Abstract
The recent identification of antisense RNA in the transcriptomes of many eukaryotes has generated enormous interest. The presence of antisense RNA in Plasmodium falciparum, the causative agent of severe malaria, remains controversial. Elucidation of the mechanism of antisense RNA in P. falciparum synthesis is critical in order to demonstrate the origin and function of these transcripts. Therefore, a systematic analysis of antisense and sense RNA synthesis was performed using direct labeling experiments. Nuclear run on experiments with single-stranded DNA probes demonstrated that antisense RNA is synthesized in the nucleus at several genomic loci. Antisense RNA synthesis is sensitive to the potent RNA polymerase II inhibitor α-amanitin. Antisense and sense transcription was also detected in nuclei isolated from synchronized parasites, suggesting concurrent synthesis. In summary, our experiments directly demonstrate that antisense RNA synthesis is a common transcriptional phenomenon in P. falciparum, and is catalyzed by RNA polymerase II.
Keywords: antisense RNA, double-stranded RNA, transcription, transcriptome, RNA polymerase II, malaria
INTRODUCTION
Antisense RNA was originally described in bacteria, where it modulates numerous processes through base-pairing including gene expression and plasmid replication (Carpousis 2003). The presence of antisense RNA in eukaryotic transcriptomes is a relatively recent discovery. Antisense RNA has been found in humans (Lehner et al. 2002; Shendure and Church 2002; Yelin et al. 2003; Rosok and Sioud 2004), mice (Okazaki et al. 2002), plants (Osato et al. 2003), and protozoan parasites (Elmendorf et al. 2001; Patankar et al. 2001; Martinez-Calvillo et al. 2003; Gunasekera et al. 2004; Monnerat et al. 2004). However, the precise role of anti-sense RNA in eukaryotes is not well defined.
Previously, serial analysis of gene expression (SAGE) was utilized to characterize the transcriptome of Plasmodium falciparum (Patankar et al. 2001; Gunasekera et al. 2004). This protozoan parasite is the causative agent of severe human malaria, and is responsible for 1–3 million deaths annually (Breman 2001). The SAGE and other experiments strongly suggested that antisense RNAs are quite common in the P. falciparum transcriptome, and constitute ~12% of the erythrocytic-stage steady-state RNA (Kyes et al. 2002; Gunasekera et al. 2004). However, the model of widespread antisense RNA in this organism has been controversial, as direct detection of most of these RNAs has not been reported (Patankar et al. 2001; Kyes et al. 2002; Gunasekera et al. 2004). Furthermore, the mechanism of antisense RNA synthesis is unknown. One model explaining the generation of antisense RNA in P. falciparum is that both strands of the P. falciparum genomic DNA are transcribed by DNA-dependent RNA polymerases to generate complementary sense and antisense transcripts. Alternatively, RNA-dependent RNA polymerases may synthesize antisense RNA as a part of an endogenous RNA interference pathway or another undefined process (Ahlquist 2002). Thus, in this study, direct RNA labeling experiments were utilized to test the model of widespread antisense RNA synthesis in P. falciparum and reveal the location and mechanism of their synthesis. This is the first study to directly analyze antisense RNA synthesis at numerous loci in the malaria parasite.
RESULTS AND DISCUSSION
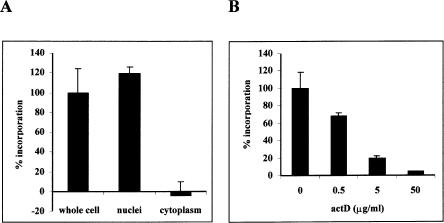
Initially, the location of RNA polymerase activity in parasite extracts was determined. Whole cell extracts from asynchronous, erythrocytic-stage parasites were prepared and tested in an RNA polymerase assay. Robust RNA polymerase activity was detected in whole cell extracts (Fig. 1A), but not in uninfected red blood cell extracts (data not shown). This RNA synthesis activity is the product of transcription, since [α-32P]UTP will not label RNA generated by nontranscriptional RNA processes such as polyadenylation and tRNA CCA addition. In order to determine the subcellular localization of this activity, extracts were further separated into both nuclear (insoluble) and cytoplasmic (soluble) fractions by low-speed centrifugation, and were tested for RNA polymerase activity. Cytoplasmic fractions contained no detectable RNA polymerase activity, whereas nuclear fractions contained robust (>99%) polymerase activity. These experiments demonstrate that most, if not all, transcription occurs in an insoluble compartment, which is most likely to be the nucleus.
FIGURE 1.
RNA polymerase activity in P. falciparum fractions. (A) RNA polymerase activity in parasite whole cell, nuclear, and cytoplasmic fractions. (B) Parasite nuclear fractions were incubated with different concentrations of actinomycin D for 15 min before the addition of labeling mix and the measurement of RNA polymerase activity. The amount of incorporation in a reaction without fraction was subtracted from each sample. The activity in the whole cell extract (A) or the fraction not subject to actinomycin D treatment (B) was designated 100% incorporation. Error bars represent one standard deviation. The error bar for activity in the fractions treated with 50 μg/mL actinomycin D in B is small, and is not visible.
In order to determine whether RNA synthesis is dependent on a DNA template, reactions containing nuclear fractions were incubated with actinomycin D, a known DNA-intercalating agent that inhibits transcription of DNA-dependent, but not RNA-dependent RNA polymerases (Reich et al. 1961; Baltimore et al. 1970; White and Wang 1990; Schiebel et al. 1993; Goodin et al. 1997). RNA synthesis in nuclear fractions was strongly inhibited by actinomycin D in a dose-dependent manner, and little RNA synthesis was observed in reactions containing high levels (50 μg/mL) of actinomycin D (Fig. 1B). Actinomycin D treatment also inhibited RNA synthesis in whole cell extracts (data not shown). These experiments implicate DNA-dependent RNA polymerases, and not RNA-dependent RNA polymerases, in transcriptional activity in P. falciparum.
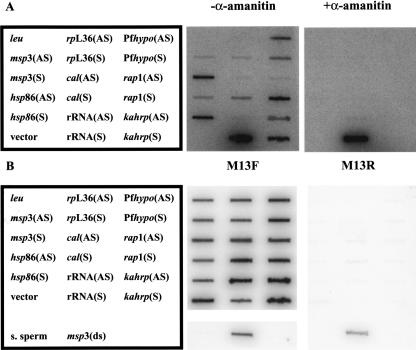
Since the majority of the RNA polymerase activity was found in the nuclear fraction, we hypothesized that both sense and antisense RNA are synthesized in this compartment. Therefore, nuclear run on experiments were used to detect synthesis of specific sense and antisense RNAs. Pulse-labeled RNA from mixed, erythrocytic-stage parasite nuclei was generated, isolated, and hybridized to filters containing M13-generated single-stranded DNA. After hybridization, the filters were treated with RNase A to ensure maximal specificity. The genes encoding merozoite surface protein 3 (msp3), calmodulin (cal), and ribosomal protein L36 (rpL36) were analyzed, as steady-state antisense RNAs were originally detected from these loci by SAGE (Patankar et al. 2001). The heat shock protein 86 (hsp86) locus was also analyzed, as steady-state antisense transcripts were not previously detected (Patankar et al. 2001). Transcription of a hypothetical gene (Pfhypo), rhoptry-associated protein 1 (rap1), and knob-associated histidine-rich protein (kahrp) genes were analyzed as examples of developmentally regulated sense transcripts. The A-type 18S ribosomal RNA (rRNA) locus was analyzed as an example of a non-protein-coding gene. Steady-state sense transcripts for these genes are found in asexual, erythrocytic-stage parasites (Bozdech et al. 2003; Le Roch et al. 2003).
As expected, synthesis of sense RNA from all loci was observed (Fig. 2A). Interestingly, antisense RNA synthesis was detected at the msp3, hsp86, cal, Pfhypo, rap1, and kahrp loci, but not typically the rpL36 locus. Antisense transcription at the hsp86 locus was unexpected, as stable antisense hsp86 transcripts were not found by SAGE (Patankar et al. 2001). The level of antisense 18S rRNA transcription was low, but detectable. There was no hybridization to the pCR2.1-topo vector and yeast leucine DNA, indicating specific hybridization. Since the single-stranded DNA targets were designed to differentiate between antisense and sense RNA transcripts, it was imperative to demonstrate that the single-stranded DNA was not contaminated with M13 replicative form (double-stranded) DNA. As expected, the M13 forward primer, but not the M13 reverse primer, was able to hybridize to the single-stranded DNA in a Southern blot (Fig. 2B). The single-stranded DNAs were >99.4% pure with respect to contamination with the complementary strand. Both primers were able to hybridize to a double-stranded msp3 PCR product generated with these primers, indicating these probes were functional. Thus, these experiments directly demonstrate that antisense RNA is synthesized in nuclear fractions, and is more common than originally expected.
FIGURE 2.
Synthesis of antisense and sense RNA in P. falciparum nuclei. (A) Nascent RNA in asynchronous, erythrocytic parasite nuclei was labeled with [α-32P]UTP in the absence and presence of α-amanitin and hybridized to filters containing single-stranded DNA. The DNAs were leu, yeast leucine biosynthetic gene; msp3, P. falciparum merozoite surface protein gene; hsp86, P. falciparum heat shock protein 86 gene; rpL36, P. falciparum ribosomal protein L36 gene; cal, P. falciparum calmodulin gene; rRNA, P. falciparum 18S rRNA gene; Pfhypo, P. falciparum hypothetical protein gene; rap1, P. falciparum rhoptry-associated protein gene; and kahrp, P. falciparum knob-associated histidine-rich protein gene. Filters were washed, treated with 10 μg/mL RNase A, and analyzed by phosphorimaging. pCR2.1-topo (vector) and a plasmid containing the yeast leucine leu gene served as negative controls for hybridization specificity. Slots labeled (AS) detect antisense transcripts and contain sense strand target DNA. Slots labeled (S) detect sense transcripts and contain antisense target DNA. (B) Single-stranded DNA was analyzed by Southern blotting with either a 5′ [32P] end-labeled M13 reverse or forward primer. S. sperm denotes salmon sperm DNA, and msp3(ds) denotes a double-stranded msp3 PCR product containing M13 reverse and forward binding sites. Filters were washed and analyzed by phosphorimaging. The M13 reverse primer is not complementary to the single-stranded DNA generated, and is not predicted to hybridize.
For most loci, there was more sense transcription than antisense transcription. This was clearly observed at the msp3 locus, where there was 60.5-fold more sense transcription than antisense transcription after correcting for thymidine bias and hybridization efficiency. However, at some loci, the amount of sense transcription was only slightly greater (kahrp; 4.6-fold) or virtually equivalent (Pfhypo; 1.7-fold) than the level of antisense transcription. Several control experiments indicated that hybridization intensity to specific probes was proportional to transcription. Dilution of the radiolabeled RNA probe proportionally reduced hybridization intensity to all targets except that detecting sense rRNA, indicating the hybridization signals are in the linear range (data not shown). The lengths of the probes used to detect antisense and sense RNA were identical for each gene. Also, the hybridization efficiency of double-stranded DNA to cognate antisense and sense targets only varied by threefold or less, indicating little to no strand hybridization bias (data not shown).
In order to determine the polymerase responsible for antisense and sense RNA synthesis, nuclear run on experiments were performed in the presence of α-amanitin. Typically, eukaryotic RNA polymerase II is extremely sensitive to α-amanitin, whereas RNA polymerase I and RNA-dependent RNA polymerases are not sensitive to this toxin (Lindell et al. 1970; Adman et al. 1972; White and Wang 1990; Schiebel et al. 1993; Goodin et al. 1997). α-Amanitin does inhibit some RNA polymerase III enzymes, but only at high concentrations (Lindell et al. 1970; Weil and Blatti 1975). Synthesis of sense 18S rRNA in P. falciparum nuclei is resistant to α-amanitin as previously demonstrated (Fig. 2A; Lanzer et al. 1992a,b). This observation is consistent with classic RNA polymerase I-mediated transcription of large ribosomal RNAs. Synthesis of both antisense and sense RNA from all protein-coding genes was sensitive to α-amanitin. This strongly suggests that both antisense and sense RNA synthesis at protein-coding loci is catalyzed by RNA polymerase II, consistent with an actinomycin D-sensitive RNA polymerase (Fig. 1B). These data are not consistent with RNA polymerase I or RNA-dependent RNA-polymerase-mediated synthesis of antisense RNA, although other α-amanitin-sensitive enzymes cannot be eliminated. RNA-dependent RNA polymerase activity in P. falciparum extracts was not detected using specific biochemical assays (White and Wang 1990), and no obvious RNA-dependent RNA polymerase homolog was found in the P. falciparum genome (data not shown). Therefore, our model is that RNA polymerase II synthesizes antisense and sense RNA at protein-coding loci in P. falciparum. Genes encoding the 12 subunits of eukaryotic RNA polymerase II have been identified in the P. falciparum genome (Coulson et al. 2004), suggesting the presence of a typical eukaryotic core enzyme.
Our data suggest that in P. falciparum nuclei, both the template and nontemplate strands of genomic DNA are transcribed by RNA polymerase II. These results provide an explanation for the high levels of steady-state antisense RNA found in asexual, erythrocytic parasites (Patankar et al. 2001; Gunasekera et al. 2004). Since many different loci have been shown to encode antisense RNA in P. falciparum (Patankar et al. 2001; Kyes et al. 2002; Gunasekera et al. 2004; this work), it is likely that antisense transcription of the P. falciparum genome is widespread. Indeed, widespread antisense transcription has been observed in humans (Lehner et al. 2002; Shendure and Church 2002; Yelin et al. 2003; Rosok and Sioud 2004), mice (Okazaki et al. 2002), plants (Osato et al. 2003), and protozoan parasites (Elmendorf et al. 2001; Patankar et al. 2001; Martinez-Calvillo et al. 2003; Gunasekera et al. 2004; Monnerat et al. 2004).
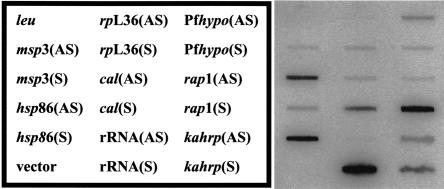
We favor a mechanism in which the chromatin context of each locus assumes a critical role in sense and antisense RNA generation (open chromatin model). Interestingly, adjacent sets of developmentally regulated genes on opposite strands have been identified in P. falciparum and suggest large regions of chromosomal DNA are simultaneously activated (Florens et al. 2002; Le Roch et al. 2003). Perhaps during activation of large or even small genomic regions, both the template and nontemplate strands are competent for transcription. It is therefore possible that the same RNA polymerase could be used for sense and antisense RNA synthesis. This prediction is consistent with the inhibition of sense and antisense RNA synthesis by α-amanitin (Fig. 2A). Thus, RNA polymerase II in P. falciparum would not have the absolute ability to distinguish between strands. This model suggests that sense and antisense transcription occurs either simultaneously, or at least in the same life-cycle stage. In support of the open chromatin model, sense and antisense transcription was detected in synchronized, schizont-stage parasites (Fig. 3). This suggests that antisense and sense transcription could occur simultaneously, and is not mutually exclusive. Consistent with this model, canonical promoter elements dictating transcriptional polarity have not been identified in P. falciparum to date. Homologs of genes encoding histone acetylases, histone deacetylases and chromatin remodeling enzymes have been found in the P. falciparum genome (Joshi et al. 1999; Coulson et al. 2004; Fan et al. 2004). Furthermore, the histone deacetylase inhibitor apicidin kills P. falciparum parasites (Darkin-Rattray et al. 1996), implicating a critical role for chromatin structure in this parasite. Our future efforts will aim to resolve questions regarding the initiation of antisense transcription in P. falciparum.
FIGURE 3.
Synthesis of antisense and sense RNA in P. falciparum nuclei from synchronous parasites. Nascent RNA in synchronous, schizont-stage erythrocytic parasite nuclei was labeled with [α-32P]UTP and hybridized to filters containing single-stranded DNA. Filters were washed, treated with 10 μg/mL RNase A, and analyzed by PhosphorImaging. The DNA targets were the same as described in the Figure 2 legend.
Interestingly, our experiments suggest that some, but not all antisense transcriptional events result in stable antisense RNA. For example, synthesis of both antisense msp3 and hsp86 RNA was detected in these experiments (Fig. 2A). However, antisense msp3 transcripts, but not antisense hsp86 transcripts, were detected in steady-state RNA (Patankar et al. 2001). Therefore, the abundance of steady-state antisense transcripts may be regulated at the level of RNA processing or stability. The mechanisms controlling processing and stability of antisense and sense RNAs have not been described in P. falciparum. Perhaps antisense RNAs that are not endowed with proper cleavage and polyadenylation signals are degraded. Alternatively, transcript-specific RNA-binding proteins may regulate processing and stability of antisense transcripts, and genes encoding numerous RNA-binding proteins have been identified in the P. falciparum genome (Coulson et al. 2004).
The function of the stable antisense transcripts in P. falciparum is currently unknown. The antisense loci analyzed in this report are not predicted to contain open reading frames as per the annotated genome sequence, and therefore it is not likely that the antisense RNAs encode proteins (Gardner et al. 2002). However, it is not possible to rule out a much higher gene density in P. falciparum than previously predicted. Therefore, what is the function of antisense RNA in P. falciparum? Antisense RNA may be a novel regulator of stage-specific gene expression in this parasite. The levels of most sense transcripts are regulated throughout the complex parasitic life cycle of P. falciparum (Bozdech et al. 2003; Le Roch et al. 2003). Based on precedence from other organisms, antisense RNA may regulate sense transcription by competition for transcription factors or modulating chromatin structure (Vanhee-Brossollet and Vaquero 1998; Carmichael 2003). Alternatively, antisense RNA in P. falciparum may base-pair with the sense RNA to regulate translation or RNA stability (Vanhee-Brossollet and Vaquero 1998; Carmichael 2003). Base-pairing interactions with the sense RNA could occur via a long antisense RNA, or a processed antisense RNA in the form of a microRNA or small interfering RNA (Eddy 2001; Carmichael 2003). The precedent for antisense RNA regulation exists in P. falciparum since episomally expressed antisense clag9 RNA has been shown to reduce clag9 RNA levels (Gardiner et al. 2000). Experimental RNA interference in P. falciparum has been reported (Kumar et al. 2002; Malhotra et al. 2002; McRobert and McConkey 2002; Mohmmed et al. 2003), although genes encoding the required RNAi machinery have not been found in the P. falciparum genome (Ullu et al. 2004). Thus, antisense RNA has the potential to regulate numerous processes in the malaria parasite, and its widespread distribution in nature suggests it plays a critical role in eukaryotic biology.
MATERIALS AND METHODS
P. falciparum culture
Asynchronous, erythrocytic, P. falciparum 3D7 parasites were cultivated as previously described (Trager and Jensen 1976). Parasites were synchronized with D-sorbitol using standard procedures (Lambros and Vanderberg 1979).
RNA polymerase activity in P. falciparum extracts
Parasites were released from red blood cells with saponin and resuspended in lysis/storage buffer (50 mM HEPES at pH 7.9, 50 mM NaCl, 1 mM EDTA at pH 8.0, 1.2 mM DTT, 10% glycerol, complete mini EDTA-free protease inhibitor cocktail tablets, Roche), homogenized six times with a size B Dounce homogenizer, and lysed by three freeze–thaw cycles. The extract was then separated into three sample groups. Whole cell extracts were not subjected to further centrifugation. Whole cell extracts were separated by low-speed centrifugation for 5 min at 1000g at 4°C to generate nuclear (insoluble) and cytoplasmic (soluble) fractions. The standard RNA polymerase assay was performed in 30 μL and contained 15 μg of fraction (roughly 6.25 × 107 cell equivalents), 50 mM HEPES (pH 7.9), 10 mM MgCl2, 1.2 mM DTT, 50 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 U/μL RNAsin, 1 mM ATP, 1 mM CTP, 1 mM GTP, and 0.5 μCi/μL [α-32P]UTP (NEN). Reactions were incubated at 37°C for 30 min and terminated by the addition of 5 mL of cold 5% tricholoracetic/100 mM disodium pyrophospate (TCA-NaPP) containing 100 μg of salmon sperm DNA. The reactions were mixed and incubated on ice for 10 min. Thereafter, samples were filtered through 25-mm Whatman GF/A filters that had been washed with 15 mL of cold TCA-NaPP using a Millipore 1225 sampling manifold. Filters were washed two times with 15 mL of cold TCA-NaPP, and then 5 mL of 95% ethanol. Filters were removed from the manifold and dried for 5 min in a 55°C oven. After drying, the filters were analyzed by liquid scintillation counting. For experiments measuring actinomycin D inhibition, actinomycin D was added to fractions and incubated for 15 min on ice before the addition of labeling mix.
Single-stranded DNA production
For single-stranded DNA production, 300–1100-bp regions of the leu, yeast leucine biosynthetic gene; msp3, P. falciparum merozoite surface protein gene (PF10_0345); hsp86, P. falciparum heat shock protein 86 gene (PF07_0029); rpL36, P. falciparum ribosomal protein L36 gene (PF11_0106); cal, P. falciparum calmodulin gene (PF14_0323); rRNA, P. falciparum 18S rRNA gene (chr5.rRNA1-18S-A); Pfhypo, P. falciparum hypothetical protein gene (PF1755c); rap1, P. falciparum rhoptry-associated protein gene (PF14_0102); and kahrp, P. falciparum knob-associated histidine-rich protein gene (PF0100c) were amplified from genomic DNA or cDNA using PCR. The numbers in parentheses represent the PlasmoDB locus (http://PlasmoDB.org). PCR products were inserted into the pCR2.1-topo vector (Invitrogen), and phagemids containing inserts in both orientations were identified by restriction mapping and DNA sequencing. Subsequently, phagemids were introduced into XL2-blue Escherichia coli (Stratagene), and single-stranded DNA was isolated using M13K07 helper phage using standard procedures (Sambrook and Russell 2001). Single-stranded DNA was confirmed by DNA sequencing. Single-stranded DNA was transferred to nylon filters in 6× SSC and fixed by UV cross-linking.
Synthesis of antisense and sense RNA in P. falciparum nuclei
Nascent RNA ~1 × 1010 infected asynchronous or synchronous erythrocytic parasite nuclei was labeled with [α-32P]UTP for 30 min at 37°C with intermittent mixing (Lanzer et al. 1992b). For experiments using α-amanitin, the toxin was incubated at 0.1 mg/mL with nuclear extracts for 15 min at 4°C prior to the addition of radioactive labeling mix (Lanzer et al. 1992a,b). Untreated nuclear fractions were also incubated for 15 min at 4°C. Reactions were processed as previously described (Farrell Jr. 1998). After purification, RNA was hybridized for 40–48 h to filters containing 1 μg of single-stranded DNA. Filters were washed, treated with 10 μg/mL RNase A, and analyzed by phosphorimaging. Levels of transcription were quantified using ImageQuant Software version 5.2 (Molecular Dynamics). Calculations of the ratio of sense to antisense transcription were corrected for small differences in hybridization efficiency and thymidine bias of each strand.
Southern blot analysis of single-stranded DNA
Single-stranded DNA was transferred to a nylon filter and analyzed by Southern blotting using the RapidHyb system according to the manufacturer’s protocol (Amersham). The probes were 5′ [32P] end-labeled M13 reverse or forward primers. Filters were washed and analyzed by phosphorimaging. Signals were quantified as described above.
Acknowledgments
We thank Johanna Daily, Lara Bethke, and Heather Sears for critical analysis of the manuscript. We also thank Swati Patankar, Sean Whelan, and Tim Pardee for critical discussions. We acknowledge Gilberto Ramirez for excellent technical support. The work presented in this manuscript was supported by NIH Postdoctoral Fellowship AI050303-01 (to K.T.M.), NIH grant GM61351-03 (to D.F.W.), and Exxon-Mobil (to D.F.W.).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7940705.
REFERENCES
- Adman, R., Schultz, L.D., and Hall, B.D. 1972. Transcription in yeast: Separation and properties of multiple RNA polymerases. Proc. Natl. Acad. Sci. 69: 1702–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296: 1270–1273. [DOI] [PubMed] [Google Scholar]
- Baltimore, D., Huang, A.S., and Stampfer, M. 1970. Ribonucleic acid synthesis of vesicular stomatitis virus, II. An RNA polymerase in the virion. Proc. Natl. Acad. Sci. 66: 572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech, Z., Llinas, M., Pulliam, B., Wong, E.D., Zhu, J., and DeRisi, J.L. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1: E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breman, J.G. 2001. The ears of the hippopotamus: Manifestations, determinants, and estimates of the malaria burden. Am. J. Trop. Med. Hyg. 64: 1–11. [DOI] [PubMed] [Google Scholar]
- Carmichael, G.G. 2003. Antisense starts making more sense. Nat. Biotechnol. 21: 371–372. [DOI] [PubMed] [Google Scholar]
- Carpousis, A.J. 2003. Degradation of targeted mRNAs in Escherichia coli: Regulation by a small antisense RNA. Genes & Dev. 17: 2351–2355. [DOI] [PubMed] [Google Scholar]
- Coulson, R.M., Hall, N., and Ouzounis, C.A. 2004. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 14: 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkin-Rattray, S.J., Gurnett, A.M., Myers, R.W., Dulski, P.M., Crumley, T.M., Allocco, J.J., Cannova, C., Meinke, P.T., Colletti, S.L., Bednarek, M.A., et al. 1996. Apicidin: A novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. 93: 13143–13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy, S.R. 2001. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2: 919–929. [DOI] [PubMed] [Google Scholar]
- Elmendorf, H.G., Singer, S.M., and Nash, T.E. 2001. The abundance of sterile transcripts in Giardia lamblia. Nucleic Acids Res. 29: 4674–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Q., An, L., and Cui, L. 2004. Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling. Eukaryot. Cell 3: 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell Jr., R.E. 1998. RNA methodologies. Academic Press, San Diego, CA.
- Florens, L., Washburn, M.P., Raine, J.D., Anthony, R.M., Grainger, M., Haynes, J.D., Moch, J.K., Muster, N., Sacci, J.B., Tabb, D.L., et al. 2002. A proteomic view of the Plasmodium falciparum life cycle. Nature 419: 520–526. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.L., Holt, D.C., Thomas, E.A., Kemp, D.J., and Trenholme, K.R. 2000. Inhibition of Plasmodium falciparum clag9 gene function by antisense RNA. Mol. Biochem. Parasitol. 110: 33–41. [DOI] [PubMed] [Google Scholar]
- Gardner, M.J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R.W., Carlton, J.M., Pain, A., Nelson, K.E., Bowman, S., et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin, M.M., Schlagnhaufer, B., Weir, T., and Romaine, C.P. 1997. Characterization of an RNA-dependent RNA polymerase activity associated with La France isometric virus. J. Virol. 71: 2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera, A.M., Patankar, S., Schug, J., Eisen, G., Kissinger, J., Roos, D., and Wirth, D.F. 2004. Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol. Biochem. Parasitol. 136: 35–42. [DOI] [PubMed] [Google Scholar]
- Joshi, M.B., Lin, D.T., Chiang, P.H., Goldman, N.D., Fujioka, H., Aikawa, M., and Syin, C. 1999. Molecular cloning and nuclear localization of a histone deacetylase homologue in Plasmodium falciparum. Mol. Biochem. Parasitol. 99: 11–19. [DOI] [PubMed] [Google Scholar]
- Kumar, R., Adams, B., Oldenburg, A., Musiyenko, A., and Barik, S. 2002. Characterisation and expression of a PP1 serine/threonine protein phosphatase (PfPP1) from the malaria parasite, Plasmodium falciparum: Demonstration of its essential role using RNA interference. Malar. J. 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes, S., Christodoulou, Z., Pinches, R., and Newbold, C. 2002. Stage-specific merozoite surface protein 2 antisense transcripts in Plasmodium falciparum. Mol. Biochem. Parasitol. 123: 79–83. [DOI] [PubMed] [Google Scholar]
- Lambros, C. and Vanderberg, J.P. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65: 418–420. [PubMed] [Google Scholar]
- Lanzer, M., de Bruin, D., and Ravetch, J.V. 1992a. A sequence element associated with the Plasmodium falciparum KAHRP gene is the site of developmentally regulated protein–DNA interactions. Nucleic Acids Res. 20: 3051–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1992b. Transcription mapping of a 100 kb locus of Plasmodium falciparum identifies an intergenic region in which transcription terminates and reinitiates. EMBO J. 11: 1949–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch, K.G., Zhou, Y., Blair, P.L., Grainger, M., Moch, J.K., Haynes, J.D., De La Vega, P., Holder, A.A., Batalov, S., Carucci, D.J., et al. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301: 1503–1508. [DOI] [PubMed] [Google Scholar]
- Lehner, B., Williams, G., Campbell, R.D., and Sanderson, C.M. 2002. Antisense transcripts in the human genome. Trends Genet. 18: 63–65. [DOI] [PubMed] [Google Scholar]
- Lindell, T.J., Weinberg, F., Morris, P.W., Roeder, R.G., and Rutter, W.J. 1970. Specific inhibition of nuclear RNA polymerase II by α-amanitin. Science 170: 447–449. [DOI] [PubMed] [Google Scholar]
- Malhotra, P., Dasaradhi, P.V., Kumar, A., Mohmmed, A., Agrawal, N., Bhatnagar, R.K., and Chauhan, V.S. 2002. Double-stranded RNA-mediated gene silencing of cysteine proteases (falcipain-1 and -2) of Plasmodium falciparum. Mol. Microbiol. 45: 1245–1254. [DOI] [PubMed] [Google Scholar]
- Martinez-Calvillo, S., Yan, S., Nguyen, D., Fox, M., Stuart, K., and Myler, P.J. 2003. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell 11: 1291–1299. [DOI] [PubMed] [Google Scholar]
- McRobert, L. and McConkey, G.A. 2002. RNA interference (RNAi) inhibits growth of Plasmodium falciparum. Mol. Biochem. Parasitol. 119: 273–278. [DOI] [PubMed] [Google Scholar]
- Mohmmed, A., Dasaradhi, P.V., Bhatnagar, R.K., Chauhan, V.S., and Malhotra, P. 2003. In vivo gene silencing in Plasmodium berghei—A mouse malaria model. Biochem. Biophys. Res. Commun. 309: 506–511. [DOI] [PubMed] [Google Scholar]
- Monnerat, S., Martinez-Calvillo, S., Worthey, E., Myler, P.J., Stuart, K.D., and Fasel, N. 2004. Genomic organization and gene expression in a chromosomal region of Leishmania major. Mol. Biochem. Parasitol. 134: 233–243. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573. [DOI] [PubMed] [Google Scholar]
- Osato, N., Yamada, H., Satoh, K., Ooka, H., Yamamoto, M., Suzuki, K., Kawai, J., Carninci, P., Ohtomo, Y., Murakami, K., et al. 2003. Antisense transcripts with rice full-length cDNAs. Genome Biol. 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patankar, S., Munasinghe, A., Shoaibi, A., Cummings, L.M., and Wirth, D.F. 2001. Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol. Biol. Cell 12: 3114–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, E., Franklin, R.M., Shatkin, A.J., and Tatum, E.L. 1961. Effect of actinomycin D on cellular nucleic acid synthesis and virus production. Science 134: 556–557. [DOI] [PubMed] [Google Scholar]
- Rosok, O. and Sioud, M. 2004. Systematic identification of sense–antisense transcripts in mammalian cells. Nat. Biotechnol. 22: 104–108. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual, 3d ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schiebel, W., Haas, B., Marinkovic, S., Klanner, A., and Sanger, H.L. 1993. RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J. Biol. Chem. 268: 11858–11867. [PubMed] [Google Scholar]
- Shendure, J. and Church, G.M. 2002. Computational discovery of sense–antisense transcription in the human and mouse genomes. Genome Biol. 3: research 0044.1–0044.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager, W. and Jensen, J.B. 1976. Human malaria parasites in continuous culture. Science 193: 673–675. [DOI] [PubMed] [Google Scholar]
- Ullu, E., Tschudi, C., and Chakraborty, T. 2004. RNA interference in protozoan parasites. Cell. Microbiol. 6: 509–519. [DOI] [PubMed] [Google Scholar]
- Vanhee-Brossollet, C. and Vaquero, C. 1998. Do natural antisense transcripts make sense in eukaryotes? Gene 211: 1–9. [DOI] [PubMed] [Google Scholar]
- Weil, P.A. and Blatti, S.P. 1975. Partial purification and properties of calf thymus deoxyribonucleic acid dependent RNA polymerase III. Biochemistry 14: 1636–1642. [DOI] [PubMed] [Google Scholar]
- White, T.C. and Wang, C.C. 1990. RNA dependent RNA polymerase activity associated with the double-stranded RNA virus of Giardia lamblia. Nucleic Acids Res. 18: 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin, R., Dahary, D., Sorek, R., Levanon, E.Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21: 379–386. [DOI] [PubMed] [Google Scholar]