Abstract
The ongoing characterization of novel species creates the need for a molecular marker which can be used for species- and, simultaneously, for mega-systematics. Recently, the use of the internal transcribed spacer 2 (ITS2) sequence was suggested, as it shows a high divergence in sequence with an assumed conservation in structure. This hypothesis was mainly based on small-scale analyses, comparing a limited number of sequences. Here, we report a large-scale analysis of more than 54,000 currently known ITS2 sequences with the goal to evaluate the hypothesis of a conserved structural core and to assess its use for automated large-scale phylogenetics. Structure prediction revealed that the previously described core structure can be found for more than 5000 sequences in a wide variety of taxa within the eukaryotes, indicating that the core secondary structure is indeed conserved. This conserved structure allowed an automated alignment of extremely divergent sequences as exemplified for the ITS2 sequences of a ctenophorean eumetazoon and a volvocalean green alga. All classified sequences, together with their structures can be accessed at http://www.biozentrum.uni-wuerzburg.de/bioinformatik/projects/ITS2.html. Furthermore, we found that, although sample sequences are known for most major taxa, there exists a profound divergence in coverage, which might become a hindrance for general usage. In summary, our analysis strengthens the potential of ITS2 as a general phylogenetic marker and provides a data source for further ITS2-based analyses.
Keywords: internal transcribed spacer 2, phylogenetics, RNA, structure
INTRODUCTION
Defining a single general marker for the taxonomic classification of an organism on all taxonomic levels is challenging. On the one hand, one needs a fast-evolving marker for classification on the species level; on the other hand this marker will be too divergent for reconstructing the phylogeny on a higher level. This problem is usually addressed by using different markers like ITS for low level and 18S or 28S rDNA for high level classification. Recently, it was suggested, that the internal transcribed spacer 2 region (ITS2) of the nuclear rDNA cistron might be a marker suitable for taxonomic classification over a wide range of levels (Coleman 2003). As the sequence of the ITS2 region evolves comparably fast, it already found a wide application for phylogenetic reconstructions on the species and genus level (Alvarez and Wendel 2003). The suggestion of a more general use is mainly based on reports that find a conservation of its structure in organisms as divergent as vertebrates and yeast (Joseph et al. 1999) or green algae and higher plants (Mai and Coleman 1997). Still, these reports used only a small number of sequences and involved manual optimizations raising the questions, whether (1) there indeed exists a conserved core structure and (2) whether it can be found without manual intervention. In this case, the structure might be used to generate reliable alignments of extremely divergent sequences automatically and the ITS2 sequence could indeed be the basis for large-scale and automatic phylogenetic reconstructions.
RESULTS AND DISCUSSION
Taxon coverage
The presence of sufficient data from close and distant relatives is a precondition for a correct phylogenetic reconstruction as otherwise it would be impossible to integrate species into existing classifications. A simple search for the string “internal transcribed spacer 2” in GenBank revealed more than 70,000 entries containing about 54,000 entries with an explicitly given position of the ITS2 sequence. The question first was, whether these are evenly distributed between all Eukaryota or whether some branches are heavily underrepresented. Indeed, we found a profound divergence in coverage between different taxa (Table 1). Whereas for example at least one ITS2 sequence is known for >40% of all fungal species listed in the NCBIs taxonomy, <4% of the Metazoa are covered. A reason might be that Metazoa tend to have sufficient visible morphological markers. Still, there might be primarily historical reasons, as for example flowering plants do have a higher coverage. To become a general tool for phylogenetics, a more evenly distributed sampling would be needed.
TABLE 1.
ITS2 sampling
| Taxon | Covered species | Species coverage (%) | Total ITS2 | Consensus fold | UGGU Motif | U-U Mismatch | UGGU and U-U |
| Rhodophyta | 99 | 7.18 | 284 | 0 | 0 | 0 | 0 |
| -Florideophyceae | 96 | 7.60 | 246 | 0 | 0 | 0 | 0 |
| Viridiplantae | 16050 | 40.45 | 25944 | 3962 | 3447 | 3373 | 2993 |
| -Chlorophyta | 317 | 24.27 | 1132 | 90 | 18 | 39 | 16 |
| -Streptophyta | 15733 | 41.01 | 24815 | 3872 | 3429 | 3334 | 2977 |
| Fungi/Metazoa group | 9074 | 14.80 | 23971 | 1046 | 316 | 615 | 232 |
| -Metazoa | 1740 | 3.98 | 5419 | 53 | 44 | 30 | 24 |
| --Eumetazoa | 1714 | 3.95 | 5367 | 52 | 43 | 30 | 24 |
| Bilateria | 1632 | 3.83 | 4525 | 46 | 40 | 25 | 22 |
| Cnidaria | 68 | 9.12 | 825 | 3 | 0 | 3 | 0 |
| -Fungi | 7328 | 41.83 | 18538 | 993 | 272 | 585 | 208 |
| --Glomeromycota | 136 | 23.53 | 574 | 63 | 19 | 58 | 19 |
| --Zygomycota | 61 | 22.18 | 296 | 8 | 0 | 5 | 0 |
| --Ascomycota | 4740 | 49.81 | 11991 | 551 | 88 | 179 | 38 |
| --Basidiomycota | 2330 | 35.45 | 5544 | 360 | 161 | 334 | 147 |
| Alveolata | 301 | 19.12 | 892 | 60 | 11 | 49 | 9 |
| -Dinophyceae | 209 | 30.42 | 545 | 59 | 20 | 48 | 8 |
| -Apicomplexa | 20 | 4.52 | 121 | 1 | 1 | 1 | 1 |
| -Ciliophora | 60 | 15.38 | 161 | 0 | 0 | 0 | 0 |
| Stramenopiles | 458 | 31.94 | 1395 | 21 | 20 | 20 | 19 |
| -Phaeophyceae | 159 | 36.98 | 479 | 0 | 0 | 0 | 0 |
| -Oomycetes | 242 | 56.94 | 775 | 21 | 20 | 20 | 19 |
Only taxa with more than 100 ITS2 sequences are shown. Subgroups are indicated by “-” in the taxon column, i.e. Bilateria and Cnidaria are subgroups of the Eumetazoa. The species coverage was calculated by dividing the number of species with known ITS2 by the total number of species annotated in the NCBIs taxonomy. As the ITS2 of a single species frequently was sampled multiple times, the number of ITS2 in each group exceeds the number of covered species. Additional rows count the number of sequences with ITS2 hallmarks as defined within the text.
Conservation of structure
For an alignment of fast-evolving sequences like ITS1 and ITS2, prediction of the correct structure is absolutely necessary (Coleman and Mai 1997; Gottschling et al. 2001). In the case of ITS2, distinct hallmarks of a core structure have been described. These are (1) four helices with (2) helix III as the longest and (3) containing an UGGU motif 5′ to the apex (deviations like UGGGU, UGG, or GGU have been described) as well as (4) a U-U mismatch in the second helix. In the case of small-scale studies, results of folding programs are usually manually evaluated and optimized. For large-scale phylogenetics, the correct structure has to be found automatically. To assess whether current structure prediction programs are capable of finding this structure automatically, we folded all 54,000 identified ITS2 sequences using RNAfold (Hofacker et al. 1994). In a second step, we checked the predicted fold for the described hallmarks with the additional constraint of an unpaired starting nucleotide. In total, 5092 sequences folded into four helices with the third as longest [features (1) and (2)]. Of these, 3255 additionally showed the postulated features (3) and (4). To test whether these structural hallmarks could be generated by chance, we randomized all 54,000 ITS2 sequences, folded them and analyzed their structure. Here, not a single structure adopted four helices with the third as the longest. Therefore, the presence of the consensus structure is highly indicative for a conserved common core within all eukaryotes even though it was identified only for a minor fraction of all ITS2 sequences. There are different reasons why this structure was not found for more sequences. First, the chosen consensus structure was very conservative to avoid false positives. For example, the structures of Drosophila melanogaster and Saccharomyces cerevisiae ITS2 show additional helices (Joseph et al. 1999; Young and Coleman 2004) and within different Scenedesmus species, helix one is always branched (van Hannen et al. 2002; Hegewald and Wolf 2003). Second, the low abundance of the consensus structure could be caused by the fold prediction itself as we considered only the minimal energy structure. This might not represent the biological active form, which could be represented by additional suboptimal structures.
Analyzing the species coverage revealed that nearly every lineage contains members of the ITS2 with the correct structure, the UGGU motif, and the U-U mismatch (Table 1). The 5000 ITS2 sequences classified as the correct fold might be a valuable resource for further phylogenetic studies. All sequences as well as their predicted fold can be retrieved from http://www.biozentrum.uni-wuerzburg.de/bioinformatik/projects/ITS2.html.
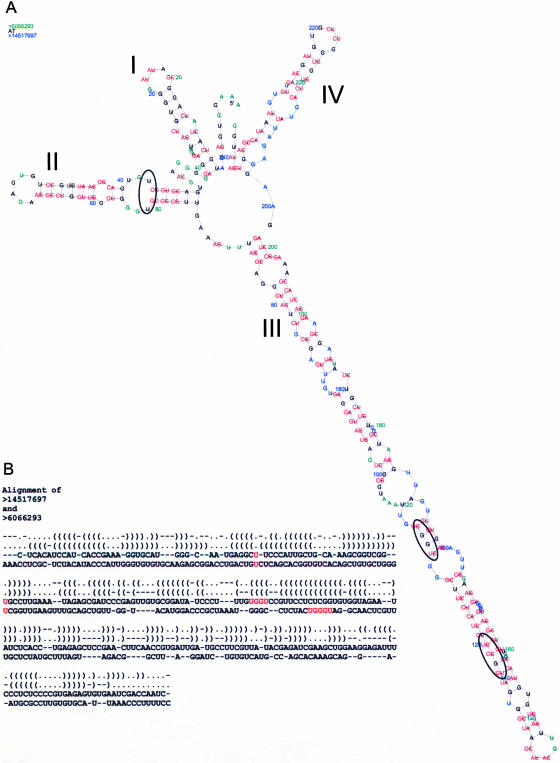
If the correct structure is given, an alignment of even extremely divergent sequences can be performed automatically using, for example, RNAforester (Höchsmann et al. 2003) or MARNA (Siebert and Backofen 2003). This was exemplified by the alignment of the ITS2 from Beroe ovata (Ctenophora, Metazoa), and Pandorina morum (Chlorophyta, Viridiplantae) (Fig. 1), accumulating additional evidence for the global conservation of the ITS2 structure.
FIGURE 1.
(A) All-in-one graphical visualization (2D-plot) of the aligned secondary structures from the ITS2 sequences of Beroe ovata (GenBank Identifier gi:14517697) and Pandorina morum (gi:6066293). Compensatory base changes are indicated in red; species-specific sites are in blue (Beroe ovata) and green (Pandorina morum). Black indicates identical nucleotides. Positions within the structures are numbered every 20 nucleotides. Helices are numbered I–IV. (B) An automatically generated global pairwise sequence alignment based on a “universally” conserved secondary structure of both sequences as obtained from RNAforester. In case of Beroe ovata a Y and a W had to be deleted before submission to RNAforester. Note the U-U mismatches as well as the UGGU respectively the UGGGU motifs highlighted in red.
Conclusion
Complementary to different small-scale manual analyses, we have shown in a large-scale automated approach, that the structure of ITS2 contains a conserved core throughout the eukaryotes. We found more than 5000 sequences with this core structure, which can be aligned automatically and might therefore be a valuable resource for further phylogenetic studies. A detailed breakdown of the taxonomy of all sequenced ITS2 revealed that, although sample ITS2s are known for most taxa, coverage is far from saturation. If it could be increased, ITS2 might indeed be a suitable marker not only for species and family level classification but also for megasystematics.
MATERIAL AND METHODS
Sequences
All entries containing the string “internal transcribed spacer 2” were retrieved from GenBank and subsequences annotated as ITS2 were extracted. These were folded using RNAfold of the Vienna package with standard parameters (Hofacker et al. 1994). A Perl script was used to identify the ITS2 hallmarks within the structures. Randomization of a sequence was performed by randomly selecting a nucleotide from the sequence n times (n = length of the sequence). Therefore, the length and the nucleotide distribution of the random sequences was identical to the ITS2 sequences.
Taxonomy
The phylogenetic information and the mapping to GenBank entries was downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy).
Acknowledgments
We thank Stefan Pinkert for providing a relational representation of the NCBIs taxonomy and Sven Rahmann for critical reading of the manuscript.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7204505.
REFERENCES
- Alvarez, I. and Wendel, J.F. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29: 417–434. [DOI] [PubMed] [Google Scholar]
- Coleman, A.W. 2003. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 19: 370–375. [DOI] [PubMed] [Google Scholar]
- Coleman, A.W. and Mai, J.C. 1997. Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J. Mol. Evol. 45: 168–177. [DOI] [PubMed] [Google Scholar]
- Gottschling, M., Hilger, H.H., Wolf, M., and Diane, N. 2001. Secondary structure of the ITS1 transcript and its application in a reconstruction of the phylogeny of Boraginales. Plant Biol. 3: 629–636. [Google Scholar]
- Hegewald, E. and Wolf, M. 2003. Phylogenetic relationships of Scenedesmus and Acutodesmus (Chlorophyta, Chlorophyceae) as inferred from 18S rDNA and ITS-2 sequence comparisons. Plant Syst. Evol. 241: 185–191. [Google Scholar]
- Höchsmann, M., Töller, T., Giegerich, R., and Kurtz, S. 2003. Local similarity in RNA secondary structure. In Proceedings of the IEEE Bioinformatics Conference 2003 (CSB 2003), pp. 159–168. IEEE Computer Society, Los Alamitos, CA. [PubMed]
- Hofacker, I.L., Fontana, W., Stadler, P.F., Bonhoeffer, L.S., Tacker, M., and Schuster, P. 1994. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 125: 167–188. [Google Scholar]
- Joseph, N., Krauskopf, E., Vera, M.I., and Michot, B. 1999. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res. 27: 4533–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai, J.C. and Coleman, A.W. 1997. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J. Mol. Evol. 44: 258–271. [DOI] [PubMed] [Google Scholar]
- Siebert, S. and Backofen, R. 2003. MARNA: A server for multiple alignment of RNAs. In Proceedings of the German Conference on Bioinformatics, pp. 135–140. Belleville Verlag Michael Farin, München, Deutschland.
- van Hannen, E.J., Fink, P., and Lurling, M. 2002. A revised secondary structure model for the internal transcribed spacer 2 of the green algae Scenedesmus and Desmodesmus and its implication for the phylogeny of these algae. Eur. J. Phycol. 37: 203–208. [Google Scholar]
- Young, I. and Coleman, A.W. 2004. The advantages of the ITS2 region of the nuclear rDNA cistron for analysis of phylogenetic relationships of insects: A Drosophila example. Mol. Phylogenet. Evol. 30: 236–242. [DOI] [PubMed] [Google Scholar]