative dependence on the concentration of RNA ascribed to intermolecular electrostatic repulsion (Fig. 4A ▶). In the presence of 20 mM MgCl2, the sedimentation coefficient shows some positive dependence on RNA concentration. Overall, the sedimentation coefficient changed by < 10% over the range of assayed RNA concentrations. This indicates some RNA self-association, although the goodness of the fit of the raw data to a single species model is consistent with a mostly unimolecular species. Furthermore, we did not observe evidence of PSIV IRES RNA multimer formation on native gels (data not shown). Also, at least one other stably folded RNA known to form a unimolecular species also shows positive dependence on RNA concentration (Deras et al. 2000). Hence, we concluded that the molecule’s deviation from ideality was low enough to allow further analysis. Also, in subsequent experiments, we used an upper magnesium concentration of 10 mM to further limit deviation from ideality.
FIGURE 4.
Analytical ultracentrifugation of the PSIV IRES. (A) Measured sedimentation coefficient, normalized to standard conditions, as a function of [RNA]. Values at 0 and 20 mM MgCl2 are shown, and the extrapolation to 0 [RNA] is depicted as a dotted line. (B) Measured sedimentation coefficient as a function of [MgCl2] for the PSIV IRES.
To obtain idealized sedimentation coefficient values, the data of Figure 4A ▶ were extrapolated to zero RNA concentration. This yields values of 4.2 and 5.1 Svedbergs (>20% change) at 0 and 20 mM MgCl2, respectively, indicating a substantial change in the hydrodynamic properties. We then analyzed the CrPV and HiPV IRESes to compare their biophysical behavior with the PSIV IRES. Both CrPV and HiPV IRESes exhibit changes similar to PSIV upon addition of magnesium (Table 1 ▶). We used a partial specific volume of 0.53 cm3 g−1 and the known molecular weight to calculate the Stokes radius (RH), the ratio of the observed frictional coefficient to the coefficient expected from a spherical molecule (f/fo), and the maximum a/b ratio of a prolate or oblate ellipsoid characterized by RH and f/fo (Table 1 ▶). The observed change in these values when magnesium is added suggests a transition from an extended conformation to a more compact shape that is conserved among all three members of the CrPV-like IRES group.
TABLE 1.
Analytical ultracentrifugation measurements
| Measured values | Calculated values | ||||||
| IRES RNA | [MgCl2] (mM) | S20,w (Svedbergs) | D20,w (×107 cm2/sec) | f/fo | RH (Å) | a/bpro | a/bobl |
| PSIVa | 0 | 3.96 ± 0.04 | 4.12 ± 0.34 | 2.79 ± 0.03 | 65.8 ± 0.62 | 44.1 ± 0.9 | 81.9 ± 2.4 |
| 10 | 5.65 ± 0.07 | 4.93 ± 0.11 | 1.95 ± 0.02 | 46.1 ± 0.54 | 18.4 ± 1.1 | 26.9 ± 1.0 | |
| CrPVb | 0 | 3.92 | 3.82 | 2.70 | 62.7 | 41.2 | 74.6 |
| 10 | 5.25 | 5.39 | 2.02 | 46.8 | 20.6 | 29.9 | |
| HiPVb | 0 | 3.86 | 3.75 | 2.75 | 63.6 | 42.6 | 78.3 |
| 10 | 5.71 | 5.10 | 1.86 | 43.0 | 16.6 | 22.8 | |
| Δregion 3 | 0 | 3.75 | 4.45 | 2.46 | 53.2 | 33.2 | 55.8 |
| 10 | 5.24 | 5.81 | 1.75 | 38.0 | 14.3 | 19.2 | |
| HCV | 0 | 5.65 | 3.93 | 2.67 | 75.3 | 39.9 | 71.5 |
| 10 | Could not fit | Could not fit | |||||
aValues for PSIV are averages of multiple measurements at similar RNA concentrations (~0.015 mg/mL). Standard errors are reported.
bValues for CrPV and HiPV are taken from a single measurement at an RNA concentration close to that of PSIV (~0.015 mg/mL).
AUC and hydroxyl radical probing suggest two transitions
To determine the concentration of MgCl2 at which the hydrodynamic properties of the CrPV-like IRES RNAs change, we measured the sedimentation coefficient of the PSIV IRES over a range of magnesium concentrations at 0.010–0.015 mg/mL [RNA] (Fig. 4B ▶). We chose PSIV as the representative IRES to allow direct comparison with the hydroxyl radical-monitored magnesium titration of Figure 3 ▶. The AUC measurements indicate a sharp transition occurring at a magnesium concentration near 0.2 mM.
The magnesium dependence of the folding transition observed by AUC seems at odds to the hydroxyl radical probing data that indicate an order of magnitude higher magnesium (~2 mM) is necessary to create the solvent-inaccessible core of the PSIV RNA (Fig. 3 ▶). We hypothesize that there are two transitions occurring, each amenable to observation by a different technique. The first is a concerted global collapse of the RNA that occurs at ~ 0.2 mM MgCl2. In this transition, secondary structures are stabilized and perhaps long-range pseudoknot interactions form, but the fold is not yet tight enough to exclude solvent. This transition is global and is observed by AUC. As the magnesium concentration increases, close backbone packing and additional stabilization occurs as the molecule forms the folded core, but the overall global conformation does not change. This transition is observed by hydroxyl radical probing. Very similar behavior has been observed with the group I intron (Takamoto et al. 2002). Magnesium and sodium titrations on the intron monitored by AUC indicate that the molecule undergoes two transitions, the first global collapse occurring in a sharp transition at 0.2 mM magnesium and the second smaller transition at higher (~9 mM) magnesium concentration. Furthermore, hydroxyl radical probing of the group I intron RNA showed that a higher magnesium concentration is needed to induce close helical packing (Latham and Cech 1989). Thus, there are parallels between the magnesium-dependent folding transitions of the CrPV-like IRESes and the group I intron. In the case of the PSIV IRES, the shape of this second transition is likely too small to be detected in our AUC measurements. Also, in the case of the HCV IRES, < 0.25 mM magnesium was needed to induce an initial stabilization of secondary structure, while 0.7–0.8 mM was needed to give rise to protection from hydroxyl radicals, again consistent with one transition that is dominated by secondary structure stabilization and a second involving close RNA backbone packing (Kieft et al. 1999).
Comparing the CrPV-like and HCV IRESes by AUC
To compare the compactly folded CrPV-like IRES RNAs to the extended HCV IRES RNA, we conducted AUC measurements on the HCV IRES as a function of magnesium. While we were able to obtain values for the sedimentation and diffusion coefficients of the HCV IRES in the absence of magnesium, we could not do so in the presence of the cation (Table 1 ▶). Addition of magnesium to the HCV IRES resulted in boundaries that were not well fit by one- or multispecies models, indicating conformational heterogeneity or substantial self-association. Previously published small-angle X-ray scattering data of the HCV IRES also revealed these characteristics (Kieft et al. 1999). Thus, while we were unable to compare changes in the unbound HCV IRES hydrodynamic parameters to those of the unbound CrPV-like IRESes quantitatively, it is clear that they are behaving differently.
Region 1 + 2 is a singular compact 40S binding domain
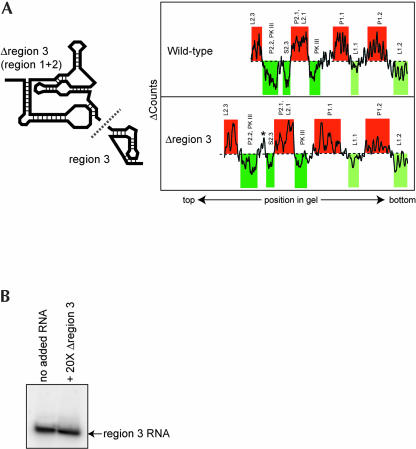
Previously, CrPV-like IRES region 1 + 2 has been shown to contain most of the affinity for the 40S subunit and to be independent of region 3 (Jan and Sarnow 2002; Nishiyama et al. 2003). We have shown that within the context of the full-length PSIV IRES, region 1 + 2 contains a hydroxyl radical protection pattern consistent with formation of a tightly folded domain. To determine whether this fold is altered when region 3 is removed, we constructed a truncated PSIV IRES mutant (Δregion 3) that contained only nucleotides 6002–6149 (Fig. 5A ▶). We first verified that this mutant bound to the 40S subunit with nearly wild-type binding affinity (data not shown) and then conducted hydroxyl radical probing on Δregion 3 to assay for formation of the folded RNA domain. The protection/enhancement pattern of Δregion 3 was similar to that observed in the context of the intact IRES RNA (Fig. 5A ▶). Differences between the two RNAs were within gel-to-gel variation and the overall pattern of protection and enhancement was maintained, indicating that removal of region 3 did not affect the global fold of the remaining PSIV IRES RNA. In addition, analytical ultracentrifugation experiments with truncated mutant Δregion 3 indicated a substantial change in its hydrodynamic properties when magnesium is added, similar to the changes observed for the intact IRES (Table 1 ▶). Specifically, the sedimentation coefficient rises from 3.75 to 5.24 (Svedbergs) and the diffusion coefficient changes from 4.45 to 5.81 (× 107 cm2/sec). These changes are even greater than that obtained with full-length PSIV IRES RNA, an expected result if region 3 is external to the domain that participates in the compact fold. Finally, we conducted in trans binding assays between Δregion 3 and an RNA that contains only region 3 (nucleotides 6149–6195) and detected no intermolecular association on a native gel (Fig. 5B ▶). Thus, we have shown that region 1 + 2 independently preforms to create the 40S subunit binding structure.
FIGURE 5.
(A) Folding of a truncated PSIV IRES RNA. On the left is a cartoon of the construction of two truncated PSIV IRES mutants: Δregion 3 (nucleotides 6002–6149) and region 3 (nucleotides 6149–6195). Comparison of the hydroxyl radical probing profile of full-length PSIV IRES and Δregion 3 is shown on the right. The analysis and color scheme is identical to that in Figure 2 ▶. An asterisk denotes the location of an apparent difference in the cleavage pattern, but this change was not observed on all gels. (B) Native gel of labeled region 3 RNA with and without 20-fold molar excess of Δregion 3. The lack of a shift upon addition of the larger RNA indicates no in trans binding.
The compact fold is necessary for 40S subunit binding
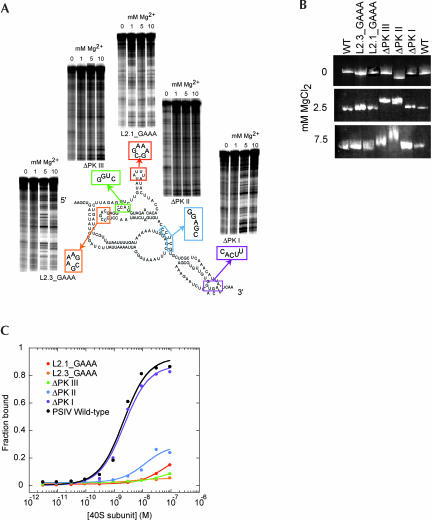
If the presence of a preformed compact fold within region 1 + 2 is necessary for 40S subunit binding, abrogation of the fold will prevent 40S subunit binding. To test this, we made a set of PSIV IRES mutants that disrupted conserved structural elements of the CrPV-like IRESes with the goal of linking formation of the compact fold and ribosome binding. The disrupted elements included the three pseudoknots and two stem–loops (Fig. 6A ▶). Similar mutants have been examined by others, showing that the targeted RNA elements are important for the ability of the IRES to bind to the ribosome and to initiate translation (Kanamori and Nakashima 2001; Jan and Sarnow 2002). Each of these mutants was examined for its (1) ability to form the prefolded compact structure and (2) ability to bind the 40S subunit in vitro.
FIGURE 6.
Analysis of the folding and 40S subunit binding of mutant IGR IRES RNAs. (A) Diagram of mutants used to link the IRES fold with 40S subunit binding. For each of the pseudoknots, we altered one side of the base-pairing interaction to its Watson–Crick complement. For the stem–loops, we substituted stable GAAA tetraloops, as these should maintain the integrity of the secondary structure. Adjacent to each of the five mutants is a portion of a hydroxyl radical probing gel of that mutant (bottom of the gel is ~nt 6084). All but mutants ΔPK III and ΔPK II show wild-type protection upon folding. (B) Native gel electrophoresis of wild-type PSIV IRES RNA and the five mutants shown in part A in the absence and in the presence of magnesium. (C) Binding curves of wild-type PSIV IRES RNA and the five mutants. Only ΔPK I binds as wild-type.
Hydroxyl radical probing revealed that only pseudoknot mutants ΔPK II and ΔPK III were unable to fold correctly, demonstrating that formation of these pseudoknots is needed to properly fold the PSIV IRES RNA compact domain (Fig. 6A ▶). All other mutants showed a cleavage protection pattern consistent with formation of the correct fold. That mutating the two stem–loops does not alter the folding of the RNA is consistent with their predicted position outside the folded core (Jan and Sarnow 2002; Nishiyama et al. 2003). Likewise, the fact that mutating PK I does not affect the fold is expected, as it resides outside the compact domain formed by region 1 + 2. For both misfolding mutants (ΔPK II and ΔPK III), we observed a complete loss of protection throughout the compact domain. Loss of either of the pseudoknot interactions did not lead to unfolding of only part of the domain; it induced a total inability to fold. Likewise, for each of the mutants that did fold correctly, a completely correct protection pattern was observed. This verifies that the ribosome-binding domain (regions 1 + 2) is a single folded tertiary entity.
The ability of each mutant to fold was confirmed using native gel electrophoresis, which is exquisitely sensitive to RNA structure; misfolded RNAs tend to migrate more slowly. In the absence of magnesium (2 mM EDTA), the mutants all run with wild-type mobility, with the exception of mutant ΔPK II, which migrates at a somewhat faster rate (Fig. 6B ▶). In the presence of magnesium both ΔPK III and ΔPK II exhibit retarded migration rates, but all of the other mutants migrate as wild-type. Thus, the native gel confirms that ΔPK III and ΔPK II are not properly folded. RNase T1 probing of all mutants further verified these results (data not shown).
We then assayed both the wild-type and mutant RNAs for their ability to bind to purified 40S subunit. Although similar PSIV and CrPV mutants have been tested by others for 40S subunit binding, we repeated these experiments to ensure consistent results with our 40S preparations and mutants (Jan and Sarnow 2002; Nishiyama et al. 2003). Wild-type PSIV IRES bound to the 40S subunit with an apparent Kd of ~2 nM in buffer containing 2.5 mM magnesium and 300 mM potassium (Fig. 6C ▶). This monovalent concentration is approximately twofold greater than physiological concentrations. However, we have previously observed that at lower cation concentrations, nonspecific binding of RNAs to purified 40S subunits can occur, and hence more stringent buffer conditions are needed to be certain of measuring specific interactions. Similar results can be obtained using non-IRES competitor RNA. Note that we measured an apparent Kd tighter than that reported for the CrPV IRES by Jan and Sarnow (2002), possibly due to the use of two different techniques to measure binding (electrophoretic mobility shift vs. filter binding). Binding assays on each of the mutants show that all but mutant ΔPK I (which folds correctly) were severely inhibited in their ability to bind to the 40S subunit (Fig. 6C ▶).
Comparing the folding and binding assays indicates our set of mutants contains two classes. The first class contains ΔPK II and ΔPK III. These mutants are unable to fold and this is likely why they are unable to bind to the 40S subunit. The existence of these mutants strongly suggests that the CrPV-like IRESes’ preformed compact conformation is a necessary precursor for ribosome binding. The second class of mutants contains the mutations to the stem–loops (L2.1_GAAA and L2.3_GAAA). These mutants fold correctly and thus loss of 40S subunit binding affinity is probably due to a loss of intermolecular interactions that directly involve these loops. This provides direct confirmation of their previously proposed role (Jan and Sarnow 2002; Nishiyama et al. 2003). It seems likely that an important role of the compact IRES is to pre-position these loops correctly for ribosome interaction. The strategy of positioning emerging stem–loops correctly in space also has been proposed as an important part of HCV IRES binding to the ribosome, although in HCV the loops are likely farther apart in space and they do not all emerge from a single folded domain (Kieft et al. 1999, 2001).
DISCUSSION
Internal initiation of translation is a mechanism in which the structure of the mRNA or viral RNA is of primary importance. The structured IRES RNA must recruit, position, and activate the protein-making machinery either through direct interaction with the ribosome, through canonical initiation factors, or through the use of trans-activating factor proteins. In some cases, the ribosome binding mechanism may use a combination of all of these. The diversity of IRES functional requirements and sequences begs the question of whether IRESes are evolutionarily disparate structures representing different solutions to a similar problem or if, at some level, IRES RNAs all share similar structural features and folding strategies. Comparing the three-dimensional RNA folding strategies of the HCV and CrPV-like IRESes with each other and with other folded RNAs shows both similarities and differences in how each IRES organizes itself for ribosome binding.
To facilitate this discussion regarding the comparison of the CrPV-like IRES RNA with other folded RNAs, we use a definition of an RNA structural “domain” based on the structures of other folded RNA domains. Examination of the structure of the P4-P6 domain of the Tetrahymena group I intron reveals that the compact fold of this RNA results in protection of ~21% of the backbone from solvent (Murphy and Cech 1993; Murphy et al. 1994; Cate et al. 1996). The crystal structure of the group I intron from Azoarcus clearly shows the packed RNA at the core of the fold (Adams et al. 2004); this fold protects ~42% of the backbone (as assayed with the Tetrahymena intron) (Latham and Cech 1989; Murphy and Cech 1993). In the case of the P4-P6 domain and the group I intron, high-resolution structures reveal that hydroxyl radical probing effectively predicts the locations of compactly folded regions (Cate et al. 1996; Adams et al. 2004). For RNase P, a high-resolution structure is not available, but hydroxyl radical probing of the RNA component from Bacillus subtilis reveals that ~34% of the folded RNA backbone is protected from solvent (Pan 1995). Hence, for this discussion we define an RNA structural domain by two criteria: First, a folded RNA domain has a solvent-inaccessible core of packed RNA that forms in the presence of cations. Second, the amount of solvent-inaccessible RNA makes up a significant portion of the total RNA (>20%). This definition allows a clear distinction between secondary structure elements and regions, and an independently folded RNA structural domain.
Applying the above definition to the PSIV and HCV IRES RNAs, it is clear that the 40S binding site (region 1 + 2) of the PSIV IRES meets the criteria of a folded compact RNA structural domain but the HCV IRES does not. Approximately 27% of the PSIV IRES RNA is protected from solvent, a greater percentage than that observed with the well-characterized and tightly folded P4-P6 domain RNA. Furthermore, this percentage is based on total RNA; if we consider only regions 1 + 2 the percent of solvent-protected backbone increases to ~34%, similar to that observed with RNase P RNA. In contrast, while the HCV IRES also folds in the presence of metal ions into a fold that is prepared to bind the ribosome, only ~10% of the backbone shows protection from solvent (Kieft et al. 1999). If we consider only that portion of the HCV IRES needed to bind to the 40S subunit (~160 nt), the percent of protected RNA rises to ~20%, but the protected regions are contained in two different independently folded domains. Both IRES RNAs contain regions of close RNA packing, but the RNA packing is giving rise to two different architectures. In the HCV IRES, the packing occurs locally, in junctions, as part of an overall extended fold that uses multiple domains to provide 40S subunit affinity. In the CrPV-like IRESes, the overall conformation is a globular one, with a single prefolded domain contributing 40S binding affinity. Hence, the ribosome binding mechanisms of the CrPV-like and HCV IRESes diverge early in the initiation pathway at the level of initial RNA folding.
The compactly folded CrPV-like IRES domain (regions 1 + 2) is a single independent structural entity, while region 3 (containing PK I) is external to this compact domain and may be performing some sort of tRNA mimicry (Jan et al. 2003). This two-part organization was predicted by Kanamori and Nakashima (2001), and here we confirm that it is inherent in the prefolded state of the RNA. The overall pattern of folding in the compact domain is shared by all of the CrPV-like IRESes that we examined, strongly suggesting it is a critical determinant of IRES function. Evolutionary pressure has been to maintain this specific conformation in different sequence contexts, probably by preserving certain conserved intramolecular contacts. Interestingly, region 3 shows greater secondary structure divergence than does the compact domain (Kanamori and Nakashima 2001; Hatakeyama et al. 2004).
The prefolded domain of the CrPV-like IRES RNAs likely serves to display critical 40S subunit recognition elements correctly. Two clearly identified recognition elements are the two stem–loops that extend into solution in the pre-folded state. Footprinting studies on the PSIV IRES by Nishiyama et al. (2003) show that all of the positions protected upon 40S subunit binding are in regions that are solvent-accessible in the prefolded molecule. The recently presented cryo-EM structure of the CrPV IRES bound to the ribosome reveals that the IRES docks into the mRNA decoding groove within the intersubunit space (Spahn et al. 2004). This reconstruction also shows two distinct areas of density that likely correspond to the two independent parts of the IRES (the compact domain and region 3). In addition, although the resolution of the cryo-EM structures is not sufficient to reveal individual helices, there is density suggestive of packed helices.
Taken together, these data suggest a role for the preorganized folded IRES RNA in the mechanism of Dicistroviridae IGR IRES binding to the ribosome. The unbound IRES RNA prefolds into a shape that is both appropriate for insertion into the mRNA decoding groove and for occupation of the intersubunit space. This compact domain also serves to display critical specific recognition surfaces such as the two emergent stem–loops. Binding of the IRES to its cognate site on the 40S subunit allows region 3 (PK I) to swing into position in the P-site of the ribosome as current models suggest. In this proposed model, the compact IRES ribosome-binding domain is similar in its three-dimensional shape both in the unbound and bound forms. Additional experiments are needed to establish the degree to which the IRES might subtly readjust structure upon binding.
IRES RNAs have evolved to use a large variety of sequences and secondary structures to achieve the goal of recruiting, placing, and activating the translation apparatus. This diversity is likely reflected in a variety of mechanisms used to recruit the ribosome to the message. At one extreme are IRES RNAs that require the action of trans-activating proteins that may act as chaperones, folding the RNA into the correct conformation for binding to the translation machinery. In these cases, the IRES RNA is a largely unfolded scaffold that requires the involvements of exterior factors for function. In the case of the Apaf-1 IRES, binding of a cofactor protein acts to unfold RNA structure, leading to IRES function (Mitchell et al. 2003). In another case, formation of the RNA structure needed for IRES function requires disruption of upstream secondary structures by a ribosome translating an upstream open reading frame (Yaman et al. 2003). Thus, these IRESes seem to require removal of inhibitory native RNA structure in order to function. At the other extreme are the CrPV-like IRESes, which self-organize and dock on the ribosome without external factors and perhaps with little structural alteration, requiring precise formation and maintenance of native RNA structure. One goal of IRES structural studies is to find secondary or tertiary structure elements common to all IRES RNAs and therefore critical to internal initiation in general. However, it is clear that similar function can arise from fundamentally different IRES RNA architectures. Hence, IRES RNAs cannot be thought of as a structural class of RNAs but as a functional class with structural diversity at the level of the unbound, folded RNAs.
MATERIALS AND METHODS
Cloning and plasmid production
Plasmids pT7CAT-5375 (encoding the PSIV IRES) and pT7CAT-HiPV6201-7259 (encoding the HiPV IRES) were a kind gift from Nobuhiko Nakashima (National Institute of Agrobiological Sciences, Japan). Plasmid pCrPV1-1 (encoding the CrPV IRES) was a kind gift of Eric Jan and Peter Sarnow (Stanford University). These plasmids were used as templates for PCR reactions to generate inserts that contain the T7 polymerase promoter and the IRES sequence flanked by cis-acting ribozymes. Inserts were ligated into the EcoRI/BamHI site of pUC-19, amplified in DH5α cells, and sequenced. Full-length IRES sequences included nucleotides 6002–6195 of PSIV, nucleotides 6029–6219 of CrPV, and nucleotides 6087–6275 of HiPV. Mutants were generated using the QuickChange mutagenesis kit (Stratagene).
Construction of the plasmid encoding nucleotides 40–372 of the HCV IRES genotype 1b has been described (Kieft et al. 1999).
RNA transcription, purification, and end labeling
RNA was generated as described previously. Briefly, plasmid DNA was linearized with BamHI and used in in vitro transcriptions as described previously (Kieft et al. 1999). RNA was purified on 6% denaturing polyacrylamide gels, passively eluted from the gel, concentrated in ultrafiltration/concentration devices (Amicon), and stored in DEPC-treated water at −20°C.
RNA was 5′ or 3′ end labeled as described (Kieft et al. 1999, 2001).
Chemical and enzymatic probing
Hydroxyl radical probing and RNase T1 probing were performed as described (Kieft et al. 1999). Buffer conditions were 30 mM HEPES-KOH (pH 7.4), 0–10 mM MgCl2, 0.1 mg/mL tRNA. Reactions were conducted at 37°C for 2 min and analyzed on 10% sequencing denaturing polyacrylamide gels. Gels were dried and imaged on a phosphorimager.
For the analysis of Figure 2 ▶, the intensity of cleavage as a function of location in the gel was determined using the “line graph” function of ImageQuant and the data were imported into Excel. To adjust for differences in lane loading on the gel, the signal was normalized using the total number of counts in each lane. Differences in the cleavage intensities between lanes were determined arithmetically and plotted as a function of gel position using KaleidaGraph.
For the analysis of Figure 3 ▶, several locations along the RNA backbone were selected based on mapped changes in the protection pattern. In each lane, the intensity of the signal (counts) at that location was measured and used to calculate the protection factor (PF), using PF = (counts at X mM MgCl2)/(counts at 0 mM MgCl2). The resultant PF was then normalized to a fraction folded scale of zero to one, assuming no folding at 0 mM MgCl2 and full folding at 10 mM MgCl2. The resultant data were fit to the Hill equation using the program KaleidaGraph.
Sequencing ladders using RNase T1 and hydrolysis ladders were generated as described (Kieft et al. 1999).
Native gel electrophoresis
Nondenaturing (native) gel electrophoresis was performed essentially as described (Kieft et al. 1999). Gels were 10% polyacrylamide (29:1 acrylamide:N-N′-methylene-bis-acrylamide ratio) in 1× TH buffer (66 mM HEPES, 33 mM Tris) and the desired amount of MgCl2 or EDTA. Gels were run at a constant power for 2–5 h (until bromophenol blue was 9 cm below the wells) in a 4°C room, followed by ethidium bromide staining and visualization on a UV transilluminator.
Analytical ultracentrifugation
To anneal and fold the RNA, it was suspended in 10 mM Na-Cacodylate buffer (pH 7.5), heated to 80°C for 1 min, cooled on the bench to room temperature, and MgCl2 was added to the desired concentration. Analytical ultracentrifugation was performed in a Beckman XL-A centrifuge with an An-60Ti rotor at 40,000 rpm at 12°C. The sample was scanned every 5 min at 260 nm. For each experiment, at least 30 boundary traces were collected; in most cases, 60 were collected.
The traces were analyzed as dc/R using the program SVED-BERG V6.39 (Philo 1997). In each case, at least 15 traces were used simultaneously. The fit directly yielded the parameters S and D. We adjusted these values to standard conditions using the program SEDNTRP V1.08. A partial specific volume of 0.53 cm3/g and the calculated molecular weight was used to obtain the parameters RH, f/fo, and a/b. This analysis is similar to that employed in the analysis of other folded RNAs (Deras et al. 2000; Takamoto et al. 2002).
40S ribosomal subunit binding assays
40S ribosomal subunit was purified from bulk rabbit reticulocyte lysate (Green Hectares) as described (Kieft et al. 2001). These subunits were used in binding assays identical to those performed on the HCV IRES using filter binding (Kieft et al. 2001). Buffer was 20 mM Tris-HCl (pH 7.0), 2.5 mM MgCl2, 100 mM K-Acetate, 200 mM KCl, 1 mM DTT.
Acknowledgments
We thank Nobuhiko Nakashima, Eric Jan, and Peter Sarnow for the gifts of plasmids. We also wish to thank Christian Spahn for his willingness to share and discuss unpublished cryo-EM results, Robert Batey and Diego Zorio for critical reading of the manuscript, and members of the Kieft Laboratory for stimulating discussion.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7184705.
REFERENCES
- Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J., and Strobel, S.A. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50. [DOI] [PubMed] [Google Scholar]
- Bonnal, S., Boutonnet, C., Prado-Lourenco, L., and Vagner, S. 2003. IRESdb: the Internal Ribosome Entry Site database. Nucleic Acids Res. 31: 427–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. [DOI] [PubMed] [Google Scholar]
- Celander, D.W. and Cech, T.R. 1991. Visualizing the higher order folding of a catalytic RNA molecule. Science 251: 401–407. [DOI] [PubMed] [Google Scholar]
- Christian, P.D. and Scotti, P.D. 1998. Picornaviruses of insects. In The insect viruses (eds. L.K. Miller and A. Ball), pp. 301–336. Plenum, New York.
- Deras, M.L., Brenowitz, M., Ralston, C.Y., Chance, M.R., and Woodson, S.A. 2000. Folding mechanism of the Tetrahymena ribozyme P4-P6 domain. Biochemistry 39: 10975–10985. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, Y., Shibuya, N., Nishiyama, T., and Nakashima, N. 2004. Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts. RNA 10: 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U. and Sarnow, P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Hershey, J.W.B. and Merrick, W.C. 2000. Pathway and mechanism of initiation of protein synthesis. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 33–88. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Jan, E. and Sarnow, P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324: 889–902. [DOI] [PubMed] [Google Scholar]
- Jan, E., Thompson, S.R., Wilson, J.E., Pestova, T.V., Hellen, C.U., and Sarnow, P. 2001. Initiator Met-tRNA-independent translation mediated by an internal ribosome entry site element in cricket paralysis virus-like insect viruses. Cold Spring Harbor Symp. Quant. Biol. 66: 285–292. [DOI] [PubMed] [Google Scholar]
- Jan, E., Kinzy, T.G., and Sarnow, P. 2003. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. 100: 15410–15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori, Y. and Nakashima, N. 2001. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7: 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Jubin, R., Murray, M.G., Lau, J.Y., and Doudna, J.A. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292: 513–529. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Jubin, R., and Doudna, J.A. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7: 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Pestova, T.V., and Hellen, C.U. 2000a. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 74: 6242–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000b. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA 6: 1791–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham, J.A. and Cech, T.R. 1989. Defining the inside and outside of a catalytic RNA molecule. Science 245: 276–282. [DOI] [PubMed] [Google Scholar]
- Mayo, M.A. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147: 1655–1663. [DOI] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Coldwell, M.J., Jackson, R.J., and Willis, A.E. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11: 757–771. [DOI] [PubMed] [Google Scholar]
- Murphy, F.L. and Cech, T.R. 1993. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry 32: 5291–5300. [DOI] [PubMed] [Google Scholar]
- Murphy, F.L., Wang, Y.H., Griffith, J.D., and Cech, T.R. 1994. Coaxially stacked RNA helices in the catalytic center of the Tetrahymena ribozyme. Science 265: 1709–1712. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Yamamoto, H., Shibuya, N., Hatakeyama, Y., Hachimori, A., Uchiumi, T., and Nakashima, N. 2003. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 31: 2434–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, T. 1995. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry 34: 902–909. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V. and Hellen, C.U. 2003. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 17: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J., and Hellen, C.U.T. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 12: [DOI] [PMC free article] [PubMed]
- Pestova, T.V., Kolupaeva, V.G., Lomakin, I.B., Pilipenko, E.V., Shatsky, I.N., Agol, V.I., and Hellen, C.U. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. 98: 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Lomakin, I.B., and Hellen, C.U. 2004. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 5: 906–913. 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo, J.S. 1997. An improved function for fitting sedimentation velocity data for low-molecular-weight solutes. Biophys. J. 72: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow, P. 2003. Viral internal ribosome entry site elements: Novel ribosome–RNA complexes and roles in viral pathogenesis. J. Virol. 77: 2801–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, J. and Nakashima, N. 1999. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 73: 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. 97: 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya, N., Nishiyama, T., Kanamori, Y., Saito, H., and Nakashima, N. 2003. Conditional rather than absolute requirements of the capsid coding sequence for initiation of methionine-independent translation in Plautia stali intestine virus. J. Virol. 77: 12002–12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova, D.V., Kolupaeva, V.G., Pestova, T.V., Shatsky, I.N., and Hellen, C.U.T. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 72: 4775–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, C.M., Kieft, J.S., Grassucci, R.A., Penczek, P.A., Zhou, K., Doudna, J.A., and Frank, J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Jan, E., Mulder, A., Grassucci, R.A., Sarnow, P., and Frank, J. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell 118: 465–475. [DOI] [PubMed] [Google Scholar]
- Stoneley, M. and Willis, A.E. 2004. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene 23: 3200–3207. [DOI] [PubMed] [Google Scholar]
- Takamoto, K., He, Q., Morris, S., Chance, M.R., and Brenowitz, M. 2002. Monovalent cations mediate formation of native tertiary structure of the Tetrahymena thermophila ribozyme. Nat. Struct. Biol. 9: 928–933. [DOI] [PubMed] [Google Scholar]
- Thompson, S.R., Gulyas, K.D., and Sarnow, P. 2001. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl. Acad. Sci. 98: 12972–12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner, S., Galy, B., and Pyronnet, S. 2001. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Pestova, T.V., Hellen, C.U., and Sarnow, P. 2000a. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520. [DOI] [PubMed] [Google Scholar]
- Wilson, J.E., Powell, M.J., Hoover, S.E., and Sarnow, P. 2000b. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell Biol. 20: 4990–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A.A., Koromilas, A.E., Zhou, L., Snider, M.D., Scheuner, D., Kaufman, R.J., et al. 2003. The zipper model of translational control: A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113: 519–531. [DOI] [PubMed] [Google Scholar]