Abstract
The DNA polymerases (gp43s) of the two related phages T4 and RB69 are DNA-binding proteins that also function as mRNA-binding autogenous translational repressors. As repressors, T4 gp43 is narrowly specific to its own mRNA whereas RB69 gp43 is equally effective against mRNA for either protein. We used in vitro RNase-sensitivity and RNA footprinting assays to identify features of the non-identical T4 and RB69 mRNA targets (translational operators) that allow for their identical binding affinities and biological responses to RB69 gp43. We observed that T4 gp43 and RB69 gp43 produce identical footprints on RNA substrates bearing the T4-derived operator, suggesting that the two gp43s make identical contacts with this operator. In contrast, the footprint produced by RB69 gp43 on its autogenous RNA target was shorter than its footprint on operator RNA from T4. As expected, we also observed only weak protection of RB69-derived operator RNA from RNase by T4 gp43; however, photocross-linking studies suggested that T4 gp43 recognizes structural features of the RB69-derived operator that are not detected by RNase- sensitivity assays. The results suggest that RB69 gp43 and T4 gp43 differ in their abilities to use RNA-sequence-independent interactions to configure potential RNA targets for translational repression.
INTRODUCTION
In the T4 family of bacteriophages, gene 43 is the structural gene for the replicative DNA polymerase, gp43 (1). Studies with T4 have shown that amounts of this enzyme in phage infected Escherichia coli hosts are subject to control at the levels of transcription (initiation and termination), post-transcriptional mRNA processing and mRNA translation (2,3). At the translational level, T4 gp43 binds and represses its own mRNA, thus regulating its own biosynthesis (4). The mRNA target, or translational operator, for the protein has been well characterized through a variety of physiological and biochemical studies (5 and references therein). It includes most or all nucleotide residues (∼35 nt) of the ribosome-binding site (RBS) and an additional ∼18-nt sequence to the immediate upstream (5′ side) of the RBS. A conspicuous feature of the upstream sequence is its propensity to exist as a structure, usually represented as an RNA hairpin, that determines specificity of the operator to the protein (5–7). The RBS portion of the operator contributes stabilizing interactions with the protein that are dependent on RNA length, but not dependent on the nucleotide sequence (5). It has been shown by in vitro assay that, together, sequence-specific (RNA hairpin) and non-specific (RNA 3′ to the hairpin) interactions between T4 gp43 and its operator contribute to a Kd value of 1–2 nM (5,7). Binding to hairpin alone, although specific, is much weaker [Kd ∼500 nM (5)]. Standard RNA footprinting assays detect the sequence-specific component (RNA hairpin) of the operator, but only a portion of the downstream RBS component (4,8).
A phylogenetic variant of T4 gp43, the enzyme from phage RB69 (9), is also an autogenous translational repressor (10), but its natural mRNA target is not as well characterized as the corresponding target for T4 gp43. Primary structure of the untranslated 5′ leader segment of RB69 gene 43-specific mRNA differs from that of the T4 counterpart (<50% nucleotide identity), although the two sequences appear to share higher-order structural features in common (10). In particular, the autogenous target for RB69 gp43 has been predicted to contain an RNA hairpin structure similar in configuration and core sequence to that of the T4 counterpart, but having a lower predicted stability and harboring some of the nucleotide determinants of the RBS [usually considered to span nucleotide positions –20 to +13 of the mRNA (11)]. We show here results of RNase-sensitivity assays that are consistent with these predictions.
We previously observed that RB69 gp43 can bind and repress gene 43-specific mRNA from either phage system, whereas T4 gp43 is narrowly specific to the natural mRNA target from T4 (10). In the study reported here, we used RNA footprinting assays to identify features of the gene 43 translational operators from the two phage systems that allow for their similar affinities to RB69 gp43 and different affinities to T4 gp43. We show by in vitro assay that T4 gp43 and RB69 gp43 produce identical footprints on RNA substrates carrying the T4 gene 43 translational operator sequence. This result may have been expected since we had previously observed that the two proteins bind this particular operator with similar affinities [Kd = 1–2 nM (10)]. In contrast, although RB69 gp43 binds its own natural target with the same high affinity as it does the translational operator from T4, its footprint on the RB69-derived RNA was observed to be ∼15% shorter than its footprint on the T4-derived RNA. T4 gp43 produced a very short, but specific, footprint on operator RNA from RB69. This footprint was centered in the hairpin structure where the nucleotide sequence is most similar to the RB69-derived RNA. Specificity of the interaction between T4 gp43 and RB69-derived operator RNA was also demonstrable by photocross-linking. The results suggest that T4 gp43 and the RB69 gp43 have similar specificities to RNA sequence, but differ in their capacities to utilize RNA-sequence-independent interactions to configure RNA for stable binding to the respective repressor.
MATERIALS AND METHODS
Preparation of RNA substrates for footprinting experiments
The RNA substrates used for RNase-sensitivity assays were prepared by in vitro transcription of designated T4 and RB69 gene 43 sequences that were cloned in the T7/SP6 expression plasmids pSP72 and pSP73 (Cat. Nos P2191 and P2221, respectively; Promega, WI). The recombinant plasmids used were pN1a33A, which encodes the wild-type T4 gene 43 translational operator sequence (12), and pCW12nd, which carries the corresponding DNA from phage RB69 (10). Figure 1 provides abbreviated physical maps of the DNA inserts in these recombinant plasmids. The plasmids were linearized by HindIII digestion and then used for in vitro transcriptions in the T7 ‘RiboMax’ system sold by Promega (Cat. No. P1300). Reaction mixtures (100 µl) contained 10 µg of DNA template, the four ribonucleoside triphosphates (ATP, CTP, UTP and GTP) at 3 mM each, and 180 U of T7 RNA polymerase. Transcription reactions were incubated for 2 h at 37°C and then quenched by the addition of 10 U of RQ1 RNase-free DNase (Cat. No. M6101; Promega) followed by an additional incubation of 15 min at 37°C. RNA products, purified through two phenol–chloroform (pH 4.5) extractions and one ethanol precipitation, were dissolved in 100 µl of nuclease-free 10 mM Tris–HC1 (pH 8.0) buffer and separated from small DNA and RNA fragments and nucleotides on ‘Quick Spin G-50’ columns (Cat. No. 100406; Boehringer Mannheim). Purity of the RNA products was evaluated by agarose gel electrophoresis in the presence of 10% urea, and RNA concentrations were determined spectrophotometrically. RNA stocks were stored at –80°C and used within 4–5 weeks of preparation.
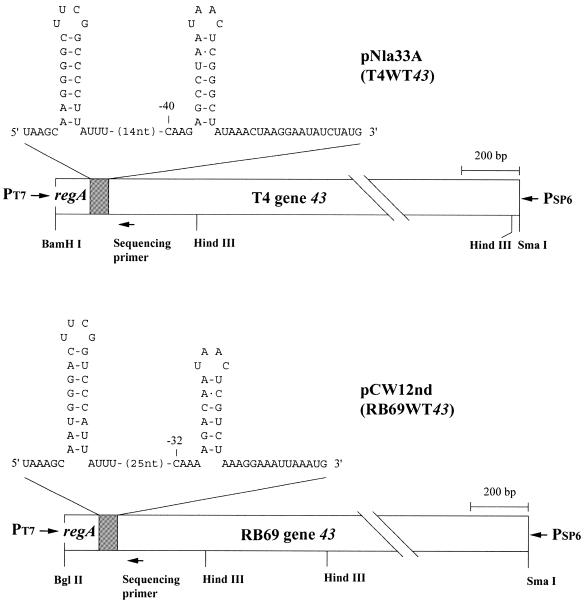
Figure 1.
The wild-type (WT43) T4 and RB69 DNA fragments used for in vitro transcription reactions. The DNA fragments had been cloned to allow expression under control of a plasmid-borne T7 Φ10 promoter in pSP73 (pNla33A; T4WT43) or pSP72 (pCW12nd; RB69WT43) as described in the Materials and Methods. The regA–43 intercistronic region is represented as the RNA copy. Drawings of the RNA hairpin structures shown are based on simulations by the ‘mfold 3.1’ program (24) and on previous studies (5,7,16) suggesting a four-base loop for the RNA hairpin of the T4 gene 43 translational operator.
RNase-sensitivity and RNA footprinting assays
For RNase-sensitivity assays, partial digestion of RNA substrates with RNases A and T1 (Cat. Nos E70194Y and E78021Y, respectively; Amersham Pharmacia Biotech) was carried out in 25 µl reaction mixtures containing 50 mM Tris–HC1 (pH 8.3), 60 mM NaCl, 5 mM DTT, 0.1 mg/ml of E.coli tRNA (Cat. No. 109550; Boehringer Mannheim) and 2.5 pmol of the RNA under examination. Incubations were carried out at 37°C for 3 min with either RNase A (used at 5 × 10–7 U per reaction) or RNase T1 (used at 0.1 U per reaction). Reactions were stopped by the addition of 25 µl of a solution containing 400 mM sodium acetate (pH 5.2) and 20 mM EDTA, and the mixtures were immediately extracted with a phenol–chloroform solution (equilibrated with sodium acetate, pH 4.5), precipitated with ethanol, pelleted by centrifugation and then dried and redissolved in 5 µl of nuclease-free TE buffer (10 mM Tris–HCl, 1 mM EDTA at pH 8.0). About 1 pmol RNA (2 µl sample) was annealed to 4 pmol of an appropriate primer (labeled with 32P at its 5′ end) for subsequent use in primer extension assays as described previously (3). Nucleotide sequences of the primers used for these assays were 5′-TTCCTTTCCATTTTCATCA-3′ (complementary to codons 20–26 of T4 gene 43 encoded mRNA) and 5′-TTCACGGCCATTAGAATCG-3′ (complementary to codons 20–26 of RB69 gene 43 encoded mRNA). Primer extension was catalyzed by AMV reverse transcriptase (Cat. No. M510A; Promega). Products of the reactions were examined by electrophoresis on 8% sequencing gels and visualized by the use of a phosphorimager (Fuji Model FLA-3000).
In RNA footprinting experiments, the RNA substrates were incubated for 15 min at room temperature with an appropriate amount of T4 gp43 or RB69 gp43 (Figs 2 and 3) before treating with RNases as described above for RNase-sensitivity assays. The gp43 preparations used in the footprinting experiments were purified as described earlier for RB69 gp43 (10).
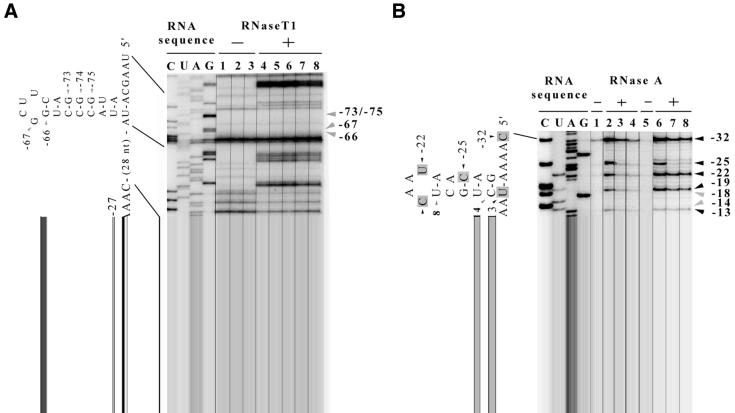
Figure 2.
RNase sensitivity and protection by gp43 (RNA footprinting) of the translational operator from RB69 gene 43. The analyses were carried out with RNA prepared by in vitro transcription of HindIII digested pCW12nd DNA (RB69WT43; Fig. 1). (A) The ‘RNA sequence’ lanes reproduce parts of an autoradiogram from an experiment in which the DNA sequence of the RB69 regA–43 intercistronic region was determined. Nucleotide sequence of the RNA is written to the left of the sequence lanes. The ‘– RNase T1’ lanes show products of cDNA synthesis from transcripts of pCW12nd, obtained with AMV-RT, in the absence of any gp43 (lane 1) or presence of RB69 gp43 (lane 2) or T4 gp43 (lane 3). The ‘– RNase’ incubations tested for possible RNase contamination in the gp43 preparations used. The ‘+ RNase T1’ lanes show the corresponding cDNA products obtained for the same in vitro transcripts following digestion of the RNA with RNase T1, in the absence of any gp43 (lane 4) or presence of RB69 gp43 [5 pmol (lane 5) and 10 pmol (lane 6)] or T4 gp43 [5 pmol (lane 7) and 10 pmol (lane 8)]. (B) Results of a footprint analysis for a portion of the RNA from the RB69 regA–43 intercistronic region. The ‘+ RNase A’ lanes show products of digestion of the RNA with RNase A in the absence of gp43 (lanes 2 and 6) or presence of RB69 gp43 [5 pmol (lane 3) and 10 pmol (lane 4)] or T4 gp43 [5 pmol (lane 7) and 10 pmol (lane 8)]. Lanes 1 and 5 of (B) show results of a similar analysis conducted without RNase digestion, but in the presence of RB69 gp43 (lane 1) or T4 gp43 (lane 5). The incubations for lanes 1 and 5 tested for RNase contamination in the gp43 preparations used. Nucleotide positions that were protected from RNase by RNA secondary structure are marked by gray arrows to the right of the autoradiogram. Nucleotide positions protected from RNase by gp43 are similarly marked, but by black arrows, and are shadowed on the RNA sequences shown to the left of the autoradiogram lanes. Nucleotide positions are numbered relative to the A of the initiator AUG. Experimental details are included in the Materials and Methods, and our interpretation of the results is summarized in Figure 5 (Discussion).
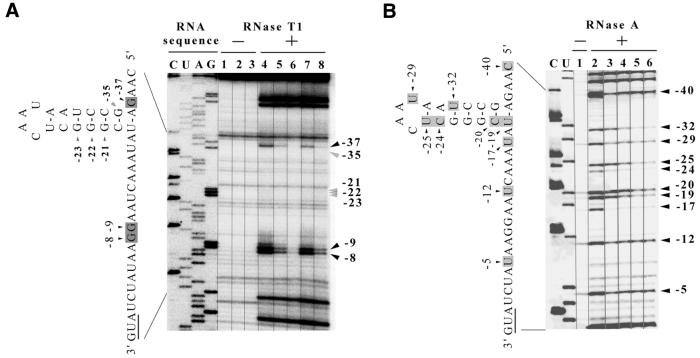
Figure 3.
RNA footprinting of the translational operator for T4 gene 43, using T4 gp43 and RB69 gp43. RNA corresponding to the T4 regA–43 intercistronic region was prepared by in vitro transcription of HindIII-digested pNla33A DNA (T4WT43; Fig. 1) and then analyzed for protection from RNases T1 and A by RB69 gp43 and T4 gp43, as described in the Materials and Methods. (A) Results of the footprinting analysis under conditions similar to those described for Figure 2A. (B) The ‘– RNase A’ lane (lane 1) shows products of cDNA synthesis from transcripts of pNla33A, obtained with AMV-RT, in the absence of any gp43. The ‘+ RNase A’ lanes show cDNA sequence patterns obtained following RNase A digestion in the absence of any gp43 (lane 2) or presence of RB69 gp43 [5 pmol (lane 3) and 10 pmol (lane 4)] or T4 gp43 [5 pmol (lane 5) and 10 pmol (lane 6)]. Nucleotide positions protected by RNA secondary structure are marked by gray arrows. Positions protected by gp43 are marked by black arrows to the right and shadowed on the RNA sequence to the left of the autoradiogram lanes. See text (Results) for comments regarding weakly and strongly protected positions in the RNA footprints. Nucleotide positions are numbered relative to the A of the initiator AUG. Other details are in the Materials and Methods and an interpretation of the results is summarized in Figure 5 (Discussion).
Conditions for gp43–RNA cross-linking with visible light (photocross-linking)
Previous studies of the gp43–operator interaction in T4 implicated the RNA hairpin component of the operator in determination of specificity of the operator to the protein (5,7,13). In the work reported here we sought to find out if the operator hairpin could be cross-linked to the cognate repressor. Towards this goal, we utilized a variety of conditions for UV-induced cross-linking and methylene blue-mediated photocross-linking with iodo-modified and unmodified RNA substrates, and with both RB69 gp43 and T4 gp43. With iodoU-substituted and unmodified RNA substrates, yields of cross-linked products did not exceed ∼1% under any condition of irradiation (results not shown). In contrast, high yields of protein–RNA cross-linked product (∼50% cross-linking) could be obtained with iodoC- substituted operator RNA substrates and exposure to visible light at high intensity in the presence of methylene blue. It is known that in the presence of this dye cross-linking is specific to base-paired regions of DNA and RNA (14). We used methylene blue in conjunction with iodoC-substituted RNA to distinguish the RNA specificities of T4 gp43 and RB69 gp43 through photocross-linking.
The RNA targets used for T4 gp43 and RB69 gp43 in the photocross-linking experiments described in this report were designed to encompass nucleotide positions –35 to +1 of the translation initiation region for RB69 gene 43 (Fig. 1). The wild-type DNA sequence for this region was cloned in the BglII–SmaI interval of pSP72 such that it could be transcribed from the T7 promoter of the recombinant plasmid. In addition, two C-to-U substitution mutants (for nucleotide positions –14 and –19, respectively) were constructed from the wild-type clone using the ‘Quick Change’® site-directed mutagenesis protocol and reagents (Cat. No. 200518; Stratagene). RNA was subsequently prepared by in vitro transcription of the three cloned DNA sequences as described above for RNase-sensitivity studies, but with 5-iodo CTP (Cat. No. 15628; Sigma) replacing CTP in the T7 RiboMax® system.
To effect gp43–RNA photocross-linking, we incubated the iodoC-substituted photoreactive RNA targets for 15 min at room temperature with either RB69 gp43 or T4 gp43 in 20 µl of a buffer containing 50 mM Tris–HCl (pH 8.3), 60 mM NaCl, 5 mM DTT in the presence of Methylene Blue (Cat. No. M9140; Sigma; used at 5 ng of dye per pmol RNA). These binding mixtures were transferred to the wells of a micro-titer plate that was placed on an ice surface and irradiated with a high-intensity visible light source (20 W, 1200 lumens lamp; EarthLight®, Philips) for 40 min. Subsequently, the irradiated solutions were made 7 M in urea, incubated at 42°C for 15 min and then analyzed by electrophoresis in 10% polyacrylamide gels containing 7 M urea. Resolved radioactive gel bands were visualized and quantitated by using the phosphorimager.
RESULTS
RNA secondary structure in the RB69 gene 43 translational operator
As suggested by previous studies (8,10), the RB69 gene 43 translational operator probably includes an RNA hairpin structure somewhat similar to that of the T4 homolog (4), but with lower predicted intrinsic stability and a different nucleotide sequence in the putative base-paired stem segment [(9); Fig. 1]. We tested for the existence of RNA secondary structure in the RB69-derived operator by comparing resistance of in vitro generated transcripts of the T4 and RB69 gene 43 translation initiation regions to RNases A and T1. Results of primer-extension analysis of partially hydrolyzed RNA samples from the analysis are shown in Figure 2 for RB69 and Figure 3 for T4. In the putative base-paired segment of the RNA target from RB69 (Fig. 2), we observed that the phosphodiester bonds between nucleotide positions –15(U) and –16(G) and between –26(A) and –27(G) are partially resistant to attack by RNase T1 (Fig. 2A, lane 4). Also, the bonds between nucleotide positions –13(U) and –14(C) and between –17(C) and –18(U) were resistant to RNase A (Fig. 2B, lane 2 or 6). These observations are consistent with computer-assisted analyses, which predict that these nucleotide positions of the RB69 gp43 operator engage in secondary structure formation, as depicted in Figure 1. Results of RNase-sensitivity assays that utilized an RNA target corresponding to the T4 translational operator (Fig. 3A, lane 4) were similar to results from previous studies with this type of RNA (4). We should also mention that our assays detected additional RNase-resistant regions in both the RB69 (Fig. 2A, positions –66, –73, –74, –75) and T4 derived RNA (not shown here, but see ref. 4). These regions correspond to the locations of predicted RNA hairpin structures that form to the downstream (3′ side) of the regA cistrons of the respective T4 and RB69 regA-43 intercistronic regions (Fig. 1). In T4, the hairpin structure near the regA gene has been implicated in transcription termination (3). The role of the corresponding hairpin in RB69 has not been determined.
The footprints of T4 gp43 and RB69 gp43 on the gene 43 translational operators from the two phage systems
Figures 2 and 3 also show results from RNA footprinting studies that were aimed at characterizing the broader RNA-binding specificity of RB69 gp43 as compared with that of T4 gp43 (10). Our main observations from the current study are summarized below.
(i) The footprints obtained with gp43 from the two phage sources on the T4-derived RNA were identical to each other, although the two proteins may differ in the degree to which they protect some bonds within their respective footprints. Results are shown in Figure 3A for experiments in which RNase T1 was used and Figure 3B for experiments in which RNase A was used. With both proteins, protection was strongest (>90% protection) near nucleotide position –17 and weakest (∼60% protection) near nucleotide position –5. At some nucleotide positions (especially near –19 and –20), protection appeared to be stronger with T4 gp43 (80–90% protection) than with RB69 gp43 (60–70% protection).
(ii) RB69 gp43 produced a shorter footprint on RNA corresponding to its natural autogenous target than on RNA corresponding to the natural target for T4 gp43. We note, however, that the RNA segments protected by RB69 gp43 on the two RNA targets were both positioned to the 5′ side of the initiation codons for the respective mRNAs. With RB69-derived RNA as target, the segment protected by RB69 gp43 encompassed nucleotide positions –3 to –32 relative to the AUG (Fig. 2A, lanes 5 and 6; Fig. 2B, lanes 3 and 4). With T4-derived RNA as target, the RB69 gp43 footprint included nucleotide positions –5 to –40 relative to the AUG (Fig. 3A, lanes 5 and 6; Fig. 3B, lanes 3 and 4). The Shine–Dalgarno sequences and RNA hairpin structures were protected by RB69 gp43 in both cases, suggesting that the RB69 protein represses its own translation by an identical autogenous control mechanism to that used by T4 gp43.
(iii) Although T4 gp43 binds RB69 operator RNA only poorly in vitro and does not repress RB69 gp43 synthesis in vivo (10), the T4 protein strongly protects the phosphodiester bond between nucleotide positions –24(A) and –25(C) and weakly protects the bonds between –18(U) and –19(C) and between –21(A) and –22(U) of the RB69 operator. The protected bonds are located within the highly conserved central 10-nt sequence of the two operator hairpins (Fig. 2B, lanes 7 and 8).
Binding of T4 gp43 to the RB69-derived operator RNA is specific
The observed footprint of T4 gp43 on RB69-derived operator RNA (Fig. 2B) and binding-affinity measurements for this cross-species protein–RNA interaction (5,10) suggest that, although weak, the interaction is specific to the nucleotide sequence that determines the hairpin structure of this RNA target. We utilized a protein–RNA photocross-linking approach to determine if residues of the RB69 operator hairpin that are not footprinted by T4 gp43 in RNase-sensitivity assays are nevertheless involved in this cross-species interaction. We show below that the base-paired cytosine residue at nucleotide position –14 of the RB69 operator hairpin can be photocross-linked to T4 gp43, as well as to RB69 gp43.
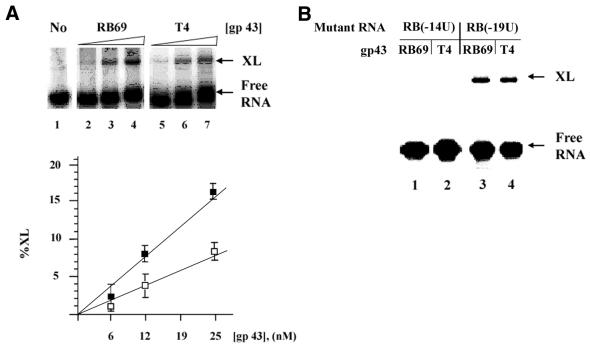
We tested the abilities of the C residues at nucleotide positions –14 (which is part of the hairpin stem) and –19 (which is part of the unpaired hairpin loop) of the RB69 operator to form cross-links with gp43 from T4 and RB69, respectively. We prepared iodoC-substituted RNA from clones of the wild-type RB69 gene 43 operator and two C-to-U mutants of this operator, one for C(–14) and one for C(–19). We subjected these RNAs to photocross-linking treatments as described in the Materials and Methods. As shown in Figure 4, and as expected from its lower affinity to the RB69 operator (5,10), T4 gp43 exhibited a lower yield of cross-linked products with the iodoC-substituted wild-type RB69-derived operator than did RB69 gp43 (Fig. 4A). The results in Figure 4B show that ability of the RB69-derived RNA target to form cross-links with either protein was abolished when C(–14) was substituted with U, i.e., with RNA containing U(–14) and iodoC(–19). The mutant RNA containing iodoC(–14) and U(–19) appeared to cross-link to protein normally. Since C(–14) is base paired whereas C(–19) is not in the RNA from RB69 (Figs 2 and 3), we interpret the photocross-linking results to mean that the base-paired C at nucleotide position –14 of the RB69 operator (as depicted in Fig. 1) contributes to recognition of this operator by either gp43.
Figure 4.
Photocross-linking of RB69-derived translational operator RNA to T4 gp43 and RB69 gp43, respectively. (A) Methylene blue-mediated photo-crosslinking of photoreactive RB69 operator RNA with RB69 gp43 and T4 gp43, respectively. Photocross-linking was carried out as described in the Materials and Methods by using iodoC-substituted RNA (25 nM) and 6, 12 or 25 nM RB69 gp43 or T4 gp43. XL, cross-linked products. Results of three independent reactions with T4 gp43 (empty squares) and RB69 gp43 (filled squares) are also presented graphically. (B) Results of photocross-linking of C-to-U mutants of the RB69 operator with gp43 from the two phage sources. The iodoC-substituted RNA substrates (used at 100 nM) from the mutants were treated for photocross-linking with T4 or RB69 gp43 (used at 400 nM) and the products of treatments were analyzed as described in the Materials and Methods.
DISCUSSION
Figure 5 summarizes the RNA footprinting results from the current study and presents a two-dimensional structure for the RNA hairpin component of the RB69 gene 43 translational operator that closely resembles the corresponding nucleotide-sequence-specific component of the T4 gene 43 translational operator. In both cases, existence of some base pairing in the mRNA targets for gp43 can be inferred from RNase- sensitivity assays and computer-assisted predictions (Figs 2 and 3) (4,8). It should be emphasized, however, that these types of analyses do not take into account the role of protein-induced RNA conformations that shape the final structure of a repressed mRNA target (15). Evidence that favors our interpretation of the structural framework depicted for the gp43-bound T4 operator hairpin shown in Figure 5 has been derived from a variety of studies. These include mutational analyses (5,8,12), in vitro selection of RNA-binding ligands to the protein (SELEX; 7), NMR studies with RNA targets from natural and SELEX generated sources (13,16), and the photocross-linking studies described here (Fig. 4). Based on our current studies, it appears that operator RNA from T4 achieves the same structure with either the T4 or RB69 gp43 as repressor, since the two proteins produced identical footprints on this RNA; however, the two paths leading to this structure may or may not be identical to each other. Possibly, the two proteins overlap in their specificity to a shared nucleotide-sequence-dependent feature of the hairpin, but differ in their abilities to drive potential RNA targets bearing this feature to the final RNA structure in a repressed complex. In this context, intrinsic structure of the natural target for T4 gp43 may represent one of the intermediate structures that are recognized by RB69 gp43 when the RB69 operator site is converted to the repressed state. Unlike RB69 gp43, T4 gp43 may require the presence of the pre-formed intermediate in order to establish translational repression with a potential target. This explanation is consistent with the observed in vitro binding affinities and in vivo repressor activities of T4 gp43 and RB69 gp43 on operator RNA variants (summarized in 5). We note in particular that the proposed RNA hairpins for the T4 and RB69 gene 43 operators bear nearly identical nucleotide sequences for the central 10 nt of the RNA structure, including the 5′-AC-3′ in the unpaired loop. This 2-nt sequence is invariant among T4 gp43 repressible RNA targets (5,7). The only difference between the 10-nt sequences of the T4 and RB69 derived RNAs is a U (at position –32) in T4 for a C (at position –25) in RB69. Interestingly, this nucleotide position in the RB69 target is footprinted by the T4 protein (Fig. 2). So, at least for this portion of the translational operator, purine versus pyrimidine positioning is identical and nucleotide sequence is nearly identical for the two phage systems. Where the two operators diverge more widely from each other is in the segment that contributes nucleotide-sequence-independent interactions with the repressor, i.e., the RNA length 3′ to the hairpin structure. Such interactions must therefore play an important role in configuring the mRNA target for repression by a gp43. Given a certain core sequence in a potential RNA target, RB69 gp43 may have a greater capacity than T4 gp43 to utilize sequence-independent interactions to produce a repressible configuration from the target. Likewise, the specific RNA induces conformational changes in its repressor (17), which may mean that the RNA target also plays an active role in determining its own structural destiny in the gp43– operator complex.
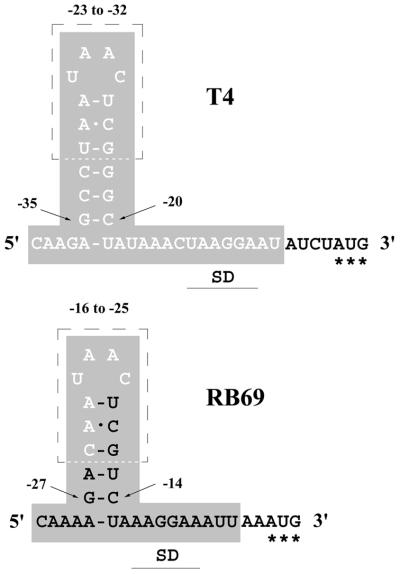
Figure 5.
Summary of the RNase-sensitivity and RNA footprinting analyses described in Figures 2 and 3, and in the text. The chart depicts the nucleotide sequences of the RNA targets used for T4 gp43 and RB69 gp43 in our studies. The Shine–Dalgarno (SD) sequence complementarities and initiator AUGs (***) are marked. Note that the boxed 10-nt sequences are nearly the same for the two translational operators studied here. The shaded sequences mark segments of the two RNA targets that were protected from RNase attack by either gp43. T4 gp43 and RB69 gp43 protect identical segments of the T4 gene 43 translational operator (upper panel). With RB69-derived operator RNA, the T4 gp43 footprint is limited to the ∼7-nt segment of the RNA sequence marked by hollow letters. Also, note that RB69 gp43 protects a shorter segment of RNA when its natural target is used than with operator RNA from T4.
The overlap we observe between the determinants for initiation and repression of translation in the T4 and RB69 orthologs of gene 43 may reflect an evolutionary selection for close linkage between the mRNA target for gp43 and the structural gene for this replication protein. In T4, the RNA hairpin component of the translational operator (nucleotide positions approximately –20 to –40 from the AUG) lies largely outside the RBS component [nucleotide positions –20 through +13 relative to the AUG (11)]. By comparison, in RB69, the hairpin includes RBS determinants (i.e., nucleotide positions –13 to –20; Fig. 1). Interestingly, in both T4 and RB69, operator constitutive phage mutants in which the hairpin is disrupted are viable and appear to replicate normally in laboratory strains of E.coli. That is, translational repression of gp43 biosynthesis does not seem to be essential for phage replication. Thus, it is not surprising that the sequence of the RNA hairpin component of the translational operator had undergone divergence between T4 and RB69 during evolution. Even greater divergence appears to have been allowed in the nucleotide determinants of the operator that are not sequence specific. However, despite this divergence, the ability to bind specific RNA has been conserved in both T4 gp43 and RB69 gp43. That the RNA-binding functions of the two proteins share common origins is indicated by the strong cross-reactivity we observe between RB69 gp43 and the T4-derived operator, which co-evolved with another repressor (T4 gp43) that exhibits poor cross-reactivity with the RNA from RB69.
The selective pressures that helped to maintain mutual recognition between each gp43 and its translational operator during evolution may have been based in overlaps between some of the protein’s RNA recognition determinants and its determinants for other, perhaps more essential, functions, e.g., the ability to serve as DNA replication enzymes. That is, despite transient losses in translational regulation that may have occurred through operator inactivating mutations in an evolving gene 43, there may have been subsequent returns to regulation through compensatory mutations in either partner of the RNA–protein interaction, but particularly in the divergence-prone mRNA target. This scenario contrasts with another example of translational repression by an RNA-sequence-specific translational repressor that also exhibits an essential DNA-binding function (18,19). The single-strand-binding (Ssb) protein gp5 of several M13-like phages (Ff phage family) represses translation of the mRNA for gp2 (an endonuclease), which is also essential for phage DNA replication. A natural variant of the M13 phage family, phage IKe, encodes a gp5 that does not serve as a translational repressor of gp2 synthesis while still serving its essential Ssb role in replication (20). With regard to gp43, it remains to be seen if T4-like phages exist in nature where DNA polymerase biosynthesis is not autogenously regulated at the translational level, and if the gp43 variants of these phages lack specific RNA-binding properties altogether or use specific RNA for functions other than translational repression. It also remains to be seen if the gp43-like DNA polymerases of other organisms, e.g., the archaea (21–23), contain gp43-like RNA-binding activities.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr James Nolan for helpful comments and suggestions on the manuscript and Dr Andrey Pavlov for many discussions. We also thank Jill Barbay for help with manuscript preparation. This work was supported by grant GM54627 from the NIGMS.
REFERENCES
- 1.Karam J.D. and Konigsberg,W.H. (2000) DNA polymerase of the T4 related bacteriophages. In Moldave,K. (ed.), Progress in Nucleic Acids Research and Molecular Biology. Academic Press, San Diego, Vol. 64, pp. 65–96. [DOI] [PubMed]
- 2.Miller E.S., Karam,J.D. and Spicer,E. (1994) Control of translation initiation: mRNA structure and protein repressors. In Karam,J.D., Drake,J.W., Kreuzer,K.N., Mosig,G., Hall,D.H., Eiserling,F.A., Black,L.W., Spicer,E.K., Kutter,E., Carlson,K. and Miller,E.S. (eds), Molecular Biology of Bacteriophage T4. ASM Press, Washington, DC, pp. 193–208.
- 3.Hsu T. and Karam,J.D. (1990) Transcriptional mapping of a DNA replication gene cluster in bacteriophage T4. Sites for initiation, termination, and mRNA processing. J. Biol. Chem., 265, 5303–5316. [PubMed] [Google Scholar]
- 4.Andrake M., Guild,N., Hsu,T., Gold,L., Tuerk,C. and Karam,J. (1988) DNA polymerase of bacteriophage T4 is an autogenous translational repressor. Proc. Natl Acad. Sci. USA, 85, 7942–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavlov A.R. and Karam,J.D. (2000) Nucleotide-sequence-specific and nonspecific interactions of T4 DNA polymerase with its own mRNA. Nucleic Acids Res., 28, 4657–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrake M. (1991) Translational repression of bacteriophage T4 DNA polymerase biosynthesis. PhD Thesis, Medical University of South Carolina, Charleston.
- 7.Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 8.Tuerk C., Eddy,S., Parma,D. and Gold,L. (1990) Autogenous translational operator recognized by bacteriophage T4 DNA polymerase. J. Mol. Biol., 213, 749–761. [DOI] [PubMed] [Google Scholar]
- 9.Wang C.C., Yeh,L.S. and Karam,J.D. (1995) Modular organization of T4 DNA polymerase. Evidence from phylogenetics. J. Biol. Chem., 270, 26558–26564. [DOI] [PubMed] [Google Scholar]
- 10.Wang C.C., Pavlov,A. and Karam,J.D. (1997) Evolution of RNA-binding specificity in T4 DNA polymerase. J. Biol. Chem., 272, 17703–17710. [DOI] [PubMed] [Google Scholar]
- 11.Gold L. (1988) Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem., 57, 199–233. [DOI] [PubMed] [Google Scholar]
- 12.Andrake M.D. and Karam,J.D. (1991) Mutational analysis of the mRNA operator for T4 DNA polymerase. Genetics, 128, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirmira S.R. and Tinoco,I.,Jr (1996) A quadruple mutant T4 RNA hairpin with the same structure as the wild-type translational repressor. Biochemistry, 35, 7675–7683. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z.R., Wilkie,A.M., Clemens,M.J. and Smith,C.W. (1996) Detection of double-stranded RNA-protein interactions by methylene blue-mediated photo-crosslinking. RNA, 2, 611–621. [PMC free article] [PubMed] [Google Scholar]
- 15.Williamson J.R. (2000) Induced fit in RNA-protein recognition. Nature Struct. Biol., 7, 834–837. [DOI] [PubMed] [Google Scholar]
- 16.Mirmira S.R. and Tinoco,I.,Jr (1996) NMR structure of a bacteriophage T4 RNA hairpin involved in translational repression. Biochemistry, 35, 7664–7674. [DOI] [PubMed] [Google Scholar]
- 17.Petrov V.M., Ng,S.-s. and Karam,J.D. (2002) Protein determinants of RNA binding by DNA polymerase of the T4-related bacteriophage RB69. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 18.Fulford W. and Model,P. (1988) Bacteriophage f1 DNA replication genes. II. The roles of gene V protein and gene II protein in complementary strand synthesis. J. Mol. Biol., 203, 39–48. [DOI] [PubMed] [Google Scholar]
- 19.Michel B. and Zinder,N.D. (1989) Translational repression in bacteriophage f1: characterization of the gene V protein target on the gene II mRNA. Proc. Natl Acad. Sci. USA, 86, 4002–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaman G.J., Kaan,A.M., Schoenmakers,J.G. and Konings,R.N. (1992) Gene V protein-mediated translational regulation of the synthesis of gene II protein of the filamentous bacteriophage M13: a dispensable function of the filamentous-phage genome. J. Bacteriol., 174, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopfner K.P., Eichinger,A., Engh,R.A., Laue,F., Ankenbauer,W., Huber,R. and Angerer,B. (1999) Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl Acad. Sci. USA, 96, 3600–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez A.C., Park,H.W., Mao,C. and Beese,L.S. (2000) Crystal structure of a pol alpha family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9 degrees N-7. J. Mol. Biol., 299, 447–462. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y., Jeruzalmi,D., Moarefi,I., Leighton,L., Lasken,R. and Kuriyan,J. (1999) Crystal structure of an archaebacterial DNA polymerase. Struct. Fold Des., 7, 1189–1199. [DOI] [PubMed] [Google Scholar]
- 24.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]