Abstract
Elongator has been reported to be a histone acetyltransferase complex involved in elongation of RNA polymerase II transcription. In Saccharomyces cerevisiae, mutations in any of the six Elongator protein subunit (ELP1–ELP6) genes or the three killer toxin insensitivity (KTI11–KTI13) genes cause similar pleiotropic phenotypes. By analyzing modified nucleosides in individual tRNA species, we show that the ELP1–ELP6 and KTI11–KTI13 genes are all required for an early step in synthesis of 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5) groups present on uridines at the wobble position in tRNA. Transfer RNA immunoprecipitation experiments showed that the Elp1 and Elp3 proteins specifically coprecipitate a tRNA susceptible to formation of an mcm5 side chain, indicating a direct role of Elongator in tRNA modification. The presence of mcm5U, ncm5U, or derivatives thereof at the wobble position is required for accurate and efficient translation, suggesting that the phenotypes of elp1–elp6 and kti11–kti13 mutants could be caused by a translational defect. Accordingly, a deletion of any ELP1–ELP6 or KTI11–KTI13 gene prevents an ochre suppressor tRNA that normally contains mcm5U from reading ochre stop codons.
Keywords: 5-methoxycarbonylmethyl-2-thiouridine, 5-methoxycarbonylmethyluridine, 5-carbamoylmethyluridine, Elongator complex, KTI genes
INTRODUCTION
In the RNA polymerase II transcription cycle, various accessory factors are needed for efficient and selective initiation and elongation of transcription on DNA templates. Some of these accessory factors, such as histone acetyltransferases, modify chromatin to make DNA accessible for transcription (Roth et al. 2001). The Saccharomyces cerevisiae Elongator complex, first described to include three proteins (Elp1, Elp2, and Elp3), was found to be associated with the hyperphosphorylated form of RNA polymerase II (Otero et al. 1999). Subsequently, Elp4, Elp5, and Elp6 proteins were identified to be part of the Elongator complex as a subcomplex (Krogan and Greenblatt 2001; Winkler et al. 2001). In vitro, the six-subunit Elongator complex has histone H3 and H4 acetyltransferase activity, and this activity was associated with the Elp3 protein in particular (Wittschieben et al. 1999; Winkler et al. 2002). Orthologs of Elp1–Elp4 can be found in humans as part of a six-subunit Elongator complex, which has in vitro histone H3 and H4 acetyltransferase activity (Winkler et al. 2001; Hawkes et al. 2002; Kim et al. 2002). Whether the remaining two components of human Elongator are orthologs of Elp5 and Elp6 proteins has not yet been determined (Hawkes et al. 2002).
Mutations in the human Elp1 ortholog IKAP (I kappa B kinase complex-associated protein) can result in the severe human hereditary disorder familial dysautonomia, a neurodegenerative disease (Anderson et al. 2001; Slaugenhaupt et al. 2001). Null alleles of the S. cerevisiae Elongator subunit genes cause pleiotropic phenotypes, including resistance to the Kluyveromyces lactis killer toxin (Otero et al. 1999; Frohloff et al. 2001; Jablonowski et al. 2001; Krogan and Greenblatt 2001). The three-subunit killer toxin secreted from K. lactis will irreversibly arrest sensitive S. cerevisiae strains in the G1 phase of the cell cycle (Butler et al. 1991). In addition to the elp1–elp6 mutants, a set of mutants has been identified to be killer toxin insensitive (KTI), including kti11, kti12, and kti13 (Frohloff et al. 2001; Fichtner and Schaffrath 2002). Mutations in any of the KTI11–KTI13 genes generate pleiotropic phenotypes similar to those caused by mutations in the ELP1–ELP6 genes (Frohloff et al. 2001; Jablonowski et al. 2001; Fichtner and Schaffrath 2002). Furthermore, KTI11 and KTI12 gene products physically interact with subunits of the Elongator (Fichtner et al. 2002, 2003). Unexpectedly, we have found that the six subunits in the Elongator complex and the Kti11–Kti13 proteins are all required for the biosynthesis of modified uridine nucleosides present at the wobble position in tRNA.
Transfer RNAs are adapter molecules, which decode mRNA into protein and thereby play a central role in gene expression. The primary tRNA transcript undergoes a series of processing and modification events in order to generate a mature tRNA (Hopper and Phizicky 2003). In this maturation process, specific subsets of the four normal nucleosides adenosine (A), guanosine (G), cytidine (C), and uridine (U) are modified. The modifications are introduced post-transcriptionally, and the formation of a modified nucleoside may require one or several enzymatic steps (Björk 1995). Of the 50 modified nucleosides so far identified in eukaryotic tRNAs, 25 are present in cytoplasmic tRNAs from S. cerevisiae (Sprinzl et al. 1998; Rozenski et al. 1999). In the anticodon region of tRNA, especially in positions 34 and 37, nucleosides are frequently modified. Their function seems to be primarily in the decoding process of mRNA, i.e., reading frame maintenance and/or restriction or improvement of codon–anticodon interactions (Agris 1991; Lim 1994; Björk 1995; Yokoyama and Nishimura 1995). They also act as identity elements in aminoacyl-tRNA synthetase recognition (Giege et al. 1998).
In this study, we show that nine proteins, Elp1 to Elp6 and Kti11 to Kti13, are required for the formation of 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5) groups on uridines at the wobble position in tRNA. Absence of these modifications influences decoding, providing a likely explanation for the pleiotropic phenotypes of elp and kti mutants.
RESULTS
The Elp3 protein homolog Sin3p influences modification of wobble uridines
In Schizosaccharomyces pombe, a sin3-193 mutant showed reduced levels of the modified nucleoside 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) (see Fig. 1 for structure; Heyer et al. 1984; Grossenbacher et al. 1986), which is located at the wobble position (base 34) in a subset of tRNAs. The sin3-193 mutation caused an antisuppressor phenotype, i.e., the ochre serine tRNA suppressor encoded by the sup3-18 gene was no longer able to suppress the ade7-413 ochre allele (Thuriaux et al. 1976). The cell volume, length, and amount of dead cells increased slightly in the sin3-193 mutant. This was independent of the presence or absence of the ochre tRNA suppressor, suggesting that the Sin3 protein affected cell cycle regulation (Heyer et al. 1984; Grossenbacher et al. 1986).
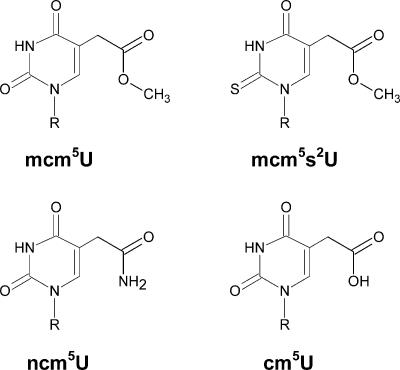
FIGURE 1.
Structure of 5-methoxycarbonylmethyluridine (mcm5U), 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), 5-carbamoyl-methyluridine (ncm5U), and 5-carboxymethyluridine (cm5U). R represents ribose.
We have identified the sin3+ gene as the uncharacterized open reading frame (ORF) SPAC29A4.20 and found that a null allele caused a lack of mcm5s2U in tRNAGlu mcm5s2UUC and 5-methoxycarbonylmethyluridine (mcm5U) (see Fig. 1 for structure) in the sup3-18-encoded ochre suppressor tRNASer (data not shown; see Materials and Methods). Lack of mcm5U at the wobble position in the suppressor tRNASer correlated with an inability to suppress the ade7-413 ochre allele (data not shown). The sin3+ gene encodes a highly conserved protein showing 77.0% identity on the amino acid level to the S. cerevisiae Elp3 protein, which has been extensively studied (Wittschieben et al. 1999, 2000; Frohloff et al. 2001; Jablonowski et al. 2001). Elp3p has in vitro histone H3 and H4 acetyltransferase activity and is a subunit of the Elongator complex, implicated in elongation of RNA polymerase II transcription (Otero et al. 1999; Wittschieben et al. 1999; Krogan and Greenblatt 2001; Winkler et al. 2001).
Deletion of the ELP3 gene influences ochre tRNATyr suppression
In S. cerevisiae, eight genes encode tRNATyr GφA, all of which can exist as suppressor derivatives. The SUP4 allele codes for a tRNATyr suppressor with a G34-to-U34 exchange in the anticodon, resulting in a tRNA reading UAA ochre stop codons. The wild-type sup4+ gene was replaced with the SUP4 allele in a strain containing the two ochre alleles ade2-1 and can1-100. The ADE2 gene encodes an enzyme participating in adenine biosynthesis, and suppression of the ade2-1 mutation allows the strain to grow on medium lacking adenine. The CAN1 gene encodes an arginine permease, and suppression of the can1-100 mutation results in no growth of the strain on medium lacking arginine and containing the toxic arginine analog canavanine (for suppression assay, see Materials and Methods). The SUP4 strain was able to grow on plates lacking adenine, but not on media lacking arginine and containing canavanine, showing suppression of the ade2-1 and can1-100 alleles (Fig. 2). Since unmodified uridines are very rare wobble nucleosides in cytosolic tRNAs (Sprinzl et al. 1998), it was of interest to identify the nucleoside present in the SUP4 encoded tRNA. We purified tRNATyr species from the wild type and SUP4 strains using an oligonucleotide coupled to magnetic beads. As the oligonucleotide was complementary to both tRNATyrGΨA and the suppressor derivative, only a fraction of the isolated tRNAs from the SUP4 strain represents the suppressor tRNA. The purified tRNAs were degraded to nucleosides and the composition analyzed by HPLC. We found that mcm5U was present in tRNATyr species from the SUP4 strain, whereas it was not detected in tRNATyr isolated from wild type (data not shown). Thus, the SUP4 encoded tRNATyr contains mcm5U (tRNATyr mcm5UΨA).
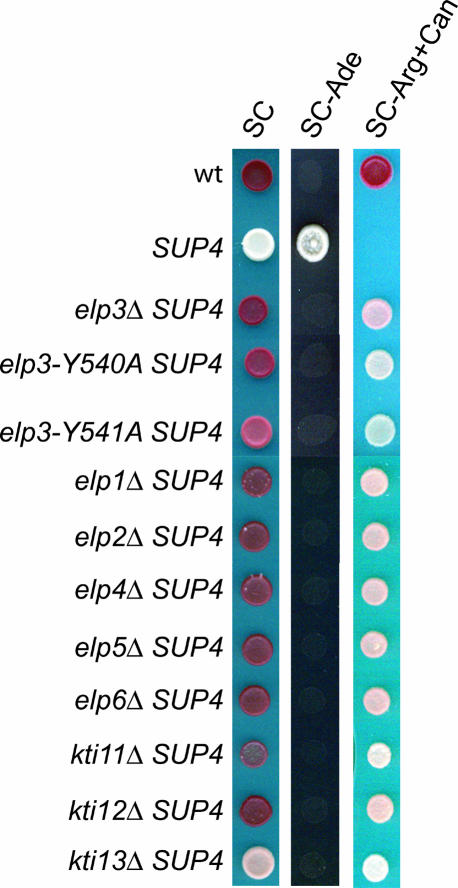
FIGURE 2.
Strains with elp and kti mutations confer a loss-of-suppression phenotype. Approximately 104 cells of wild type (W303-1B), SUP4 (UMY2893), SUP4 elp1-elp6 (UMY2916, UMY3039, UMY3040, UMY2912, UMY2914, UMY2918, UMY2920, UMY2922), and SUP4 kti11-kti13 (UMY2936, UMY2938, UMY2940) strains were spotted on SC, SC-Ade, or SC-Arg+Can plates and incubated for 3 d at 30°C.
As a sin3 mutation in S. pombe renders the sup3-18 encoded ochre tRNASer suppressor deficient in mcm5U and unable to decode ochre codons, we assumed that a deletion of the ELP3 gene would cause a loss-of-suppression phenotype in S. cerevisiae. Accordingly, introduction of an elp3::KanMX4 deletion into the SUP4 strain abolished the formation of mcm5U in the ochre suppressor tRNATyr and simultaneously nullified suppression of the ade2-1 and can1-100 ochre alleles (Fig. 2; data not shown). The elp3 deletion strain was transformed with a plasmid carrying the wild-type ELP3 gene, and the ability to suppress the ade2-1 and can1-100 alleles was restored (data not shown). Thus, lack of the mcm5 side chain at the wobble U in the sup3-18 and SUP4 encoded suppressor tRNAs prevents them from reading ochre stop codons.
An elp3-null mutant is deficient in formation of mcm5U, mcm5s2U, and ncm5U in tRNA
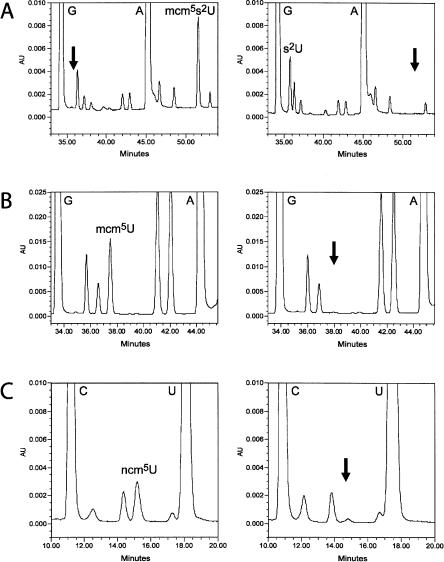
Loss of suppression in the sin3- and elp3-null mutants correlates with lack of mcm5U in the sup3-18 and SUP4 encoded suppressor tRNAs, respectively (data not shown). Furthermore, the sin3 mutant lacked mcm5s2U in tRNAGlu mcm5s2UUC (data not shown). In S. cerevisiae, mcm5U34 is present in tRNAArg mcm5UCU (Kuntzel et al. 1975), and mcm5s2 U34 is present in tRNAGlumcm5s2UUC and tRNALysmcm5s2UUU (Smith et al. 1973; Kobayashi et al. 1974). Another wobble nucleoside, 5-carbamoylmethyluridine (ncm5U34), present in tRNAProncm5UGG and tRNAValncm5UAC (Yamamoto et al. 1985; Keith et al. 1990), has a structure related to mcm5U (Fig. 1). We reasoned that an early step(s) is common in the synthesis of these modified nucleosides. Therefore, we determined whether the elp3 deletion affected the levels of mcm5s2U, mcm5U, and ncm5U in purified tRNAGlu mcm5s2UUC, tRNAArgmcm5UCU, and tRNAProncm5UGG, respectively. In the wild-type strain, but not in the elp3 mutant, mcm5s2U, mcm5U, and ncm5U nucleosides were present in tRNAGlumcm5s2UUC, tRNAArgmcm5UCU, and tRNAProncm5UGG (Table 1; Fig. 3). Introduction of a plasmid carrying the wild-type ELP3 gene into the elp3 mutant restored the ability to form these nucleosides (Table 1). No intermediates of mcm5U and ncm5U were observed in tRNAArg and tRNAPro from the elp3-null mutant. However, in the mutant an additional compound was detected in tRNAGlu (Table 1; Fig. 3). This compound was identified as 2-thiouridine (s2U), based on its spectrum, retention time, and comigration with synthetic s2U (data not shown). Thus, the elp3 deletion abolished the formation of the mcm5 or ncm5 side chains on the uridine, whereas the thiolation at position 2 was still present in tRNAGlu (tRNAGlus2UUC).
TABLE 1.
Content of modified nucleosides in tRNAGlumcm5s2UUC , tRNAArgmcm5UCU , and tRNAProncm5UGG isolated from elp1–elp6 and kti11–kti13 mutants
| tRNA species | Modified nucleoside | SUP4 | elp1Δa | elp2Δ | elp3Δ | elp3-Y540A | elp3-Y541A | elp3Δ/pELP3 | elp4Δ | elp5Δ | elp6Δ | kti11Δ | kti12Δ | kti13Δ |
| tRNAGlumcm5s2UUC | Ψ | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| m5C | 1.00 | 0.89 | 0.94 | 0.71 | 0.79 | 0.92 | 0.67 | 0.92 | 1.01 | 0.89 | 0.79 | 0.82 | 0.83 | |
| m5U | 1.00 | 0.99 | 1.01 | 0.95 | 0.96 | 1.00 | 0.81 | 1.00 | 1.00 | 0.94 | 0.89 | 0.99 | 0.96 | |
| s2U | −c | +d | + | + | + | + | − | + | + | + | + | + | + | |
| mcm5s2U | 1.00 | <0.09b | <0.07 | <0.02 | <0.05 | 0.13 | 1.22 | <0.07 | <0.03 | <0.04 | <0.05 | <0.06 | 0.18 | |
| tRNAArgmcm5UCU | Ψ | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| m1A | 1.00 | 0.93 | 0.90 | 1.24 | 1.20 | 1.17 | 1.12 | 0.89 | 1.14 | 1.04 | 1.03 | 0.85 | 0.85 | |
| m5U | 1.00 | 1.00 | 0.98 | 1.01 | 1.07 | 1.05 | 1.09 | 1.00 | 1.05 | 1.03 | 1.04 | 0.98 | 1.02 | |
| m1G | 1.00 | 1.04 | 1.08 | 1.07 | 0.88 | 0.96 | 0.79 | 1.04 | 0.94 | 0.98 | 0.90 | 1.10 | 1.04 | |
| m2G | 1.00 | 1.04 | 1.08 | 1.10 | 0.87 | 0.97 | 0.79 | 1.05 | 0.87 | 0.94 | 0.88 | 1.11 | 1.04 | |
| m22G | 1.00 | 1.05 | 1.09 | 1.09 | 0.88 | 0.97 | 0.79 | 1.06 | 0.88 | 0.95 | 0.89 | 1.11 | 1.05 | |
| t6A | 1.00 | 1.05 | 1.05 | 1.29 | 1.11 | 1.12 | 1.11 | 1.14 | 0.85 | 0.98 | 1.04 | 1.14 | 1.14 | |
| mcm5U | 1.00 | <0.05b | <0.04 | <0.01 | <0.04 | 0.12 | 0.78 | <0.05 | <0.04 | <0.04 | <0.05 | <0.05 | 0.15 | |
| tRNAProncm5UGG | Ψ | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| m1A+m5Ce | 1.00 | 1.01 | 0.98 | 0.96 | 0.96 | 0.91 | 1.07 | 0.99 | 0.99 | 1.00 | 1.04 | 0.98 | 1.05 | |
| Cm | 1.00 | 0.93 | 0.95 | 1.03 | 0.83 | 0.87 | 0.67 | 0.91 | 0.94 | 0.93 | 0.77 | 0.99 | 0.95 | |
| m5U+m7Gf | 1.00 | 1.09 | 1.03 | 1.02 | 1.01 | 0.94 | 1.10 | 1.03 | 1.05 | 1.04 | 1.15 | 1.01 | 1.07 | |
| m1G | 1.00 | 0.94 | 0.92 | 0.90 | 0.92 | 0.89 | 0.96 | 0.93 | 1.01 | 0.98 | 0.89 | 0.96 | 1.00 | |
| ncm5U | 1.00 | <0.03b | <0.05 | <0.03 | <0.04 | <0.03 | 0.60 | <0.04 | <0.04 | <0.04 | <0.04 | <0.04 | 0.16 |
Pseudouridine (Ψ), present three times in tRNAGlumcm5s2UUC , three times in tRNAArgmcm5UCU , and four times in tRNAProncm5UGG , was used as the internal standard. The numbers given are the ratios of the various modified nucleosides (modified nucleoside/Ψ) in respective tRNA isolated from the SUP4, SUP4 elp, or SUP4 kti mutants normalized to the ratio in the SUP4 strain. Nucleosides s2U, mcm5s2U, mcm5U and ncm5U are shown in bold.
aDeletion with a KanMX cassette.
bDetection limit.
cNo detection of s2U.
dPresence of s2U.
em1A comigrated with m5C.
fm5U comigrated with m7G.
Abbreviations: (Ψ) pseudouridine; (m5C) 5-methylcytidine; (m5U) 5-methyluridine; (s2C) 2-thiouridine; (mcm5s2U) 5-methoxycarbonylmethyl-2-thiouridine; (m1A) 1-methyladenosine; (m1G) 1-methylguanosine; (m2G) N2-methylguanosine; (m22G) N2, N2-dimethylguanosine; (t6A) N6-threonylcarbamoyladenosine; (mcm5U) 5-methoxycarbonylmethyluridine; (Cm) 2′-O-methylcytidine; (m7G) 7-methylguanosine; and (ncm5U) 5-carbamoylmethyluridine.
FIGURE 3.
An elp3-null mutant is lacking mcm5 and ncm5 side chains at wobble uridines. HPLC analysis of modified tRNA nucleosides from SUP4 (UMY2893, left panels) and SUP4 elp3::KanMX4 (UMY2916, right panels) strains. (A) Analysis of modified nucleosides in tRNAGlumcm5s2UUC. The part of the chromatogram between retention times 33 and 54 min is shown. Arrows indicate the expected retention time of s2U (left panel) and mcm5s2U (right panel). (B) Analysis of modified nucleosides in tRNAArgmcm5UCU. The part of the chromatogram between retention times 33 and 45.5 min is shown. The arrow indicates the expected retention time of mcm5U (right panel). (C) Analysis of modified nucleosides in tRNAPro ncm5UGG. The part of the chromatogram between retention times 10 and 20 min is shown. The arrow indicates the expected retention time of ncm5U (right panel). The small peak at this position represents an unrelated compound with a spectrum different from that of ncm5U.
The postulated acetyl-CoA binding domain of Elp3p is critical for mcm5U, mcm5s2U, and ncm5U formation
The Elp3 protein has histone H3 and H4 acetyltransferase activity in vitro (Wittschieben et al. 1999; Winkler et al. 2002). In the C-terminal part of the Elp3 protein there is a potential acetyl-CoA binding domain consisting of motifs D, A, and B (Wittschieben et al. 2000). Two tyrosines in the B motif have been suggested to be important for the histone acetyltransferase activity (Wittschieben et al. 2000). To examine if tyrosine Y540 and Y541 are required for tRNA modification activity in vivo, we replaced the wild-type allele of ELP3 with elp3-Y540A or elp3-Y541A. Such strains showed no suppression in the plate assay (Fig. 2). Replacement Y540A abolished the formation of mcm5s2U in tRNAGlumcm5s2UUC, mcm5U in tRNAArgmcm5UCU, and ncm5U in tRNAProncm5UGG (Table 1). However, small amounts of mcm5s2U and mcm5U were present in the elp3-Y541A mutant, whereas no ncm5U could be detected in tRNAPro ncm5UGG (Table 1). As in the case of the elp3-null allele, tRNAGlu isolated from the elp3-Y540A and elp3-Y541A mutants contained s2U (Table 1). We conclude that the Y540A and Y541A amino acid substitutions generate a loss-of-suppression phenotype and affect the formation of the mcm5 and ncm5 groups at U34.
All six components of the Elongator complex are required for the synthesis of mcm5s2U, mcm5U, and ncm5U
Because an elp3-null allele generated a loss-of-suppression phenotype and abolished the formation of mcm5s2U, mcm5U, and ncm5U, it seemed possible that mutations in the five other genes encoding components of the Elongator complex would cause similar phenotypes. Each of the ELP genes was independently deleted in the SUP4 ade2-1 can1-100 strain. All the elp deletion strains showed the same phenotypes as the elp3-null mutant, i.e., no suppression of the ade2-1 and can1-100 ochre alleles, and absence of mcm5U, mcm5s2U, and ncm5U in tRNAArgmcm5UCU, tRNAGlumcm5s2UUC, tRNAProncm5UGG, respectively (Fig. 2; Table 1). In all of these mutants, tRNAGlu contained s2U (Table 1), demonstrating that these genes affect the synthesis of the side chain at position 5, but that the thiolation at position 2 is independent. Each mutant was transformed with a plasmid containing the corresponding wild-type ELP gene, and in all cases the suppression was restored (data not shown). We conclude that all six Elongator subunits are required for formation of the mcm5 and ncm5 groups at U34 in tRNA.
The Elp1 and Elp3 proteins coimmunoprecipitate tRNAGluUUC
Two discrete subcomplexes consisting of Elp1–Elp3 and Elp4–Elp6 proteins form the Elongator (Winkler et al. 2001). To investigate whether Elongator binds tRNA, we constructed strains bearing genomically (HA)3-tagged versions of ELP1, ELP3, and ELP5. Extracts from these strains and a strain with no tag were incubated with 32P-labeled T7-transcribed tRNAGluUUC, which should be a substrate for formation of the mcm5 group at U34. Following UV-cross-linking, proteins were immunoprecipitated, and the presence of coimmunoprecipitated tRNA was investigated. The analysis revealed that Elp1 and Elp3 protein coimmunoprecipitated tRNAGluUUC, whereas the signal in the Elp5 immunoprecipitate was at background level (Fig. 4A). To assess the specificity of the interaction, we performed an experiment using a genomically tagged (HA)3 version of TIF4632 as the control. TIF4632 encodes one of two isoforms of translation initiation factor eIF4G and is not directly involved in tRNAMeti binding (Prevot et al. 2003). The signals from the Tif4632p and Elp5p immunoprecipitates were similar, i.e., background level, whereas the Elp1 and Elp3 proteins coimmunoprecipitated tRNAGluUUC (Fig. 4A,B). To exclude unspecific tRNA binding of the Elp1 and Elp3 proteins, we used as a control 32P-labeled T7-transcribed tRNAMeti, which has a C at position 34. No difference in tRNAMeti signal was observed in the four immunoprecipitates (Fig. 4B). These data suggest that the subcomplex consisting of Elp1–Elp3 proteins associates with tRNA that should obtain an mcm5 group at U34.
FIGURE 4.
Elp1p and Elp3p coimmunoprecipitate tRNAGluUUC. (A) T7-transcribed 32P-labeled tRNAGlu UUC was mixed with extracts from wild-type, ELP1-(HA)3, ELP3-(HA)3, or ELP5-(HA)3 strains. After UV-cross-linking, immunoprecipitation using agarose conjugated anti-HA antibodies was performed. The immunoprecipitated material was digested with proteinase K and analyzed on a denaturing polyacrylamide gel. The signals were quantified and normalized to the background signal in the control, which was set to 1. (B) T7-transcribed 32P-labeled tRNAGluUUC or tRNAMeti was mixed with extracts from TIF4632-(HA)3, ELP1-(HA)3, ELP3-(HA)3, or ELP5-(HA)3 strains. The experiment and quantifications were performed as described in A.
Lack of mcm5s2U, mcm5U, and ncm5U correlates with resistance to K. lactis killer toxin
Strains with null alleles in any of the ELP genes show resistance to the K. lactis killer toxin (Frohloff et al. 2001; Jablonowski et al. 2001). The killer toxin insensitive kti11–kti13 mutants displayed similar pleiotropic phenotypes as strains with mutations in the ELP genes (Frohloff et al. 2001; Fichtner and Schaffrath 2002). We confirmed the resistance of elp1–elp6 and kti11–kti13 mutants using the killer toxin eclipse assay (Kishida et al. 1996; data not shown). In this assay the elp3-Y540A and elp3-Y541A strains showed the same resistance as the elp3-null mutant (data not shown).
To investigate whether the Kti11–Kti13 proteins affected the synthesis of mcm5U, mcm5s2U, and ncm5U, we independently deleted the genes encoding these proteins in the SUP4 ade2-1 can1-100 strain. The kti11 and kti12 deletions prevented the suppression of the ade2-1 and can1-100 ochre alleles and the synthesis of mcm5U in tRNAArg mcm5UCU, mcm5s2U in tRNAGlu mcm5s2UUC, and ncm5U in tRNAProncm5UGG, (Fig. 2; Table 1). The kti13 deletion strain showed no suppression in the plate assay, but ~15% of mcm5U, mcm5s2U, and ncm5U was present in tRNAArgmcm5UCU, tRNAGlumcm5s2UUC, and tRNAProncm5UGG, respectively (Fig. 2; Table 1). In tRNAGlu from the kti11–kti13 mutants, s2U was detected (Table 1). Each mutant was transformed with a plasmid containing the corresponding wild-type KTI gene, which restored suppression (data not shown). We conclude that reduced levels of mcm5U, mcm5s2U, and ncm5U in tRNA correlate with resistance to K. lactis killer toxin.
DISCUSSION
The Elongator complex in yeast, consisting of Elp proteins 1–6, was suggested to participate in elongation of transcription (Otero et al. 1999; Krogan and Greenblatt 2001; Winkler et al. 2001). Mutations in any of the KTI11–KTI13 genes generate pleiotropic phenotypes similar to those caused by mutations in the ELP1–ELP6 genes, and Kti11 and Kti12 proteins interact with subunits of the Elongator complex (Frohloff et al. 2001; Jablonowski et al. 2001; Fichtner et al. 2002, 2003; Fichtner and Schaffrath 2002). Unexpectedly, we have found that ELP1–ELP6 and KTI11–KTI13 genes are required for the formation of the modified nucleosides mcm5U, mcm5s2U, and ncm5U (see Fig. 1 for structures). These modified nucleosides are located at position 34 (the wobble position) in a subset of tRNA species (Sprinzl et al. 1998). In all the elp- and kti-null mutants studied here, except kti13, formation of mcm or ncm side chains at position 5 of the uridine was abolished (Table 1; Fig. 3). A kti13-null allele significantly reduced the levels of mcm5U, mcm5s2U, and ncm5U, suggesting a regulatory role of Kti13p in formation of these nucleosides. In tRNAGlumcm5s2UUC, s2U is still present in the elp1–elp6 and kti11–kti13 mutants (Table 1; Fig. 3), suggesting that the thiolation at position 2 is unaffected and that the gene products affect the synthesis of either the first step or of an unstable intermediate(s) of the mcm5 and ncm5 side chains. Consistently, no intermediates of mcm5U and ncm5U were observed in tRNAArgmcm5s2UUC and tRNAProncm5UGG from the elp1–elp6 and kti11–kti13 mutants (Fig. 3; data not shown).
Biosynthesis of mcm5 and ncm5 side chains at the wobble position
Elongator consists of two subcomplexes, Elp1p–Elp3p and Elp4p–Elp6p (Winkler et al. 2001). We have shown that Elp1p and Elp3p coimmunoprecipitated in vitro transcribed tRNAGluUUC, which should be a substrate in the formation of the mcm5 group at U34. In contrast, there was no apparent coimmunoprecipitation of tRNAMeti that is not a substrate for formation of mcm5U or ncm5U derivatives (Fig. 4). These data indicate specificity of the Elp1–Elp3 subcomplex in tRNA binding and a direct involvement of Elongator in wobble uridine modification. Differential labeling experiments showed that both carbons of acetate were utilized with equal efficiency to form the 5-carboxymethyl (cm5) part of mcm5U (Tumaitis and Lane 1970). Based on structural similarities, ncm5U is likely to be a derivative of 5-carboxymethyluridine (cm5U) (Fig. 1). Mutations in the putative acetyl-CoA binding site of Elp3 (Y540A or Y541A) result in a reduced histone acetyltransferase activity in vitro (Wittschieben et al. 2000). These mutations abolished (Y540A) or severely reduced (Y541A) synthesis of the mcm5U, mcm5s2U, and ncm5U nucleosides in tRNA (Table 1), implying that acetyl-CoA could be a donor in the tRNA modification reaction. However, the labeling experiment showed that acetate or acetyl-CoA is not an immediate donor in formation of the cm5 group of mcm5U. Acetate had to be metabolized before the carbons are incorporated into the cm5 side chain and the actual donor is not known (Tumaitis and Lane 1970). In the tRNA modification reaction, Elongator components could be involved in generating the donor. In addition to the acetyl-CoA binding domain, the Elp3 protein shows homology to the catalytic domain of S-adenosylmethionine (SAM) radical enzymes (Chinenov 2002). Radical SAM enzymes contain a labile iron–sulfur cluster and require SAM to generate 5′-deoxyadenosyl radicals that can initiate an enzyme reaction by abstracting a hydrogen atom from the substrate (Layer et al. 2004). Thus, a radical mechanism could be involved in the formation of cm5U. To date, two iron–sulfur cluster containing RNA-modifying enzymes are known, MiaBp and RumAp (Agarwalla et al. 2002; Pierrel et al. 2002). Of these, the MiaB protein is a bifunctional radical SAM enzyme involved in formation of 2-methylthio-N6-isopentenyladenosine (ms2i6 A37) in tRNA by introducing the methylthio group at position 2 of the adenosine (Pierrel et al. 2004). In formation of cm5U in tRNA, the donor is not known and we have so far not been able to establish an in vitro assay.
The esterified methyl constituent of mcm5 originates from SAM and is transferred by the mcm5U/mcm5s2U tRNA carboxyl methyltransferase, Trm9p (Tumaitis and Lane 1970; Kalhor and Clarke 2003). No candidate gene product(s) has been identified for the final step(s) in formation of ncm5U. One of the Elp1–Elp6 or Kti11–Kti12 proteins could potentially catalyze this step. If so, the integrity of the Elongator complex and the associated Kti11–Kti12 proteins must be important as no nucleoside intermediates are observed in any mutant.
Dual function of the Elongator complex?
The Elongator complex was found to cofractionate with the hyperphosphorylated form of RNA polymerase II, suggested to be the elongating form of the polymerase (Otero et al. 1999). Elongator has histone acetyltransferase activity in vitro and was proposed to assist RNA polymerase II during transcription through chromatin (Wittschieben et al. 1999). Human Elongator showed a weak stimulation of transcription by human RNA polymerase II in vitro and human Elp1 and Elp3 associated with the pS2 gene promoter in chromatin immunoprecipitation (ChIP) experiments (Kim et al. 2002; Metivier et al. 2003). In contrast, the yeast Elongator did not stimulate elongation by RNA polymerase II in vitro and was not occupying open reading frames in ChIP experiments analyzed for specific genes or in a genome-wide manner (Krogan et al. 2002; Pokholok et al. 2002; Gilbert et al. 2004). The association of Elongator with RNA polymerase II has been questioned as no RNA polymerase II copurified with tagged Elp1p, Elp2p, Elp3p, or Elp5p (Krogan et al. 2002). However, the hyperphosphorylated form of RNA polymerase II has been observed to coimmunoprecipitate with tagged Elp2p and Elp5p (Frohloff et al. 2003). Localization studies showed that Elongator subunits are primarily cytosolic, both in yeast and HeLa cells (Kim et al. 2002; Pokholok et al. 2002; Huh et al. 2003). This localization is consistent with a role in wobble base tRNA modification as modifications in the anticodon region normally take place in the cytosol (Hopper and Phizicky 2003). Recently, Elp1 and Elp3 proteins were found to coimmunoprecipitate unspliced and spliced mRNA (Gilbert et al. 2004; Petrakis et al. 2004), implying that a small fraction of Elongator localizes to the nucleus and participates in RNA metabolism. In summary, Elongator has been extensively studied, but the experimental data are not conclusive.
The lack of mcm5 and ncm5 side chains in elp1–elp6 mutants could formally be explained by a defect in transcription of a gene(s) encoding the tRNA-modifying enzyme(s). As the elp1–elp6 mutants show a complete lack of mcm5 and ncm5 side chains at wobble uridines, it would indicate that the tRNA-modification gene is not at all expressed, which we find unlikely to be a consequence of a defect in elongation of transcription. In fact, only a small set of genes show reduced mRNA levels in Elongator mutants (Krogan and Greenblatt 2001). However, it cannot be excluded that Elongator has a dual function, as some tRNA-modifying enzymes have other substrates than tRNA. It is known that the pseudouridine synthases Pus1p and Pus7p in addition to tRNA have U2 snRNA as a substrate (Massenet et al. 1999; Ma et al. 2003). Recently, the mammalian Pus1p (mPus1p) was identified as a coactivator for retinoic acid receptor mRARγ-dependent RNA polymerase II transcription (Zhao et al. 2004). In this process, mPus1p is a component of the coactivator complex and pseudouridylation of the RNA component, Steroid Receptor RNA Activator, is required for transcriptional activation (Zhao et al. 2004). Thus, Elongator might participate both in transcription and tRNA modification.
Phenotypes and translational decoding in elp1–elp6 and kti11–kti13 mutants
Null mutations in Elongator subunit genes cause delayed transcriptional activation of PHO5, GAL1, GAL10, and ENA1 upon a shift to conditions that should activate transcription of these genes (Otero et al. 1999; Wittschieben et al. 1999). It was also shown that elp mutants are slow growing and slow to adapt to various growth conditions (Otero et al. 1999; Frohloff et al. 2001; Jablonowski et al. 2001). Furthermore, elp1–elp6 and kti11–kti13 mutants are resistant to K. lactis killer toxin, temperature-sensitive, sensitive to caffeine and calcofluor white, and mildly sensitive to 6-azauracil (Otero et al. 1999; Frohloff et al. 2001; Jablonowski et al. 2001; Fichtner and Schaffrath 2002). In addition, elp1–elp3, elp5, and kti12 mutants show a significant delay in G1-to-S transition (Frohloff et al. 2001). Thus, mutations in the ELP1–ELP6 and KTI11–KTI13 genes cause pleiotropic phenotypes.
In S. cerevisiae 13 of the 42 cytoplasmic tRNA species have a U at the wobble position (Percudani et al. 1997). Of these 13 tRNAs, the identity of the nucleoside at position 34 is known in eight species, and of these, six are mcm5U or ncm5U derivatives (Smith et al. 1973; Kobayashi et al. 1974; Kuntzel et al. 1975; Yamamoto et al. 1985; Keith et al. 1990; Glasser et al. 1992). The two others have an unmodified U (tRNALeuUAG) and a pseudouridine (Ψ, tRNAIleΨAΨ) at position 34 (Randerath et al. 1979; Szweykowska-Kulinska et al. 1994). The nature of the U34 nucleoside in the five remaining tRNAs had not been established. We investigated those and found that all contained mcm5U, ncm5U, or derivatives thereof (data not shown). In the elp3 deletion mutant the formation of the mcm5 or ncm5 group at U34 was abolished in the 11 tRNA species having such derivatives (Table 1; data not shown).
What function would mcm5 and ncm5 groups have in decoding? According to the original wobble hypothesis a U at position 34 will read A and G in the decoding process (Crick 1966). Since then the wobble hypothesis has been revised and states that an unmodified U34 recognizes U and C in addition to A and G, whereas a modified uridine will restrict wobble recognition (Agris 1991; Lim 1994; Yokoyama and Nishimura 1995). The presence of mcm5 or ncm5 groups at U34 limits pairing to A- and G-ending codons (Lim 1994; Yokoyama and Nishimura 1995). Lack of these groups at U34 in tRNAs decoding split codon boxes, with codons for more than one amino acid, could result in mistranslation. Additionally, lack of mcm5 or ncm5 groups at U34 is likely to cause a general reduction in the efficiency of decoding A- and G-ending codons (Lim 1994; Yokoyama and Nishimura 1995). Consistent with this, the SUP4 and sup3-18 encoded ochre tRNA suppressors require the mcm5 group to efficiently decode UAA ochre codons (Fig. 2; data not shown). By using a reporter construct, the ability of the SUP4 encoded suppressor tRNA to decode UAA was threefold decreased in an elp3 mutant (data not shown). In addition to affecting the decoding properties, modifications at U34 are known to act as tRNA identity elements and influence the efficiency of the aminoacylation reaction (Giege et al. 1998). Although the pleiotropic phenotypes of elp1–elp6 mutants have been attributed to a transcriptional defect, our results provide an alternative explanation. We suggest that the phenotypes of the elp1–elp6 and kti11–kti13 mutants could equally well be a consequence of less efficient translation and/or mistranslation, caused by lack of mcm5 and ncm5 groups at the wobble position in 11 out of 42 tRNA species in yeast. In fact, lack of the modified nucleosides queuosine (Q34) or ms2i6A37 in Shigella flexneri cause a reduced posttranscriptional expression of the transcriptional activator VirFp (Durand et al. 1994, 1997). This reduction of VirF protein renders a decreased transcription of target genes, showing that translational defects caused by hypomodified tRNAs can affect transcription. Moreover, lack of modified nucleosides in the anticodon region has been shown to cause pleiotropic phenotypes in Salmonella enterica (Ericson and Björk 1986; Björk and Nilsson 2003).
Potential target of K. lactis killer toxin
Strains with null mutations in any of the ELP1–ELP6 or KTI11–KTI13 genes show resistance to K. lactis killer toxin (Frohloff et al. 2001; Jablonowski et al. 2001; Fichtner and Schaffrath 2002). Interestingly, high dosage of a gene encoding tRNAGlu mcm5s2UUC suppresses the killer toxin sensitivity of a wild-type strain (Butler et al. 1994), suggesting that tRNAGlu mcm5s2UUC could be the target of killer toxin. The over-expressed tRNAGlu mcm5s2UUC might be hypomodified, if the modification enzyme(s) is not able to cope with increased amounts of substrate. Thus, if the target of the toxin is tRNAGlu mcm5s2UUC, a hypomodified derivative may be resistant. Alternatively, increased amounts of fully modified tRNAGlu mcm5s2UUC might sequester the killer toxin present in the cell, leaving a pool of tRNAGlumcm5s2UUC functional in translation.
Conclusions
We have shown that the Elp1–Elp6 and Kti11–Kti13 proteins are all required for formation of mcm5 and ncm5 side chains at U34 in tRNA. It is remarkable that an early step(s) in the formation of mcm5 and ncm5 side chains is dependent on at least nine proteins. So far no more than two gene products have been implicated in the synthesis of a modified nucleoside in yeast (Gerber and Keller 1999; Anderson et al. 2000; Alexandrov et al. 2002). The Kti13 protein seems to have a regulatory role because a null mutant still contains ~15% of these nucleosides. The Kti11 and Kti12 proteins have also been suggested to act as regulators, since loss of the Kti11p enhances the proteolytic processing of the Elp1 protein, and Kti12p is important for the phosphorylation status of Elp1p (Fichtner et al. 2003; Jablonowski et al. 2004). We find it likely that the Kti11–Kti13 proteins regulate Elongator in the wobble uridine modification process and that the activity of Elongator modulate translational efficiency.
MATERIALS AND METHODS
Yeast strains, media, and genetic procedure
The yeast strains used are listed in Table 2. S. cerevisiae and S. pombe transformation, media, and genetic procedures have been described (Moreno et al. 1991; Burke et al. 2000).
TABLE 2.
Strains used in this study
| Yeast strain | Genotype | Source |
| S. pombe | ||
| BP231 | h+ura4-D18 | J. Keeney, pers. comm. |
| h+ade7-413 sup3-18 | J. Kohli, pers. comm. | |
| h+ade7-413 | J. Kohli, pers. comm. | |
| h+ade7-413 sup3-18 sin3-193 | J. Kohli, pers. comm. | |
| UMY3041 | h+ade7-413 sup3-18 sin3::KanMX6 | This study |
| UMY2821 | h+ade7-413 sup3-18 sin3-193 ura4-D18 | This study |
| S. cerevisiae | ||
| 354 | MATα SUP4 trp5-38 his5-2 lys1-1 ade2-1 ura3 can1 met | F. Sherman, pers. comm. |
| W303-1A | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | Fiorentini et al. 1997 |
| W303-1B | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | Fiorentini et al. 1997 |
| UMY2893 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | This study |
| UMY2894 | MATa SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | This study |
| UMY2912 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 elp1::KanMX4 | This study |
| UMY2914 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 elp2::KanMX4 | This study |
| UMY2916 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 elp3::KanMX4 | This study |
| UMY2918 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 elp4::KanMX4 | This study |
| UMY2920 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 elp5::KanMX6 | This study |
| UMY2922 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 elp6::KanMX4 | This study |
| UMY2936 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 kti11::KanMX6 | This study |
| UMY2938 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 kti12::KanMX4 | This study |
| UMY2940 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 kti13::KanMX4 | This study |
| UMY3039 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 elp3-Y540A | This study |
| UMY3040 | MATα SUP4 leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 elp3-Y541A | This study |
| UMY3114 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 ELP1-(HA)3::KanMX6 | This study |
| UMY3116 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 ELP3-(HA)3::KanMX6 | This study |
| UMY3118 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 ELP5-(HA)3::KanMX6 | This study |
| UMY3094 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 his3-11,15 TIF4632-(HA)3::KanMX6 | This lab |
The sup3-18 and ade7-413 alleles in the S. pombe strain h+ ade7-413 sup3-18 sin3-193 were amplified by PCR and sequenced. The sup3-18 gene was confirmed to encode a tRNASer UUA ochre-suppressing tRNA (Amstutz et al. 1985). The mutation in ade7-413 creates an ochre stop codon after the 74th codon. To generate strain UMY2821, the ura4+ gene in the ade7-413 sup3-18 sin3-193 strain was replaced with an ura4-D18 DNA fragment PCR-amplified from strain BP231. To delete the sin3+ gene, a two-round PCR procedure was used (Krawchuk and Wahls 1999), generating a sin3::KanMX6 fragment. Strain UMY3041 was constructed by transforming the sin3::KanMX6 fragment into strain h+ ade7-413 sup3-18 and selecting for G418 resistance. The deletion was confirmed by PCR.
The wild-type sup4+ locus was replaced with the SUP4 allele in strain W303-1B using the two-step method (Scherer and Davis 1979). To target the SUP4 allele to the correct chromosomal location, plasmid pRS306-SUP4 was linearized with MunI and transformants selected on synthetic complete (SC) medium lacking uracil. Colonies able to grow on SC plates lacking adenine (SC-Ade) and unable to grow on SC medium lacking arginine and containing 60 μg/mL canavanine (SC-Arg+Can), were streaked on 5-fluoro-orotic acid (5-FOA)-containing medium. The SUP4 locus from 5-FOAR, Ade+, and CanS colonies was amplified by PCR and sequenced, confirming the gene replacement. Strains UMY2893 and UMY2894 were obtained from a tetrad in a cross between one of the generated SUP4 strains and W303-1A.
To construct the elp5::KanMX6 and kti11::KanMX6 deletions, oligonucleotides containing 45-nt homology to the flanking sequences of the ELP5 or KTI11 genes were used to amplify the KanMX6 cassette (Longtine et al. 1998). To delete ELP1, ELP2, ELP3, ELP4, ELP6, and the KTI12, KTI13 genes, chromosomal DNA from the corresponding null mutants in the yeast deletion collection (Research genetics) was used as templates. Appropriate primers were used to amplify DNA fragments containing the KanMX4 cassette and 300–400 nt of flanking sequences. Each PCR product was transformed into strain UMY2893 and transformants selected on YEPD plates containing 200 μg/mL of G418. Deletions of the ELP or KTI genes were confirmed by PCR followed by a backcross, to verify 2:2 segregation of the KanMX4/KanMX6 allele and cosegregation of the G418R and loss-of-suppression phenotypes. The strains generated were UMY2912 (elp1::KanMX4), UMY2914 (elp2::KanMX4), UMY2916 (elp3::KanMX4), UMY2918 (elp4::KanMX4), UMY2920 (elp5::KanMX6), UMY2922 (elp6::KanMX4), UMY2936 (kti11::KanMX6), UMY2938 (kti12::KanMX4), and UMY2940 (kti13::KanMX4).
The wild-type ELP3 gene was replaced by mutant elp3 alleles in strain UMY2893 using the two-step method (Scherer and Davis 1979). To target the elp3 alleles to the correct chromosomal location, plasmids pRS306-elp3-Y540A or pRS306-elp3-Y541A were linearized with BclI and transformants selected on SC medium lacking uracil. After selection for 5-FOAR segregants, PCR amplification, and DNA sequencing confirmed that red colonies contained the elp3 mutations. The generated strains, UMY2939 (elp3-Y540A) and UMY2946 (elp3-Y541A), were transformed with pRS315-ELP3, which restored suppression. Strains containing C-terminal (HA)3-tagged Elp1, Elp3, and Elp5 proteins, UMY3114, UMY3116, and UMY3118, were derived from W303-1A and constructed by the PCR-mediated strategy using plasmid pFA6a-3HA-kanMX6 as the template (Longtine et al. 1998). The strains were confirmed by PCR and by Western blot analysis using anti-HA antibodies. Strains with tagged Elp proteins were killer toxin sensitive, confirming the functionality of the tagged proteins.
Identification of the sin3+ gene and phenotypes of sin3 mutants
To clone the wild-type sin3+ gene, a URA3-based S. pombe genomic library was introduced into the ade7-413 sup3-18 sin3-193 ura4-D18strain (UMY2821) and two plasmids were identified that restored the suppression of the ade7-413 allele (data not shown). DNA sequencing revealed that the two plasmids had the uncharacterized open reading frame SPAC29A4.20 in common. In order to find the sin3-193 mutation, we PCR-amplified and sequenced the SPAC29A4.20 open reading frame from sin3+ and sin3-193 strains. The sin3-193 mutation generated an arginine-to-histidine substitution at position 220 (R220H) of the Sin3p (data not shown).
Introduction of a sin3::KanMX6 allele into an ade7-413 sup3-18 strain abolished the suppression of the ade7-413 allele (data not shown). Analysis of tRNA revealed that tRNAGlu mcm5s2UUC (Wong et al. 1979) from the sin3+ strain contained mcm5s2U, whereas it was reduced in the sin3-193 (5% of wild type) and not detectable in the sin3::KanMX6 derivative (data not shown). Furthermore, analysis of the ochre suppressor tRNASer revealed the presence of mcm5U in the ade7-413 sup3-18 strain, but not in the sin3-193 and sin3::KanMX6 derivatives or the wild-type ade7-413 strain (data not shown).
Suppression assay
To investigate the effect of the elp1-6- and kti11-13-null alleles on tRNA suppression, we used the ade2 can1 system (Hopper et al. 1980). Strains W303-1A and W303-1B contain the two ochre alleles, ade2-1 (UAA) and can1-100 (UAA). ADE2 encodes an enzyme participating in adenine biosynthesis. Strains with an ade2 mutation have a requirement for adenine and accumulate a red pigment. Mutations in the CAN1 gene that codes for an arginine permease generate resistance to canavanine, an arginine analog toxic to yeast cells. The efficiency of an ochre suppressor tRNA to decode the ade2-1 (UAA) and can1-100 (UAA) codons can be investigated on plates. If the suppressor tRNA is functional, the strains with the ade2-1 (UAA) and can1-100 (UAA) alleles will grow on SC-Ade, but not on SC-Arg+Can plates. If the suppressor tRNA is nonfunctional, the strains will grow on SC-Arg+Can, but not on SC-Ade medium.
Plasmid constructions
DNA manipulations, plasmid preparations, and bacterial transformations were performed according to standard protocols. Genes were PCR-amplified using Pwo DNA polymerase or the Expand Long Template PCR system (Roche Applied Science). An integrative vector carrying the SUP4 locus was constructed by cloning a BamHI/SacI fragment PCR-amplified from strain 354 into the corresponding sites of pRS306 (Sikorski and Hieter 1989), generating plasmid pRS306-SUP4. DNA sequencing confirmed that the anticodon was TTA. The oligonucleotides used for the PCR were 5′-TTTTGGATCCGTCCAGATGCCTTTACGAGT-3′and 5′-ATATGAGCTCGTTTCGGCTCTAATCCACTG-3′.
A low-copy LEU2 plasmid carrying ELP3 (pRS315-ELP3) was constructed by cloning a BamHI/XhoI fragment, PCR-amplified from strain W303-1B into the corresponding sites of pRS315 (Sikorski and Hieter 1989). The oligonucleotides used were 5′-TT GTGGATCCTTGGCTTCAGGTGTCATTC-3′ and 5′-ATCTCTC GAGTATCTGGGGCCAATTGT-3′.
Plasmid pRS306-ELP3 was constructed by cloning a BamHI/XhoI ELP3 fragment from pRS315-ELP3 into the corresponding sites of plasmid pRS306. Mutant alleles of ELP3 were obtained using the QuickChange protocol (Stratagene), generating pRS306-elp3-Y540A and pRS306-elp3-Y541A.
Plasmid pRS315-ELP5 was constructed by cloning a SalI/SpeI fragment, PCR-amplified from strain W303-1B into pRS315. The oligonucleotides used were 5′-ATTTGTCGACCACTTGGAGTTC ACGATGTTA-3′ and 5′-GAGAACTAGTGGGGATCGACACCTA AATTCA-3′. PCR fragments containing ELP1, ELP2, ELP4, or ELP6 amplified from strain W303-1B were cloned into the pGEMR-T Easy Vector (Promega) following addition of an A overhang by Taq DNA polymerase (Roche Applied Science). The oligonucleotides used were 5′-TTCGACGTTTTCATGGACCA-3′ and 5′-CGCCAACAAACTCTAGCTCAT-3′ (ELP1), 5′-AATGA TGGGGAGAGTGACGTA-3′ and 5′-GTTTCGCTTTACGAGAAA AGG-3′ (ELP2), 5′-AAATATCGCATCGAATGGAA-3′ and 5′-TGAGTTTGAAGCTGAACCGT-3′ (ELP4), or 5′-AGCAAGGTG TCGAATCAAGTT-3′ and 5′-ATCACCCATAAAGGCAGGAA-3′ (ELP6).
Plasmids pRS315-ELP1, pRS315-ELP2, pRS315-ELP4, and pRS315-ELP6 were constructed by cloning SacI/SacII (ELP1), ApaI/SacI (ELP2 and ELP4), and SacII/SalI (ELP6) fragments from the respective pGEMR-T Easy vector into plasmid pRS315. Plasmids pRS315-KTI11, pRS315-KTI12, and pRS315-KTI13 were constructed by cloning BamHI/XhoI fragments, PCR-amplified from strain W303-1B into the corresponding sites of pRS315. The oligonucleotides used were 5′-CTTAGGATCCGCACATACTTTT TGTCCTGGTG-3′ and 5′-CTTTCTCGAGCATATACACCTACC CAGTATCC-3′ (KTI11), 5′-GAGAGGATCCGTGGACGTGGAT GTAGAATCG-3′ and 5′-CTTTCTCGAGCTGCCTCTCTTGGTA CGACA-3′ (KTI12) or 5′-TGTTGGATCCGCTGCTGTAGAGCG TTTGGTG-3′ and 5′-GTTTCTCGAGGTATGAGCTCCTTCGCTGGG-3′ (KTI13). A tRNAGluUUC gene under the T7 promoter gene was constructed by ligating three pairs of oligonucleotides (Sampson and Uhlenbeck 1988) into the EcoRI/BamHI sites of pUC18 (Roche Applied Science), generating p1537. The tRNAGluUUC gene carried two mutations (G1-C72) to improve the efficiency of T7 transcription.
tRNA methods and HPLC analysis
Cells were grown at 30°C in 2-L YEPD or appropriate selective media. The cells were harvested at OD600 = 1.0 and single tRNA species prepared as previously described (Björk et al. 2001). The purified tRNAs were digested to nucleosides and analyzed by HPLC using a Develosil C-30 reverse-phase column (Gehrke et al. 1982; Gehrke and Kuo 1990). The presence of only negligible amounts of other nucleosides than expected demonstrated the purity of the tRNAs analyzed. Dihydrouridine (D) was not detected at 254 nm and is not included in Table 1. Nucleosides mcm5s2U, mcm5U, ncm5U, and s2U were identified by their spectrum, retention time, and by comigration with corresponding synthetic nucleosides. The ncm5U nucleoside was obtained by conversion of mcm5U as described previously (Fissekis and Sweet 1970). The T7-transcribed radiolabeled tRNAMeti and tRNAGlu UUC were prepared by using MvaI-linearized p2142 (Åström and Byström 1994) or p1537, 5′-[α-32P]CTP (400 Ci/mmol; Amersham Biosciences), and the Riboprobe in vitro transcription system (Promega). The radiolabeled transcripts were purified as previously described (Johansson and Byström 2004).
UV cross-linking and immunoprecipitation
Cells were grown in 50 mL of YEPD at 30°C to an OD600 between 0.6 and 0.8. The cells were harvested, washed with cold breaking buffer (10 mM HEPES at pH 7.3, 50 mM KCl, 10 mM MgAc, 5 mM DTT, 5% glycerol), resuspended in 300 μL of cold breaking buffer supplemented with protease inhibitor mixture (Otero et al. 1999), and crude extracts were prepared by vortexing in the presence of glass beads. The glass beads were extracted twice with 300 μL of cold breaking buffer, and the extract was cleared by two centrifugations in a microfuge. Radiolabeled tRNA transcripts (1 ng, ~100,000 dpm) and 20 U of RNasin were added to 300 μL of the extracts. Fractions (20 μL) were transferred to a 96-well microtitre plate, which was incubated at room temperature for 10 min. The microtitre plate was exposed five times to 120 mJ/cm2 of UV using a Stratalinker (Stratagene), before the fractions were pooled and diluted to 600 μL with cold breaking buffer. Immunoprecipitation was performed for at least 2 h at 8°C in a rotating chamber by using 50 μL of anti-HA agarose conjugate (Sigma). After five washes with 1 mL of cold wash buffer (10 mM HEPES at pH 7.3, 50 mM KCl, 10 mM MgAc, 0.1% Triton X-100), proteins were eluted by addition of 100 μL of TES (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, 1% SDS) and a 20-min incubation at 65°C. The beads were washed once with 50 μL of TES. To confirm the presence of the immunoprecipitated protein, a fraction of the pooled eluate was analyzed by Western blot analysis, using anti-HA antibodies. Based on this analysis, the immunoprecipitations were equally efficient in all protein extracts. The remaining eluate was treated with Proteinase K, precipitated, and separated on an 8% polyacrylamide, 8 M urea gel. The gel was dried and the signals were visualized and quantified by PhosphorImager analysis.
Acknowledgments
We acknowledge J. Kohli, J. Keeney, L. Symington, F. Sherman, and N. Gunge for yeast strains. M. Poitelea is acknowledged for the S. pombe genomic library. A. Malkiewicz is acknowledged for providing mcm5s2U, mcm5U, and s2U; and D. Johnels for valuable advice concerning conversion of mcm5U to ncm5U. G.R. Björk, M. Pollard, H. Wolf-Watz, and B-E. Uhlin are acknowledged for valuable discussions and K. Jacobsson for technical assistance. This work was financially supported by the Swedish Research Council (621-2001-1890) and the Swedish Cancer Society (3516-B03-10XAB).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7247705.
REFERENCES
- Agarwalla, S., Kealey, J.T., Santi, D.V., and Stroud, R.M. 2002. Characterization of the 23 S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J. Biol. Chem. 277: 8835–8840. [DOI] [PubMed] [Google Scholar]
- Agris, P.F. 1991. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: A modified-wobble hypothesis. Biochimie 73: 1345–1349. [DOI] [PubMed] [Google Scholar]
- Alexandrov, A., Martzen, M.R., and Phizicky, E.M. 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8: 1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amstutz, H., Munz, P., Heyer, W.D., Leupoid, U., and Kohli, J. 1985. Concerted evolution of tRNA genes: Intergenic conversion among three unlinked serine tRNA genes in S. pombe. Cell 40: 879–886. [DOI] [PubMed] [Google Scholar]
- Anderson, J., Phan, L., and Hinnebusch, A.G. 2000. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 97: 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, S.L., Coli, R., Daly, I.W., Kichula, E.A., Rork, M.J., Volpi, S.A., Ekstein, J., and Rubin, B.Y. 2001. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 68: 753–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åström, S.U. and Byström, A.S. 1994. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell 79: 535–546. [DOI] [PubMed] [Google Scholar]
- Björk, G.R. 1995. Biosynthesis and function of modified nucleosides. In tRNA: Structure, biosynthesis, and function (eds. D. Söll and U. L. RajBhandary), pp. 165–205. ASM Press, Washington, DC.
- Björk, G.R. and Nilsson, K. 2003. 1-Methylguanosine-deficient tRNA of Salmonella enterica serovar Typhimurium affects thiamine metabolism. J. Bacteriol. 185: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk, G.R., Jacobsson, K., Nilsson, K., Johansson, M.J., Byström, A.S., and Persson, O.P. 2001. A primordial tRNA modification required for the evolution of life? EMBO J. 20: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D., Dawson, D., and Stearns, T., eds. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Butler, A.R., White, J.H., and Stark, M.J. 1991. Analysis of the response of Saccharomyces cerevisiae cells to Kluyveromyces lactis toxin. J. Gen. Microbiol. 137: 1749–1757. [DOI] [PubMed] [Google Scholar]
- Butler, A.R., White, J.H., Folawiyo, Y., Edlin, A., Gardiner, D., and Stark, M.J. 1994. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol. 14: 6306–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov, Y. 2002. A second catalytic domain in the Elp3 histone acetyltransferases: A candidate for histone demethylase activity? Trends Biochem. Sci. 27: 115–117. [DOI] [PubMed] [Google Scholar]
- Crick, F.H. 1966. Codon–anticodon pairing: The wobble hypothesis. J. Mol. Biol. 19: 548–555. [DOI] [PubMed] [Google Scholar]
- Durand, J.M., Okada, N., Tobe, T., Watarai, M., Fukuda, I., Suzuki, T., Nakata, N., Komatsu, K., Yoshikawa, M., and Sasakawa, C. 1994. vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of Escherichia coli K-12. J. Bacteriol. 176: 4627–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, J.M., Björk, G.R., Kuwae, A., Yoshikawa, M., and Sasakawa, C. 1997. The modified nucleoside 2-methylthio-N6-isopentenyladenosine in tRNA of Shigella flexneri is required for expression of virulence genes. J. Bacteriol. 179: 5777–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson, J.U. and Björk, G.R. 1986. Pleiotropic effects induced by modification deficiency next to the anticodon of tRNA from Salmonella typhimurium LT2. J. Bacteriol. 166: 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner, L. and Schaffrath, R. 2002. KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol. Microbiol. 44: 865–875. [DOI] [PubMed] [Google Scholar]
- Fichtner, L., Frohloff, F., Burkner, K., Larsen, M., Breunig, K.D., and Schaffrath, R. 2002. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol. Microbiol. 43: 783–791. [DOI] [PubMed] [Google Scholar]
- Fichtner, L., Jablonowski, D., Schierhorn, A., Kitamoto, H.K., Stark, M.J., and Schaffrath, R. 2003. Elongator’s toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol. Microbiol. 49: 1297–1307. [DOI] [PubMed] [Google Scholar]
- Fiorentini, P., Huang, K.N., Tishkoff, D.X., Kolodner, R.D., and Symington, L.S. 1997. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 17: 2764–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fissekis, J.D. and Sweet, F. 1970. Synthesis of 5-carboxymethyluridine. A nucleoside from transfer ribonucleic acid. Biochemistry 9: 3136–3142. [DOI] [PubMed] [Google Scholar]
- Frohloff, F., Fichtner, L., Jablonowski, D., Breunig, K.D., and Schaffrath, R. 2001. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 20: 1993–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohloff, F., Jablonowski, D., Fichtner, L., and Schaffrath, R. 2003. Subunit communications crucial for the functional integrity of the yeast RNA polymerase II elongator (γ-toxin target (TOT)) complex. J. Biol. Chem. 278: 956–961. [DOI] [PubMed] [Google Scholar]
- Gehrke, C.W. and Kuo, K.C.T. 1990. Chromatography and modification of nucleosides. Elsevier, Amsterdam.
- Gehrke, C.W., Kuo, K.C., McCune, R.A., Gerhardt, K.O., and Agris, P.F. 1982. Quantitative enzymatic hydrolysis of tRNAs: Reversed-phase high-performance liquid chromatography of tRNA nucleosides. J. Chromatogr. 230: 297–308. [PubMed] [Google Scholar]
- Gerber, A.P. and Keller, W. 1999. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286: 1146–1149. [DOI] [PubMed] [Google Scholar]
- Giege, R., Sissler, M., and Florentz, C. 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26: 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, C., Kristjuhan, A., Winkler, G.S., and Svejstrup, J.Q. 2004. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 14: 457–464. [DOI] [PubMed] [Google Scholar]
- Glasser, A.L., el Adlouni, C., Keith, G., Sochacka, E., Malkiewicz, A., Santos, M., Tuite, M.F., and Desgres, J. 1992. Presence and coding properties of 2′-O-methyl-5-carbamoylmethyluridine (ncm5Um) in the wobble position of the anticodon of tRNALeu (U*AA) from brewer’s yeast. FEBS Lett. 314: 381–385. [DOI] [PubMed] [Google Scholar]
- Grossenbacher, A.M., Stadelmann, B., Heyer, W.D., Thuriaux, P., Kohli, J., Smith, C., Agris, P.F., Kuo, K.C., and Gehrke, C. 1986. Antisuppressor mutations and sulfur-carrying nucleosides in transfer RNAs of Schizosaccharomyces pombe. J. Biol. Chem. 261: 16351–16355. [PubMed] [Google Scholar]
- Hawkes, N.A., Otero, G., Winkler, G.S., Marshall, N., Dahmus, M.E., Krappmann, D., Scheidereit, C., Thomas, C.L., Schiavo, G., Erdjument-Bromage, H., et al. 2002. Purification and characterization of the human elongator complex. J. Biol. Chem. 277: 3047–3052. [DOI] [PubMed] [Google Scholar]
- Heyer, W.D., Thuriaux, P., Kohli, J., Ebert, P., Kersten, H., Gehrke, C., Kuo, K.C., and Agris, P.F. 1984. An antisuppressor mutation of Schizosaccharomyces pombe affects the post-transcriptional modification of the “wobble” base in the anticodon of tRNAs. J. Biol. Chem. 259: 2856–2862. [PubMed] [Google Scholar]
- Hopper, A.K. and Phizicky, E.M. 2003. tRNA transfers to the limelight. Genes & Dev. 17: 162–180. [DOI] [PubMed] [Google Scholar]
- Hopper, A.K., Schultz, L.D., and Shapiro, R.A. 1980. Processing of intervening sequences: A new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell 19: 741–751. [DOI] [PubMed] [Google Scholar]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O’Shea, E.K. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Jablonowski, D., Frohloff, F., Fichtner, L., Stark, M.J., and Schaffrath, R. 2001. Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol. Microbiol. 42: 1095–1105. [DOI] [PubMed] [Google Scholar]
- Jablonowski, D., Fichtner, L., Stark, M.J., and Schaffrath, R. 2004. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol. Biol. Cell 15: 1459–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, M.J. and Byström, A.S. 2004. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA 10: 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor, H.R. and Clarke, S. 2003. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 23: 9283–9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, G., Desgres, J., Pochart, P., Heyman, T., Kuo, K.C., and Gehrke, C.W. 1990. Eukaryotic tRNAsPro: Primary structure of the anticodon loop; presence of 5-carbamoylmethyluridine or inosine as the first nucleoside of the anticodon. Biochim. Biophys. Acta 1049: 255–260. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Lane, W.S., and Reinberg, D. 2002. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. 99: 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida, M., Tokunaga, M., Katayose, Y., Yajima, H., Kawamura-Watabe, A., and Hishinuma, F. 1996. Isolation and genetic characterization of pGKL killer-insensitive mutants (iki) from Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 60: 798–801. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Irie, T., Yoshida, M., Takeishi, K., and Ukita, T. 1974. The primary structure of yeast glutamic acid tRNA specific to the GAA codon. Biochim. Biophys. Acta 366: 168–181. [DOI] [PubMed] [Google Scholar]
- Krawchuk, M.D. and Wahls, W.P. 1999. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15: 1419–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J. and Greenblatt, J.F. 2001. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 8203–8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., Kim, M., Ahn, S.H., Zhong, G., Kobor, M.S., Cagney, G., Emili, A., Shilatifard, A., Buratowski, S., and Greenblatt, J.F. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol. Cell. Biol. 22: 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntzel, B., Weissenbach, J., Wolff, R.E., Tumaitis-Kennedy, T.D., Lane, B.G., and Dirheimer, G. 1975. Presence of the methylester of 5-carboxymethyl uridine in the wobble position of the anticodon of tRNAIII Arg from brewer’s yeast. Biochimie 57: 61–70. [DOI] [PubMed] [Google Scholar]
- Layer, G., Heinz, D.W., Jahn, D., and Schubert, W.D. 2004. Structure and function of radical SAM enzymes. Curr. Opin. Chem. Biol. 8: 468–476. [DOI] [PubMed] [Google Scholar]
- Lim, V.I. 1994. Analysis of action of wobble nucleoside modifications on codon–anticodon pairing within the ribosome. J. Mol. Biol. 240: 8–19. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Ma, X., Zhao, X., and Yu, Y.T. 2003. Pseudouridylation (Ψ) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 22: 1889–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet, S., Motorin, Y., Lafontaine, D.L., Hurt, E.C., Grosjean, H., and Branlant, C. 1999. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell. Biol. 19: 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier, R., Penot, G., Hubner, M.R., Reid, G., Brand, H., Kos, M., and Gannon, F. 2003. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115: 751–763. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Otero, G., Fellows, J., Li, Y., de Bizemont, T., Dirac, A.M., Gustafsson, C.M., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3: 109–118. [DOI] [PubMed] [Google Scholar]
- Percudani, R., Pavesi, A., and Ottonello, S. 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 268: 322–330. [DOI] [PubMed] [Google Scholar]
- Petrakis, T.G., Wittschieben, B.O., and Svejstrup, J.Q. 2004. Molecular architecture, structure–function relationship, and importance of the Elp3 subunit for the RNA binding of holo-elongator. J. Biol. Chem. 279: 32087–32092. [DOI] [PubMed] [Google Scholar]
- Pierrel, F., Bjork, G.R., Fontecave, M., and Atta, M. 2002. Enzymatic modification of tRNAs: MiaB is an iron–sulfur protein. J. Biol. Chem. 277: 13367–13370. [DOI] [PubMed] [Google Scholar]
- Pierrel, F., Douki, T., Fontecave, M., and Atta, M. 2004. MiaB Protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J. Biol. Chem. 279: 47555–47563. [DOI] [PubMed] [Google Scholar]
- Pokholok, D.K., Hannett, N.M., and Young, R.A. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9: 799–809. [DOI] [PubMed] [Google Scholar]
- Prevot, D., Darlix, J.L., and Ohlmann, T. 2003. Conducting the initiation of protein synthesis: The role of eIF4G. Biol. Cell 95: 141–156. [DOI] [PubMed] [Google Scholar]
- Randerath, E., Gupta, R.C., Chia, L.L., Chang, S.H., and Randerath, K. 1979. Yeast tRNA Leu UAG. Purification, properties and determination of the nucleotide sequence by radioactive derivative methods. Eur. J. Biochem. 93: 79–94. [DOI] [PubMed] [Google Scholar]
- Roth, S.Y., Denu, J.M., and Allis, C.D. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70: 81–120. [DOI] [PubMed] [Google Scholar]
- Rozenski, J., Crain, P.F., and McCloskey, J.A. 1999. The RNA modification database: 1999 update. Nucleic Acids Res. 27: 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, J.R. and Uhlenbeck, O.C. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 85: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, S. and Davis, R.W. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. 76: 4951–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt, S.A., Blumenfeld, A., Gill, S.P., Leyne, M., Mull, J., Cuajungco, M.P., Liebert, C.B., Chadwick, B., Idelson, M., Reznik, L., et al. 2001. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 68: 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.J., Teh, H.S., Ley, A.N., and D’Obrenan, P. 1973. The nucleotide sequences and coding properties of the major and minor lysine transfer ribonucleic acids from the haploid yeast Saccharomyces cerevisiae S288C. J. Biol. Chem. 248: 4475–4485. [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweykowska-Kulinska, Z., Senger, B., Keith, G., Fasiolo, F., and Grosjean, H. 1994. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNAIle. EMBO J. 13: 4636–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux, P., Minet, M., Hofer, F., and Leupold, U. 1976. Genetic analysis of antisuppressor mutants in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 142: 251–261. [DOI] [PubMed] [Google Scholar]
- Tumaitis, T.D. and Lane, B.G. 1970. Differential labelling of the carboxymethyl and methyl substituents of 5-carboxymethyluridine methyl ester, a trace nucleoside constituent of yeast transfer RNA. Biochim. Biophys. Acta 224: 391–403. [DOI] [PubMed] [Google Scholar]
- Winkler, G.S., Petrakis, T.G., Ethelberg, S., Tokunaga, M., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. 2001. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276: 32743–32749. [DOI] [PubMed] [Google Scholar]
- Winkler, G.S., Kristjuhan, A., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. 99: 3517–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben, B.O., Otero, G., de Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., Li, Y., Allis, C.D., Tempst, P., and Svejstrup, J.Q. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4: 123–128. [DOI] [PubMed] [Google Scholar]
- Wittschieben, B.O., Fellows, J., Du, W., Stillman, D.J., and Svejstrup, J.Q. 2000. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J. 19: 3060–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, T.W., McCutchan, T., Kohli, J., and Soll, D. 1979. The nucleotide sequence of the major glutamate transfer RNA from Schizosaccharomyces pombe. Nucleic Acids Res. 6: 2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, N., Yamaizumi, Z., Yokoyama, S., Miyazawa, T., and Nishimura, S. 1985. Modified nucleoside, 5-carbamoylmethyluridine, located in the first position of the anticodon of yeast valine tRNA. J. Biochem. (Tokyo) 97: 361–364. [DOI] [PubMed] [Google Scholar]
- Yokoyama, S. and Nishimura, S. 1995. Modified nucleosides and codon recognition. In tRNA: Structure, biosynthesis, and function (eds. D. Söll and U.L. RajBhandary), pp. 207–223. ASM Press, Washington, DC.
- Zhao, X., Patton, J.R., Davis, S.L., Florence, B., Ames, S.J., and Spanjaard, R.A. 2004. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol. Cell 15: 549–558. [DOI] [PubMed] [Google Scholar]