Abstract
SR proteins regulate alternative splicing by binding to exonic sequences where, via an arginine/serine-rich splicing activation domain, they enhance the binding of the spliceosome to the adjacent splice sites. Here, a system is described in which a nontoxic derivative of the small molecule rapamycin is used to control pre-mRNA splicing in vitro. This involves the rapamycin-dependent recruitment of a splicing activation domain located on one protein to a second protein bound to the pre-mRNA. These results provide a new approach to explore for regulating gene expression in vivo with small molecules by controlling pre-mRNA splicing.
Keywords: SR proteins, splicing, rapamycin, small molecule
The ability to regulate gene expression with small molecules has numerous experimental and therapeutic applications. Many systems have been developed to regulate gene expression by using small molecules to activate transcription (Pollock et al. 2002). One small molecule that has been widely used for these approaches is rapamycin. Rapamycin can simultaneously interact with two proteins called FK506 binding protein (FKBP) (Standaert et al. 1990) and the rapamycin binding domain (FRB) of FRAP (FKBP rapamycin-associated protein) (Chen et al. 1995), which do not normally interact with one another. By fusing other proteins to FKBP and FRB, the association of the proteins can be controlled with rapamycin. For example, rapamycin has been used to regulate transcription by modulating the association of a DNA binding domain with a transcription activation domain, each of which were fused to either FKBP or FRB (Ho et al. 1996; Rivera et al. 1996; Liberles et al. 1997).
One of the main problems with using rapamycin to control gene expression in vivo is that it is toxic to cells due to its ability to inhibit cell proliferation. The interaction of rapamycin with FRAP inhibits the kinase activity of FRAP, which in turn inhibits the translation of specific mRNAs and arrests cells at the G1 phase of the cell cycle (Brown and Schreiber 1996). To circumvent this, a nontoxic derivative of rapamycin, called rapamycin*, has been synthesized (Liberles et al. 1997). Rapamycin* contains a methylallyl group at the C16 position and is not toxic because it does not bind to FRAP (Liberles et al. 1997). A compensatory mutant of FRB that can bind to rapamycin* was isolated in a three-hybrid screen and is referred to as FRB* (Liberles et al. 1997). Thus, by using rapamycin* and FRB*, one can circumvent the toxicity issues surrounding rapamycin.
Alternative splicing is a prevalent mechanism by which metazoans regulate gene expression (Black 2000; Graveley 2001). In fact, as many as 74% of human genes may encode alternatively spliced mRNAs (Johnson et al. 2003). An important group of splicing regulators is the serine/arginine-rich (SR) protein family (Graveley 2000). SR proteins regulate alternative splicing by binding to exons where they promote the binding of the general splicing machinery to flanking introns. All SR proteins contain one or two N-terminal RNA binding domains and a variable length C-terminal domain rich in arginine and serine residues called the RS domain. The RNA binding and RS domains are modular and can be interchanged between different SR proteins (Chandler et al. 1997). Moreover, when fused to a heterologous RNA binding domain, an RS domain is sufficient to activate splicing of a pre-mRNA containing a binding site for the hybrid protein (Graveley and Maniatis 1998). Thus, RS domains function as modular splicing activation domains.
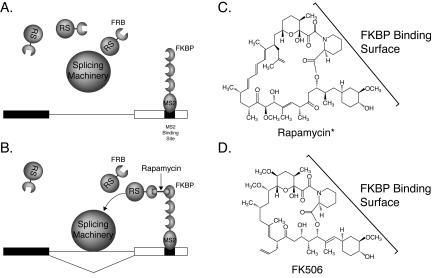
The modular nature of SR proteins and the ability of rapamycin to function as a heterodimerizer suggested that a system could be developed in which rapamycin could control splicing. In this system, rapamycin would regulate the association of one protein containing an SR protein RS domain fused to FRB with a second protein containing an RNA binding domain fused to FKBP (Fig. 1A,B ▶). By using a pre-mRNA substrate containing an enhancer-dependent intron and a binding site for the heterologous RNA binding protein, splicing could be rendered rapamycin dependent.
FIGURE 1.
Experimental design. (A) In the absence of rapamycin*, the FRB*-RS fusion protein is not recruited to the RNA. As a result, splicing of the intron does not occur. (B) In the presence of rapamycin*, the FRB*-RS fusion protein is recruited to the RNA via the MS2-F3 protein and splicing of the intron occurs. (C) The structure of rapamycin*. (D) The structure of FK506. The surface of each molecule that interacts with FKBP is indicated.
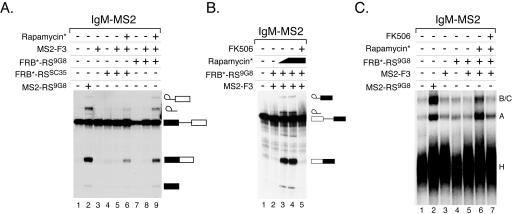
To test this, the MS2 coat protein, an RNA binding domain with a high degree of specificity to a small RNA hairpin loop (Graveley and Maniatis 1998) was fused to three FKBP domains (MS2-F3) and separately, the RS domains from two different human SR proteins (9G8 or SC35) were fused to FRB* (FRB*-RS9G8 and FRB*-RSSC35). The MS2-F3 and FRB*-RS proteins were expressed in and purified from Escherichia coli and baculovirus-infected Sf9 cells, respectively. To test whether rapamycin* could control pre-mRNA splicing, in vitro splicing assays were conducted in HeLa cell nuclear extract using a pre-mRNA substrate (IgM-MS2) containing an intron from the human IgM pre-mRNA and a single MS2 binding site in the downstream exon (Graveley and Maniatis 1998). This pre-mRNA is not spliced in nuclear extract alone (Fig. 2A ▶, lane 1), but is efficiently spliced (30% splicing) when a control protein containing a potent RS domain from the SR protein 9G8 directly fused to MS2 (MS2-RS9G8) is added to the reaction (lane 2). Addition of MS2-F3, FRB*-RS9G8, or FRB*-RSSC35 alone or in combination did not activate splicing (lanes 3–5,7,8). However, in the presence of rapamycin*, MS2-F3, and either FRB*-RS protein, splicing of the IgM-MS2 pre-mRNA was observed (lanes 6,9). The efficiency of splicing observed with FRB*-RSSC35 (6.5%) and FRB*-RS9G8 (18%) correlates well with the previous observation that the RS domain of 9G8 is more potent than the SC35 RS domain (Graveley et al. 1998). Interestingly, the efficiency of splicing that occurs in the presence of MS2-F3, FRB*-RS9G8, and rapamycin* (18%) is lower than that observed in the presence of MS2-RS9G8 (30%). This is likely due to the fact that three molecules (FRB*-RS9G8, MS2-F3, and rapamycin*) need to simultaneously interact with one another and bind the RNA to activate splicing whereas the MS2-RS9G8 protein simply needs to bind RNA. Although the MS2 protein has three FKBP modules fused to it, it is not clear whether all three modules can simultaneously be bound by rapamycin* and FRB*-RS9G8. Thus, it is possible that the three FKBP modules simply increase the probability of a productive recruitment of an RS domain rather than recruiting multiple RS domains. Splicing of the pre-mRNA is dependent upon dimerization of the MS2-F3 and FRB*-RS proteins because a preincubation with the small molecule FK506 (Fig. 1D ▶), which acts as a competitive inhibitor by binding to FKBP but not FRB*, inhibits the ability of rapamycin* to activate splicing (Fig. 2B ▶, lanes 4,5). Analysis of the splicing reactions by native gel electrophoresis demonstrates that the addition of rapamycin* enhances the early stages of spliceosome assembly, but that this effect can be inhibited by FK506 (Fig. 2C ▶). Together, these results demonstrate that rapamycin* can be used to control splicing by regulating the recruitment of a splicing activation domain to the pre-mRNA, which in turn enhances spliceosome assembly.
FIGURE 2.
Small-molecule-dependent pre-mRNA splicing. The IgM-MS2 pre-mRNA was incubated in the presence or absence of MS2-RS9G8, FRB*-RS9G8, FRB*-RSSC35, MS2-F3, rapamycin*, or FK506 as indicated and resolved on either a denaturing polyacrylamide gel (A) or a native polyacrylamide gel (B). The concentrations of rapamycin* and FK506 used in the experiments are as follows: (A, lanes 6,9) 200 nM rapamycin*; (B, lane 3) 200 nM rapamycin*, (lane 4) 500 nM rapamycin*, (lane 5) 5 μM FK506 and 500 nM rapamycin*; (C, lane 6) 500 nM rapamycin*, (lane 7) 5 μM FK506 and 500 nM rapamycin*.
The ability to manipulate gene expression has a large number of experimental and clinical uses. A tremendous amount of work has focused on the use of antisense (Estibeiro and Godfray 2001; Petersen and Wengel 2003; Vacek et al. 2003), ribozyme (Lewin and Hauswirth 2001) and, more recently, RNA interference (Clayton 2004) to modulate gene expression. While all of these have been shown to be useful, they are primarily used to inactivate gene expression. In contrast, the ability to regulate gene expression with small molecules at the level of transcription, and now pre-mRNA splicing, can be used to activate and fine-tune the expression of the target gene. These approaches have tremendous experimental and clinical potential. For example, in transgenic animals that express DNA binding and transcription activation domains separately fused to proteins that bind to small molecules, the expression of transgenes can be reversibly regulated by oral or intravenous administration of the small molecule (Rivera et al. 1996; Pollock et al. 2002). Such systems can be used for experimental purposes by designing them to examine gene function at a specific time in development. Alternatively, similar approaches can be used to fine-tune the expression levels of proteins in gene therapy settings. The demonstration that pre-mRNA splicing can be controlled in vitro with rapamycin* provides a new approach to explore for regulating gene expression in vivo with small molecules.
MATERIALS AND METHODS
The MS2-RS9G8 protein was described previously (Graveley and Maniatis 1998) and was purified from baculovirus-infected Sf9 cells. To produce the FRB*-RS9G8 and FRB*-RSSC35 proteins, recombinant baculoviruses were generated that encoded amino acids 2025–2114 of human FRAP (Chen et al. 1995) containing mutations that can accommodate rapamycin* (Liberles et al. 1997) and the RS domains of either 9G8 or SC35 (Graveley and Maniatis 1998). The proteins were His-tagged at the N terminus. Sf9 cells were infected with the viruses and the proteins purified under native conditions as described (Graveley and Maniatis 1998).
To produce the MS2-F3 protein an expression vector was generated by ligating a XhoI–SalI fragment encoding three FKBP domains from plasmid pBJ5-E′F3E (Belshaw et al. 1996) into pRSET-H6-MS2 (Graveley and Maniatis 1998). The plasmid was transformed into E. coli (BL21(DE3)) and expression induced with IPTG. The protein was purified under native conditions as described (Graveley and Maniatis 1998).
The IgM-MS2 pre-mRNA was described previously (Graveley and Maniatis 1998) and was synthesized with SP6 RNA polymerase in the presence of 32P-UTP. The RNA was gel purified prior to use. In vitro splicing was performed essentially as described (Graveley and Maniatis 1998) except that the reactions were supplemented with rapamycin* and or FK506 at the designated concentrations. The splicing reactions were resolved on either denaturing polyacrylamide or native polyacrylamide gels.
Acknowledgments
I thank Tom Maniatis for his support during the early stages of this project, Stuart Schreiber and Stephen Liberles for the generous gifts of rapamycin*, FK506, and plasmids, and for their continual advice throughout the course of this work, and Gordon Carmichael, Kristen Lynch, and Rob Reenan for their comments on the manuscript. This work was supported by NIH grants GM62516 and AR46026 to B.R.G.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7229705.
REFERENCES
- Belshaw, P.J., Ho, S.N., Crabtree, G.R., and Schreiber, S.L. 1996. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc. Natl. Acad. Sci. 93: 4604–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103: 367–370. [DOI] [PubMed] [Google Scholar]
- Brown, E.J. and Schreiber, S.L. 1996. A signaling pathway to translational control. Cell 86: 517–520. [DOI] [PubMed] [Google Scholar]
- Chandler, S.D., Mayeda, A., Yeakley, J.M., Krainer, A.R., and Fu, X.-D. 1997. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc. Natl. Acad. Sci. 94: 3596–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Zheng, X.F., Brown, E.J., and Schreiber, S.L. 1995. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. 92: 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, J. 2004. RNA interference: The silent treatment. Nature 431: 599–605. [DOI] [PubMed] [Google Scholar]
- Estibeiro, P. and Godfray, J. 2001. Antisense as a neuroscience tool and therapeutic agent. Trends Neurosci. 24: S56–62. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17: 100–107. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. and Maniatis, T. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1: 765–771. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R., Hertel, K.J., and Maniatis, T. 1998. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 17: 6747–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N., Biggar, S.R., Spencer, D.M., Schreiber, S.L., Crabtree, G.R. 1996. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature 382: 822–826. [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., and Shoemaker, D.D. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144. [DOI] [PubMed] [Google Scholar]
- Lewin, A.S. and Hauswirth, W.W. 2001. Ribozyme gene therapy: Applications for molecular medicine. Trends Mol. Med. 7: 221–228. [DOI] [PubMed] [Google Scholar]
- Liberles, S.D., Diver, S.T., Austin, D.J., and Schreiber, S.L. 1997. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc. Natl. Acad. Sci. 94: 7825–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M. and Wengel, J. 2003. LNA: A versatile tool for therapeutics and genomics. Trends Biotechnol. 21: 74–81. [DOI] [PubMed] [Google Scholar]
- Pollock, R., Giel, M., Linher, K., and Clackson, T. 2002. Regulation of endogenous gene expression with a small-molecule dimerizer. Nat. Biotechnol. 20: 729–733. [DOI] [PubMed] [Google Scholar]
- Rivera, V.M., Clackson, T., Natesan, S., Pollock, R., Amara, J.F., Keenan, T., Magari, S.R., Phillips, T., Courage, N.L., Cerasoli Jr., F., et al. 1996. A humanized system for pharmacologic control of gene expression [see comments]. Nat. Med. 2: 1028–1032. [DOI] [PubMed] [Google Scholar]
- Standaert, R.F., Galat, A., Verdine, G.L., and Schreiber, S.L. 1990. Molecular cloning and overexpression of the human FK506-binding protein FKBP. Nature 346: 671–674. [DOI] [PubMed] [Google Scholar]
- Vacek, M., Sazani, P., and Kole, R. 2003. Antisense-mediated redirection of mRNA splicing. Cell Mol. Life Sci. 60: 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]