Abstract
Sequence-specific RNA–protein interactions underlie regulation of many mRNAs. Here we analyze the RNA sequence specificity of Caenorhabditis elegans FBF-1, a founding member of the PUF protein family. Like other PUF proteins, FBF-1 binds to the 3′ UTR of target mRNAs and decreases expression of those target genes. Here, we show that FBF-1 and its close relative, FBF-2, bind with similar affinity to multiple RNA sites. We use mutagenesis and in vivo selection experiments to identify nucleotides that are essential for FBF-1 binding. The binding elements comprise a “core” central region and flanking sequences. The core region is similar but distinct from the binding sites of other PUF proteins. We combine the identification of binding elements with informatics to predict new FBF-1 binding sites in a C. elegans 3′ UTR database. These data identify a set of new candidate mRNA targets of FBF-1 and FBF-2.
Keywords: RNA/protein interactions, PUF protein, three-hybrid system, FBF
INTRODUCTION
mRNA translation, stability, and localization are regulated by sequence-specific RNA binding proteins. Their selectivity of these proteins determines which mRNAs are subject to control. The binding sites for these regulatory proteins often reside between the termination codon and poly(A) tail—in the 3′ untranslated region (3′ UTR). In some mRNAs, these binding sites possess secondary and tertiary structure (Allain et al. 1996; Hentze and Kühn 1996). More commonly, the 3′ UTR binding sites for regulatory proteins lack apparent structure, yet achieve protein interactions that are specific and of high affinity.
The PUF protein family provides a paradigm for 3′ UTR regulatory proteins. They exist throughout eukaryotes, including Drosophila, Caenorhabditis elegans, Arabidopsis, Saccharomyces cerevisiae, and Homo sapiens (Wickens et al. 2002). PUF proteins bind 3′ UTR elements and reduce expression, either by repressing translation or causing mRNA instability (Wickens et al. 2002). PUF proteins contain eight repeats of ~ 40 amino acids, called Puf repeats. Each Puf repeat contains a diagnostic central consensus sequence (Zamore et al. 1997; Zhang et al. 1997; Wickens et al. 2002). Structures of two closely related PUF proteins have been solved, one bound to an RNA ligand (Edwards et al. 2001; Wang et al. 2001, 2002). Each Puf repeat forms three α-helices. These repeats lie on one another to form an arc of roughly 120°. The α-helices that bind RNA all lie on one face of the structure, and contain the “core” consensus sequences (Fig. 1A). Each consecutive helix interacts with one of eight bases in the RNA recognition site by hydrogen bonding, while one amino acid is stacked between adjacent bases (Fig. 1A; Wang et al. 2002). Two 5′ terminal bases loop back to make additional contacts (Wang et al. 2002). These structural data reveal how one PUF protein, human Pumilio1, interacts with a likely target site. However, different PUF proteins bind and regulate different mRNAs, implying that they bind distinct sequences even in a single organism (Olivas and Parker 2000; Tadauchi et al. 2001; Gerber et al. 2004).
FIGURE 1.
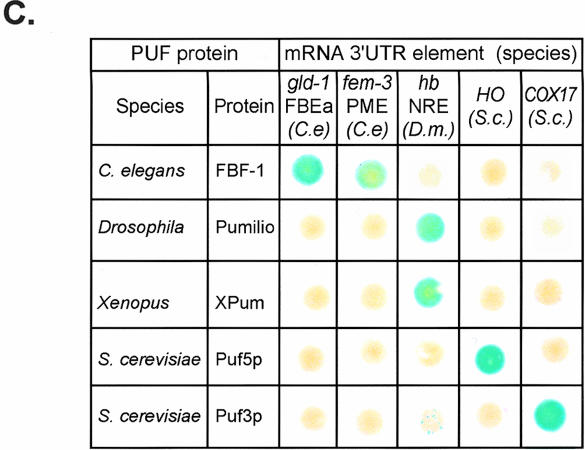
PUF/RNA interactions are specific. (A) A PUF protein binding RNA. A depiction of the interaction between human Pumilio1 and the NRE (crystal coordinates from Wang et al. 2002). RNA binding helices are red cylinders, RNA binding amino acid side chains are green, and the RNA is blue. (Left) A side view of the RNA binding face. (Right) The same image rotated 90° to present a straight-on view of the RNA binding face. Amino (N) and carboxy (C) termini of the protein are indicated, along with the 5′ and 3′ ends of the RNA. The region corresponding to what we define as the core binding element is that depicted in blue. RNA binding amino acid side chains are not depicted. (B) The yeast three-hybrid assay. A fusion protein consisting of the LexA DNA binding protein linked to MS2 coat protein tethers a hybrid RNA to the promoter region upstream of a reporter gene (e.g., LacZ). Interaction between a segment of the hybrid RNA (“RNA X”) and the PUF protein activates transcription of the reporter via a transcription activation domain (AD). (C) β-Galactosidase expression in yeast expressing combinations of PUF proteins and RNA binding sites. PUF proteins were tested in combination with their own RNA targets and those of other PUF proteins. Shown are the results of a single set of LacZ assays using colonies grown on a plate and transferred to a filter. Citations to each PUF–RNA combination are as follows: C. elegans FBF/gld-1 RNA (Crittenden et al. 2002); C. elegans FBF/fem-3 RNA (Zhang et al. 1997); Drosophila Pumilio/hunchback (hb) NRE (Zamore et al. 1997; Sonoda and Wharton 1999); Xenopus Pumilio/hb RNA (Nakahata et al. 2003); S. cerevisiae Puf5p/HO RNA (Tadauchi et al. 2001); S. cerevisiae Puf3p/COX17 RNA (Olivas and Parker 2000; Jackson et al. 2004).
To date, all RNA targets of PUF proteins analyzed contain a UGU trinucleotide that is critical for binding (Zamore et al. 1997; Zhang et al. 1997; Souza et al. 1999; Olivas and Parker 2000; Nakahata et al. 2001; Tadauchi et al. 2001; Crittenden et al. 2002; Wang et al. 2002; Eckmann et al. 2004; Gerber et al. 2004; Jackson et al. 2004; Lamont et al. 2004). Thus the determinants of PUF specificity must lie elsewhere. Understanding the basis of selectivity will require mutational and structural analysis of other PUF–RNA complexes and comparisons of their protein–RNA contacts.
Two closely related PUF proteins, FBF-1 and FBF-2 (collectively referred to as FBF), function redundantly to regulate critical events in the C. elegans germline (Zhang et al. 1997; Crittenden et al. 2002; Lamont et al. 2004). They do so by binding to the 3′ UTRs of specific mRNAs and controlling their expression (Ahringer et al. 1992; Zhang et al. 1997; Crittenden et al. 2002; Lamont et al. 2004). Regulation of gld-1 and gld-3 mRNAs underlies FBF’s role in stem cell proliferation (Crittenden et al. 2002; Eckmann et al. 2004), while control of fem-3 mRNA is critical for FBF’s role in the switch from spermatogenesis to oogenesis (Zhang et al. 1997). fbf-1 and fbf-2 mRNAs are regulated by each other’s proteins to control germline patterning (Lamont et al. 2004). In addition, FBF is required to produce functional sperm (Luitjens et al. 2000).
Here we focus on the RNA binding specificities of FBF-1 and FBF-2. We combine yeast three-hybrid and biochemical assays to analyze a battery of mutations in the binding site. These data reveal nucleotides that are critical for FBF-1 binding, and provide a molecular explanation for how two natural binding sites differ in their affinity for FBF-1. Using this mutational data, we derive a reference sequence and search parameters that enable us to successfully search a C. elegans 3′ UTR database for potential FBF binding sites. We identify 18 additional sites in the 3′ UTRs of C. elegans mRNAs that bind FBF-1. Our approach accurately predicts FBF-1 binding sites and strongly suggests a means to identify new mRNA targets.
RESULTS
PUF proteins discriminate distinct RNA sequences
Comparison of PUF protein amino acid sequences suggests that PUF domains have similar structures. The eight imperfect repeats of the PUF domain form hydrogen bonds and stacking interactions with RNA (Fig. 1A; Wang et al. 2002). These PUF repeats are highly conserved among family members (Wickens et al. 2002). In particular, as judged by the cocrystal of human Pumilio1 and RNA, the amino acids in each repeat that directly contact RNA are present in virtually all PUF proteins (green residues in Fig. 1A).
To examine the specificity of the interactions between PUF proteins and RNA, the binding of a set of PUF proteins to a set of known PUF binding sites was tested using the yeast three-hybrid assay (Fig. 1B). In this assay, the hybrid RNA is tethered to the promoter of a reporter gene via a chimeric LexA/MS2 coat protein fusion. A test protein is produced as a transcription activation domain fusion (e.g., PUF/AD). If the RNA–protein interaction occurs, the reporter gene (HIS3 or LacZ) is transcribed. The affinity of the interaction is directly related to the level of reporter gene expression (Hook et al. 2005).
Each PUF protein was tested for binding with its natural binding site and with the sites of other PUF family members. Each RNA contained 20–30 nt of 3′ UTR sequence, including the UGU motif (Zamore et al. 1997; Zhang et al. 1997; Wickens et al. 2002). FBF-1 interacted only with C. elegans fem-3 and gld-1 elements while Drosophila Pumilio, and a close vertebrate relative (Xpum), bound only to sites in hb (hunchback) mRNA. S. cerevisiae Puf3p and Puf5p interacted solely with COX-17 and HO sites, respectively (Fig. 1C). We conclude that PUF proteins recognize distinct binding elements, despite the high conservation of amino acids thought to contact RNA.
We hypothesized that PUF proteins bind RNA in a manner related to human Pumilio1, which binds with maximum affinity to a short sequence containing UGU (Fig. 1A; Wang et al. 2002). The FBF binding region in the gld-1 and fem-3 3′ UTRs each contain a UGU trinucleotide and an AU pair 3 nt downstream (Fig. 2A). We define an 11-nt region corresponding to the minimal binding site of human Pumilio1, as analyzed previously (Wang et al. 2002; see Fig. 1A), to be the “core element” (Fig. 2A, black box).
FIGURE 2.
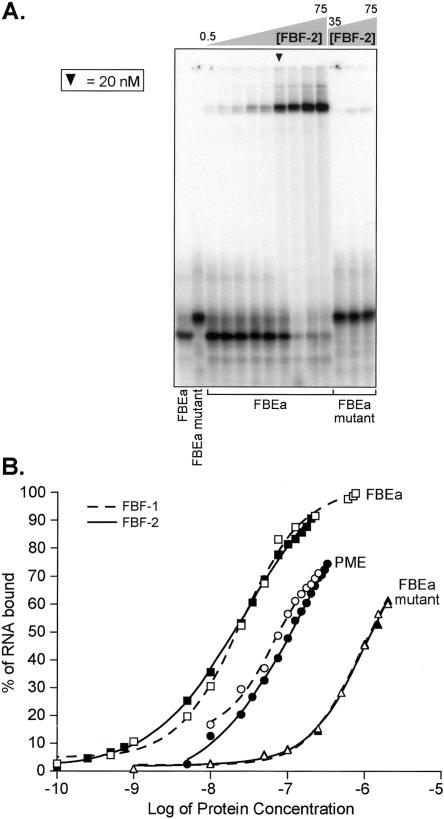
FBF-1 binding to the gld-1 FBEa and the fem-3 PME. (A) Binding sites. The sequence of two FBF binding sites are shown. The predicted core binding elements (see text) are highlighted in black. Numbering begins with +1 at the first uridine of the conserved UGU trinucleotide. (B) Affinities. Various concentrations of purified, recombinant FBF-1 were incubated with labeled FBEa and PME RNAs in vitro and analyzed by electrophoretic mobility shift assays. Protein concentrations are indicated above the gel; for ease of comparison, arrowheads indicate 20 nM FBF-1. Concentrations were 0.01, 0.05, 0.1, 0.5, and 1, then doubled beginning at 5 nM; the 0.01, 0.05, and 0.1 points were omitted for the PME. The leftmost lane lacks protein. Apparent Kd was estimated by fitting a curve to the percentages of total shifted material. Also presented are phenotypes observed in the yeast three-hybrid assay. LacZ activity was monitored in a luminometer using a linked assay (Hook et al. 2005); error terms represent the standard deviation of six repetitions. HIS3 activity was monitored by growth on increasing concentrations of 3-aminotriazole, a competitive inhibitor of His3p (Bernstein et al. 2002).
Differential binding to the gld-1 FBEa and the fem-3 PME
The affinities of FBF-1 for the elements in gld-1 and fem-3 mRNAs were compared in both the three-hybrid assay and in vitro. In the three-hybrid assay, the gld-1 FBEa interacts more strongly than the fem-3 PME: The difference in reporter gene activation is dramatic, judged by β-galactosidase activity and HIS3 phenotypes (Fig. 2B). The difference was quantified in vitro, using recombinant FBF-1 and radiolabeled 28-nt RNA oligonucleotides. In electrophoretic mobility shift assays, FBF-1 bound the FBEa with an apparent Kd of 23 ± 3 nM, and the PME with a Kd of 160 ± 90 nM (Fig. 2B). A version of the FBEa containing a UGU to ACA mutation (FBEa mutant) bound with an apparent Kd > 1500 nM (Fig. 2B). At high protein concentrations, this apparently non-sequence-specific binding generated complexes of slower electrophoretic mobility, both with the mutant FBEa and with the PME. Additional analyses will be needed to determine the composition of the multiple PME-containing complexes.
Similar experiments were performed with recombinant FBF-2. FBF-1 and FBF-2 are 91% identical in amino acid sequence (Zhang et al. 1997), regulate common mRNAs, and overlap in biological functions (Crittenden et al. 2002; Lamont et al. 2004). GST/FBF-2 bound the FBEa, PME, and FBEa mutant RNAs with affinities very similar to those of GST/FBF-1 (Fig. 3). We conclude that FBF-1 and FBF-2 have similar RNA binding characteristics in vitro.
FIGURE 3.
FBF-1 and FBF-2 exhibit similar binding characteristics. (A) FBF-2 binding the FBEa. Various concentrations of purified, recombinant FBF-2 were incubated with labeled FBEa and FBEa mutant RNAs in vitro and analyzed by electrophoretic mobility shift assays. FBF-2 concentrations tested with the FBEa were 0.5, 0.75, 1, 5, 10, 25, 35, 50, and 75 nM; the FBEa mutant was combined with 35, 50, and 75 nM protein. Arrowhead indicates 25 nM FBF-2. The two leftmost lanes lack protein. (B) FBF-1 versus FBF-2. Binding curves comparing the affinity of FBF-1 (dashed line) and FBF-2 (solid line) for three RNAs. Bound fraction of the RNA was calculated from densitometry readings of the shifted and unshifted RNA in each lane.
The core binding element
To identify nucleotides within the core binding element that are specifically recognized by FBF-1, we first used a screen in which the RNA had been randomized at positions +4 through +9 (the first U of UGU is designated “+1”; Fig. 4). The library of randomized core binding elements was prepared in a suitable DNA vector and introduced into yeast. In the library, the core sequence was flanked by alternating CA, not present in natural binding sites (Fig. 4). Yeast transformants were identified that contained RNAs that bound FBF-1 using the three-hybrid assay.
FIGURE 4.
A randomization-selection screen to identify optimal core binding sites. (Top) Sequence of the wild-type FBEa (“WT”) and the randomized library (“n” indicates all four bases were present equally). The sequences of 17 recovered RNAs that bind FBF-1 are indicated. (Bottom) A summary sequence derived from the selection experiments.
To do so, ~ 14,000 yeast transformants were screened on media lacking histidine and containing 5 mM 3-aminotriazole (3-AT), a competitive inhibitor of the HIS3 enzyme. Seventeen independent clones were recovered. These 17 sequences yield a consensus that closely resembles the tight-binding FBEa site, except for the G to A transition at position +4 (Fig. 4). No colonies were observed at higher concentrations of 3-AT using these artificial sequences, even though cells expressing the 28-nt wild-type FBEa sequence are viable under more stringent conditions. Together, these data suggest that nucleotides within the core element are essential, and that the FBEa site is nearly optimal. In addition, the failure to identify RNAs that bind as tightly as the wild-type site raises two possibilities: Either the introduced CA repeats inhibit FBF-1 binding or the sequences outside the core binding element are necessary for maximal binding.
To examine core sequences systematically, two series of mutations were analyzed (Fig. 5). In the first, adjacent di-nucleotide pairs were substituted (Fig. 5A); in the second, each position in the core was changed individually to the other 3 nt, generating 33 point mutations (Fig. 5B). Effects were quantified using the three-hybrid assay by measuring β-galactosidase activity. A 100-fold change in relative LacZ activity corresponds to an ~10-fold difference in Kd (Hook et al. 2005).
FIGURE 5.
Mutational analysis of the core binding element. (A) Double nucleotide substitutions in the FBEa. Substitutions are indicated in lowercase; core element is boxed. β-Galactosidase values are relative to the wild-type FBEa; error bars represent the standard deviation of six repetitions. (B) Single nucleotide substitutions in the FBEa. “x” indicates position of the substitutions; every nucleotide identity was assayed at each position. (Box) The core element. β-Galactosidase values are relative to the wild-type FBEa; error bars represent the standard deviation of six repetitions. The wild-type identity is labeled at each position. (C) Summary. A summary sequence of the FBF-1 core binding element based on the experiments in A and B.
Double mutations replacing either nucleotides within the UGUG (+1 to +4) or the AU dinucleotide (+7/+8) abolished the interaction (Fig. 5A). Dinucleotide mutations at the 5′ and 3′ ends of the core element (−2/−1 and +9/+10) had little effect (Fig. 5A).
Analysis of single mutations corroborate and extend these findings (Fig. 5B). The identity of positions +1, +2, +3, +7 and +8 are particularly constrained; at these positions, all nucleotide substitutions reduce LacZ reporter activity. A guanine to adenine transition at position +4 increased affinity for FBF. Surprisingly, a G to U transversion at position +4 was tolerated. No known PUF protein binding element contains a pyrimidine at this position (Wickens et al. 2002). Upstream of the UGU, only the change of −2C to −2G significantly reduced binding. Similarly, individual changes at positions −2 and −1, and +9 and +10 did not greatly reduce the binding of FBF-1 (Fig. 5B).
From these data, we compiled an optimal FBF-1 core element (Fig. 5C). This optimal sequence corresponds closely with the summary sequence obtained in the selection experiments (Fig. 4). The UGU at positions +1 through +3 and the AU at positions +7 and +8 are required for a strong interaction. The base identities at other positions of the core element are more flexible, though specific nucleotide substitutions substantially affect binding.
Sequences flanking the core element enhance binding
The previous data show that specific nucleotides in the core element are necessary for FBF-1 binding in the three-hybrid assay. To test whether these effects were direct, we used an electrophoretic mobility shift assay with purified recombinant FBF-1 and various chemically synthesized, 32P-labeled oligoribonucleotides (Fig. 6).
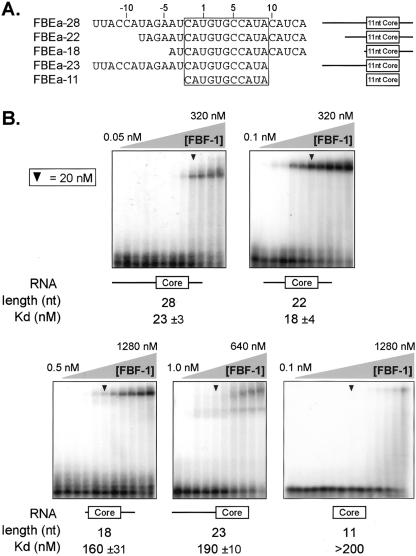
FIGURE 6.
The core binding element is insufficient for high-affinity binding. (A) Constructs. Sequences and schematic diagrams of oligoribonucleotides used. (Box) The core element. (B) Electrophoretic mobility shift assays using purified, recombinant FBF-1. Arrowheads indicate 20 nM FBF-1. Nucleotide length corresponds to oligoribonucleotides in A. Protein concentrations were 0.05, 0.1, 0.5, and 1, then doubled beginning at 5 nM; concentration ranges are indicated. The leftmost lane lacks protein. Apparent Kd was estimated by fitting a curve to the percentages of total shifted RNA, calculated from densitometry readings.
The longest RNA tested, FBEa-28, possesses 14 nt upstream of the UGU and 11 nt downstream. This RNA bound to FBF-1 with an apparent Kd of ~ 23 nM. An RNA with only 8 nt 5′ of the UGU and 11 nt downstream bound comparably (FBEa-22). However, further encroachment on either the 5′ or 3′ side reduced affinities fivefold to 10-fold. Moreover, an 11-nt RNA containing only the core element (−2 to +9) bound very poorly (apparent Kd > 200 nM). These data demonstrate directly that optimal FBF-1 binding requires RNA outside the core element, and thus differs from that of human Pumilio1 (Wang et al. 2002).
FBF-1 requires specific nucleotides 5′ of the core element
To examine the contribution of upstream nucleotides to FBF-1 binding, we created both single and double mutations from −4 to −14 of the FBEa (Fig. 7A,B). Several substitutions had deleterious effects. Most dramatically, changes from −7A to −7U and −5A to −5C reduced reporter activation in the three-hybrid assay more than two orders of magnitude (Fig. 7B).
FIGURE 7.
FBF-1 is sensitive to mutation upstream of the core binding element. (A) Double nucleotide substitutions upstream of the core binding element. Substitutions are indicated in lowercase; core element is boxed. β-Galactosidase values are relative to the wild-type FBEa; error bars represent the standard deviation of six repetitions. (B) Single nucleotide substitutions upstream of the core binding element. “x” indicates the position of the substitutions; every nucleotide identity was assayed at each position. (Box) The core element. β-Galactosidase values are relative to the wild-type FBEa; error bars represent the standard deviation of six repetitions. The wild-type identity is labeled at each position. (C) Affinities. A table of apparent binding constants of FBEa RNA oligonucleotides carrying substitutions at either position -8 or -7; the lowercase letter indicates the substitution. Apparent Kd was estimated by fitting a curve to the percentages of total shifted RNA, calculated from densitometry readings. (D) Example gel mobility shift assays. Two mutant FBEa oligoribonucleotides carrying substitutions at position -7, shifted with purified recombinant FBF-1. Arrowheads indicate 20 nM FBF-1. Protein concentrations were doubled beginning at 5 nM in lane 3 of each gel; the leftmost lane lacks protein and the adjacent lane contains 1 nM FBF-1.
To determine whether these effects were direct, we analyzed six RNAs in vitro, comprising all possible nucleotide changes at positions -7 and -8 (Fig. 7C,D). The in vitro affinities recapitulate the three-hybrid results. For example, mutation of position -7A to U decreased binding approximately 10-fold while a C substitution had little effect (Fig. 7D). We conclude that specific nucleotide identities in the region upstream of the core binding element are critical for interaction with FBF-1.
FBF-1 requires nucleotides 3′ of the core element
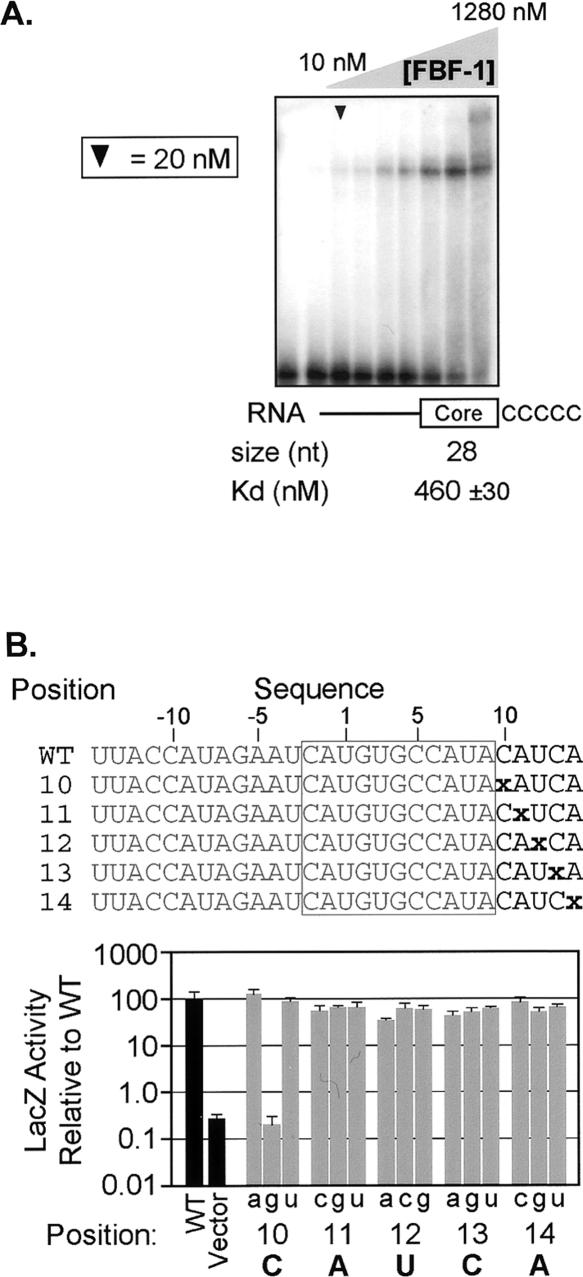
An RNA that lacks nucleotides downstream of the core element binds poorly to FBF-1 (Fig. 6B). To test whether this effect was due to a requirement for specific sequences, we prepared an RNA in which five Cs were placed downstream of the core element. This RNA interacted very weakly with FBF-1 in vitro (460 ± 30 nM), suggesting that specific sequences are important (Fig. 8A).
FIGURE 8.
Nucleotides downstream of the core binding element are critical for interaction with FBF-1. (A) A 3′ requirement. An electrophoretic mobility shift assay in which the 3′ region of the FBEa has been substituted with a poly(C) track. Arrowhead indicates 20 nM FBF-1. Concentrations were doubled beginning at 10 nM; the leftmost lane lacks protein. Apparent Kd was estimated by fitting a curve to the percentages of total shifted material, calculated from densitometry readings. (B) Single nucleotide substitutions downstream of the core binding element. “x” indicates the position of the substitutions; every nucleotide identity was assayed at each position. (Box) The core element. β-Galactosidase values are relative to the wild-type FBEa; error bars represent the standard deviation of six repetitions. The wild-type identity is labeled at each position. (C) Four-nucleotide substitutions downstream of the core binding element. Lowercase letters indicate the substitution; the core element is boxed. β-Galactosidase values are relative to the wild-type FBEa; error bars represent the standard deviation of four repetitions.
To analyze this requirement in greater detail, we introduced a variety of mutations in the downstream region (Fig. 8B). Of all the possible single substitutions, only the C to G transversion at position +10 reduced the interaction with FBF-1 significantly. Similarly, double mutations did not greatly affect the interaction (data not shown). However, a 4-nt change abrogated binding (Fig. 8C).
To test the generality of this effect, we examined the PME in the fem-3 3′ UTR, which binds much more weakly to FBF-1 (see Fig. 2B). As with FBEa, substitutions of four consecutive bases downstream of the core element decreased binding (Fig. 8C) without introducing predictable secondary structure (assessed by MFOLD; Zuker 1989). In FBEa, which binds tightly, the contribution of nucleotides in the downstream region may be masked by the high affinity of the core element. Taken together, these results demonstrate that FBF-1 is sensitive to nucleotide identity downstream of the core binding element but tolerates many substitutions.
Mutations that increase PME affinity
We next examined the cause of the difference in affinity of FBF-1 for the FBEa and PME (Fig. 2). We compared the 22-nt region important for FBEa binding to the corresponding region surrounding the PME. Ten of the 22 nt differ between the sites. We tested the contribution of these nucleotides to FBF-1 binding by creating series of RNAs in which PME nucleotide identities were changed to those of the FBEa. Several of these single and double mutations increase binding (Fig. 9). Most dramatically, a double mutation (+5C/+9A) increases affinity nearly to that of the FBEa. The individual single substitutions also increase binding; their effects appear to be additive.
FIGURE 9.
The weak binding PME can be converted to a strong FBF-1 binding site. Substitutions changing the nucleotide identity from that of the PME to the FBEa are in lowercase; the core element is boxed. β-Galactosidase values are relative to the wild-type FBEa in the three-hybrid assay; error bars represent the standard deviation of four repetitions.
New FBF-1 binding sites in C. elegans 3′ UTRs
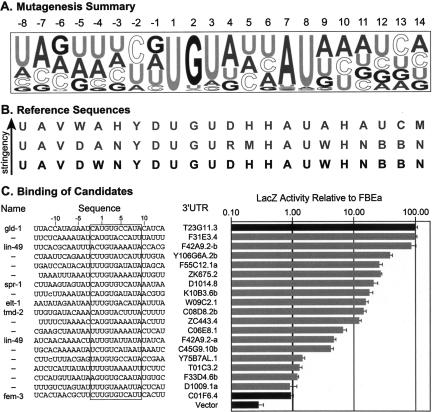
We next developed a reference sequence that could identify potential FBF mRNA targets in a sequence database with reasonable confidence. From our mutagenesis results (summarized in Fig. 10A), we derived three sequences that describe the FBF-1 binding site (Fig. 10B). These sequences represented an optimal FBF-1 binding site at a range of stringencies. A database of 10,910 C. elegans 3′ UTR sequences (as annotated by EST alignments) was constructed using an AceDB dump (http://www.acedb.org or http://www.wormbase.org/db/searches/advanced/dumper) from WormBase release WS122. This database was searched for matches to the reference sequences using PatScan (Dsouza et al. 1997).
FIGURE 10.
New FBF-1 binding sites identified in a database using a reference sequence. (A) Summary of mutagenesis. A representation of the extended FBF-1 binding site; letter height indicates binding strength of the single mutant relative to other mutants at that position. (B) Search reference sequences. Reference sequences used to search the 3′ UTR database; arrow indicates increasing stringency, bold, the consensus used for the optimized search. Reference sequence abbreviations: “B” is C, G, or U; “D” is A, G, or U; “H” is A, C, or U; “M” is A or C; “N” is A, C, G, or U; “R” is A or G; “V” is A, C, or G; “W” is A or U; “Y” is C or U. (C) New binding sites. A sampling of predicted FBF-1 binding sites that interact at least as strongly as the PME; dashes (—) distinguish multiple binding sites in a single 3′ UTR. The entire sequence tested is shown; lowercase letters indicate substitutions introduced to increase expression in yeast; the core element is boxed. β-Galactosidase values are relative to the wild-type FBEa in the three-hybrid assay; error bars represent the standard deviation of three repetitions.
Both the full (22 nt) and core element (11 nt) sequences were tested in a total of 21 database searches. By permitting various combinations of insertions, deletions, and mismatches relative to the motif pattern, we derived optimal search parameters empirically. The optimal reference sequence and search parameters were selected to yield fewer than 600 potential sites and a high number of the known targets of FBF regulation. The optimal consensus spanned 22 nt and permitted only those nucleotide identities that bound at least 65% as well as wild-type FBEa (Fig. 10B, bold). This reference sequence was used to search the database of 3′ UTR sequences, allowing one mismatch with the reference, except in the UGU trinucleotide, for which an exact match was required.
One hundred fifty possible FBF binding sites were identified. We assayed 25 randomly selected sites for interaction with FBF-1 in the three-hybrid assay; a representative group is shown in Figure 10C. Of the 25 predicted sites, 18 (72%) interacted with FBF-1 with biologically relevant affinities; all bound FBF-1 at least as well as the PME and two bound as strongly as the FBEa (Fig. 10C).
For comparison, we searched the 3′ UTR database using identical parameters with only the core reference sequence. This search identified 5107 potential FBF binding sites. We tested 16 sites and found that FBF bound only four as strongly as it does the PME (data not shown). Thus the more extended, 22-nt sequence yielded a higher fraction of true FBF binding sites than did the 11-nt sequence (p = 0.002, calculated by t test). We conclude that the 22-nt reference sequence and search parameters identify a restricted list of sites that do indeed bind FBF-1, and a low rate of false positives.
DISCUSSION
Our analysis of the RNA sequence specificity of FBF binding leads to the following main conclusions. First, FBF and other PUF proteins bind specifically to their cognate RNAs, and not to those of other PUF proteins. This finding is consistent with the observation that the five PUF proteins of S. cerivisiae appear to bind to largely nonoverlapping sets of mRNAs (Gerber et al. 2004). Second, FBF-1 and FBF-2 exhibit similar specificity and affinity for RNA. Third, high affinity interactions between FBF-1 and RNA require both a core sequence and flanking nucleotides. Thus its apparent binding site differs from that of human Pumilio1 with RNA, examined by crystallography (Wang et al. 2002). We argue below that this extended specificity may be common among PUF proteins. Fourth, we have identified new FBF binding sites among C. elegans 3′ UTRs.
The binding site and its recognition
To identify the RNA elements involved in binding FBF-1, we first analyzed base substitutions within the “core element” (Wang et al. 2002). Mutational analysis and three-hybrid selection experiments revealed strict sequence requirements at five positions (+1, +2, +3, +7, and +8). Specific identities at these positions are necessary but not sufficient for high-affinity binding, as RNAs possessing only the core binding site bind poorly (Fig. 6). In fact, the minimal RNA sequence that binds FBF-1 with maximum affinity was 22 nt in length, and contained sequence both upstream and downstream of the core element.
In natural PUF binding sites, the nucleotide following the UGU is a purine. However, our mutational analysis reveals that a U permits binding to FBF with a similar affinity. These observations suggest that a constraint other than RNA–protein binding affinity acts at this position. Most simply, a PUF protein partner may demand a purine at this position in the PUF/RNA complex.
FBF possesses a repeated amino acid sequence spaced at ~ 40 amino acid intervals, as do all other PUF proteins s(Wickens et al. 2002). In human Pumilio1, these amino acids lie within so-called RNA-recognition helices and contact the core element, including UGU (Fig. 1A; Wang et al. 2002). Almost certainly, FBF recognizes UGU using the same recognition helices; indeed, compensatory mutants in FBF-2 and the FBEa show that the eight α-helices align with the RNA core element in the same fashion as in human Pumilio1 (L. Opperman, B. Hook, M. DeFino, D. Bernstein, and M. Wickens, in prep.).
How then do flanking nucleotides affect FBF binding? In principle, they could form higher order structures that promote FBF recognition. However, stable secondary structures seem unlikely, based on folding predictions and the need for the RNA core element to be single stranded (D. Bernstein and M. Wickens, unpubl.). Alternatively flanking nucleotides may make additional and novel contacts with the RNA binding face of the PUF domain. The recognition helices form a surface wide enough to accommodate a second RNA sequence, since the core element of the RNA lies asymmetrically on the surface of α-helicies (Edwards et al. 2001; Wang et al. 2001, 2002). A conserved pattern of charged and hydrophobic amino acids in the RNA recognition helices do not contact the core element in the human Pumilio/RNA cocrystal (Wang et al. 2002). Analogous regions of FBF, which are predominantly basic, might interact with flanking nucleotides. Backbone contacts could explain the insensitivity of the region to single and double substitutions. It is also possible that flanking nucleotides contact regions of FBF outside the recognition helix face.
Flanking nucleotides in PUF–RNA interactions
The role of nucleotides outside the core element in FBF recognition is likely to reveal a common theme in PUF/ RNA recognition. Indeed, previous studies of other PUF proteins are consistent with this conclusion, though it has not been substantiated extensively. High-affinity binding of Drosophila Pumilio in vitro is sensitive to mutations upstream of the core element (Zamore et al. 1997) and requires a 25-nt RNA (Wharton et al. 1998; Zamore et al. 1999). Murine Pumilio2 recognizes specific nucleotides 3′ of the core element (White et al. 2001). Indeed, human Pumilio1 is the only PUF protein known to bind tightly to a core element alone, but the RNA analyzed is not its natural mRNA target (Wang et al. 2002).
Sequence variation outside the core element may enable differential regulation by a single species of PUF protein. For example, the ternary complex formed by Drosophila Pumilio and Nanos on the hunchback 3′ UTR is abrogated by double nucleotide changes at positions −3/−4, though the affinity of Pumilio for this RNA is unaffected (Sonoda and Wharton 1999). Furthermore, Brat protein interacts with the ternary complex formed on hunchback, but not on cyclin B mRNA (Sonoda and Wharton 2001). Thus, the identity of flanking nucleotides can contribute to regulation by modulating PUF binding or the selective binding of its protein partners.
Our analyses of the FBEa binding site reveals one solution to high affinity FBF-1 binding. The relationship between the optimal sites we deduce and binding to mRNAs in vivo may be complex. First, while we have focused on the binding specificity of FBF in isolation, FBF’s protein partners may affect binding specificity. In vivo, FBF interacts with at least three proteins, NOS-3, CPB-1, and GLD-3 (Kraemer et al. 1999; Luitjens et al. 2000; Eckmann et al. 2002). Variations among RNA binding sites may enhance binding to one FBF/partner pair, but not another. Second, the optimal binding site we deduced was based on the interaction of FBF with one particular site, the FBEa; this may not be the only sequence that yields high affinity binding. For example, some mutations to the PME binding site cause changes in affinity not predicted by the FBEa-derived optimal binding site. In addition, some sites differ in flanking sequence from the FBEa, yet bind FBF-1 well.
New FBF binding sites in C. elegans 3′ UTRs
Bioinformatic searches using a reference sequence derived from the FBEa mutagenesis and empirically determined search parameters yielded 150 candidate binding sites, 70% (105) of which likely interact with FBF-1 at biologically relevant affinities. Thus our analysis accurately models an FBF binding site and has predictive value. Importantly, however, our model does not identify all FBF binding sites. Known FBF binding sites in the 3′ UTRs of fbf-1, fbf-2, and gld-3 (Crittenden et al. 2002; Eckmann et al. 2004; Lamont et al. 2004) were not predicted. These binding sites possess the conserved core element, but were excluded in the computational search because they differ in flanking sequences. The flanking sequences in these RNAs could be recognized differently by FBF, or form structures that are similar despite their sequence divergence. We conclude that analysis identifies a subset of candidate binding sites that assemble a FBF-mediated protein complex.
The 3′ UTR database we analyzed contained a UTR for ~ 50% of C. elegans genes; since we isolated ~ 105 probable FBF binding sites, the total number of predicted targets that conform to this consensus is roughly 210. Each yeast PUF protein appears to interact with between 40 and 200 3′ UTRs (Gerber et al. 2004). Analysis of other high-affinity binding sites of FBF, with different flanking regions, would increase the number of possible mRNA targets.
Our approach to identifying new candidate target mRNAs complements those of Jacobs Anderson and Parker (2000) and Gerber et al. (2004). Jacobs Anderson and Parker searched for sequence elements present in multiple mRNAs in S. cerevisiae. They identified a short sequence motif present in many mitochondrial mRNAs, which ultimately proved to be a binding site for S. cerevisiae PUF3 (Olivas and Parker 2000; Jackson et al. 2004). Gerber et al. used DNA microarrays to identify mRNAs physically associated with tagged PUF proteins after immunoprecpitation from yeast extracts. Many mRNA species that bound a given yeast PUF protein contained a related, degenerate sequence; the sequence was diagnostic of that particular PUF protein and invariably contained a UGU trinucleotide. These findings imply a common binding site among mRNAs that bind a particular PUF protein (Gerber et al. 2004). For certain PUF proteins, no diagnostic sequence was apparent, and relative affinities to different binding sites were not examined in detail.
FBF binds to and regulates at least four mRNAs in vivo—fem-3, gld-1, gld- 3, and fbf itself (Zhang et al. 1997; Crittenden et al. 2002; Eckmann et al. 2004; Lamont et al. 2004). These regulatory events control stem cell divisions and cell fates in the germline. Our data provide a list of candidate mRNAs that may be controlled by FBF to effect these and other processes. FBF-1 protein is spatially and temporally regulated, and only some predicted mRNA targets may encounter FBF-1 protein in vivo. Moreover, some candidate mRNAs may bind another PUF protein that shares FBF’s RNA binding specificity but has a different expression pattern. However, the relatively small number of candidates can now be winnowed further using conventional molecular approaches, including RNAi. Other PUF/RNA interaction networks should be accessible through the same combination of mutagenesis and informatics approaches.
MATERIALS AND METHODS
Yeast three-hybrid assay constructs
Three-hybrid assays were performed as described (Bernstein et al. 2002). PUF proteins fused with the Gal4 activation domain were expressed from pACT and pACT2 plasmids. N-terminal truncations were expressed for FBF-1 (amino acids 121–614), Pumilio (amino acids 1093–1426), and Puf5p (amino acids 26–858). The entire ORF of Puf3p was used, while only the first four repeats of the XPum PUF domain was expressed. DNA oligonucleotides were designed to express various RNA sequences and cloned into the XmaI and SphI sites of pIIIA/MS2-2. Assays were preformed in the yeast strain YBZ-1 (Bernstein et al. 2002; Hook et al. 2005).
Yeast three-hybrid assay
β-Galactosidase activity was measured using filter lift and liquid assays (Bernstein et al. 2002). For filter lift assays, 10,000 cells were grown on selective media for 48 h, transferred to nitrocellulose membrane, and exposed to X-gal for 1 h. For liquid assays, cells were grown in selective media to an OD600 of 0.1–0.3 and mixed with an equal volume of Beta-Glo (Promega) reagent. Luminescence was measured after 1 h and normalized for cell number (Hook et al. 2005). HIS3 activation was monitored by streaking cells on increasing concentrations of 3-aminotriazole (3-AT), a competitive inhibitor of His3p.
Protein construct and purification
FBF-1 (amino acids 121–614) and FBF-2 (amino acids 121–632) were cloned into pGex6P1 (Amersham) with an N-terminal TEV protease cleavage site. The resulting GST fusion protein was expressed in BL-21 gold Escherichia coli cells (Invitrogen), grown in liquid culture to an OD600 of 0.6–0.8. Protein expression was induced overnight at 16°C by the addition of 1 mM IPTG. Cells were harvested by centrifugation and washed with Bug Wash (50 mM Tris-HCl at pH 8.0 and 10% sucrose). Cell pellets were frozen in liquid nitrogen and resuspended in Lysis Buffer (20 mM HEPES-KOH at pH 7.4, 0.5 M NaCl, 5 mM DTT, 0.02% Tween-20, and protease inhibitor [Roche]). The cells were lysed with a final lysozyme concentration of 0.5 mg/mL on ice for 30 min before freezing in liquid nitrogen. MgCl2 to a final concentration of 2 mM and DNase1 to 10 mg/mL were added to the thawed pellet and incubated on ice for 20 min. The solution was frozen in liquid nitrogen, thawed, clarified by centrifugation, and passed over glutathione sepharose resin (Amersham). Protein was cleaved with recombinant GST-TEV protease in the presence of glutathione sepharose resin. Protein purity was estimated by SDS gel electrophoresis and concentration measured by a Bradford assay. Protein used for experiments in Figure 3 was eluted from the glutathione sepharose resin with 50 mM reduced glutathione (Sigma) and dialyzed into 50 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, 1 mM DTT, and 50% glycerol instead of GST-TEV protease cleavage.
Electrophoretic mobility shift assays
One hundred femtomoles 32P-end-labeled RNA oligoribonucleotides (IDT and Dharmacon) were combined with proteins over a range of protein concentrations. Protein and RNA were incubated at room temperature for 30 min in 10 mM HEPES (pH 7.4), 1 mM EDTA, 2 μg yeast tRNA (Sigma), 50 mM KCl, 2 mM DTT, and 0.02% Tween-20. In all experiments except those shown in Figure 3, FBF protein was generated from a GST-TEV-FBF fusion protein, by cleavage with TEV protease. The uncleaved fusion proteins were used in the experiments in Figure 3.
Four microliters of loading dye (10% [v/v] Ficoll 400,000, 0.05% [w/v] xylene cyanol) were added before loading on a pre-run native polyacrylamide gel (6% [w/v] 29:1 acrylamide/bisacrylamide, 0.5× TBE). Gels were resolved at 200 V for 2 h at 4°C. The fraction of retarded RNA, relative to the total in the incubation, was determined using ImageQuant (Amersham).
Acknowledgments
We thank Judith Kimble and members of the Wickens and Kimble laboratories for helpful discussions and comments on the manuscript. We appreciate the support and advice of Richard Durbin of the Wellcome Trust Sanger Institute. We are grateful to Ann Palmenberg and Jean-Yves Sgro for their suggestions concerning the informatics analysis of FBF binding sites. Laura Vanderploeg (Media Lab, Biochemistry Department, University of Wisconsin–Madison) provided invaluable assistance preparing figures. A.H. was supported by an M.R.C. Studentship and the Wellcome Trust. This work was supported by N.I.H. research grants GM31892 and GM50942 to M.W.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7255805.
REFERENCES
- Ahringer, J., Rosenquist, T.A., Lawson, D.N., and Kimble, J. 1992. The Caenorhabditis elegans sex determining gene fem-3 is regulated post-transcriptionally. EMBO J. 11: 2303–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain, F.H., Gubser, C.C., Howe, P.W., Nagai, K., Neuhaus, D., and Varani, G. 1996. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature 380: 646–650. [DOI] [PubMed] [Google Scholar]
- Bernstein, D.S., Buter, N., Stumpf, C., and Wickens, M. 2002. Analyzing mRNA–protein complexes using a yeast three-hybrid system. Methods 26: 123–141. [DOI] [PubMed] [Google Scholar]
- Crittenden, S.L., Bernstein, D.S., Bachorik, J.L., Thompson, B.E., Gallegos, M., Petcherski, A.G., Moulder, G., Barstead, R., Wickens, M., and Kimble, J. 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663. [DOI] [PubMed] [Google Scholar]
- Dsouza, M., Larsen, N., and Overbeek, R. 1997. Searching for patterns in genomic data. Trends Genet. 13: 497–498. [DOI] [PubMed] [Google Scholar]
- Eckmann, C.R., Kraemer, B., Wickens, M., and Kimble, J. 2002. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell 3: 697–710. [DOI] [PubMed] [Google Scholar]
- Eckmann, C.R., Crittenden, S.L., Suh, N., and Kimble, J. 2004. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168: 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, T.A., Pyle, S.E., Wharton, R.P., and Aggarwal, A.K. 2001. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105: 281–289. [DOI] [PubMed] [Google Scholar]
- Gerber, A.P., Herschlag, D., and Brown, P.O. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M.W. and Kühn, L.C. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. 93: 8175–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook, B., Bernstein, D., Zhang, B., and Wickens, M. RNA–protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA 11: 227–233. [DOI] [PMC free article] [PubMed]
- Jackson Jr., J.S., Houshmandi, S.S., Lopez Leban, F., and Olivas, W.M. 2004. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA 10: 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson, J.S. and Parker, R. 2000. Computational identification of cis-acting elements affecting post-transcriptional control of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 28: 1604–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer, B., Crittenden, S., Gallegos, M., Moulder, G., Barstead, R., Kimble, J., and Wickens, M. 1999. NANOS-3 and FBF proteins physically interact to control the sperm/oocyte switch in C. elegans. Curr. Biol. 9: 1009–1018. [DOI] [PubMed] [Google Scholar]
- Lamont, L.B., Crittenden, S.L., Bernstein, D., Wickens, M., and Kimble, J. 2004. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell 7: 697–707. [DOI] [PubMed] [Google Scholar]
- Luitjens, C., Gallegos, M., Kraemer, B., Kimble, J., and Wickens, M. 2000. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes & Dev. 14: 2596–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata, S., Katsu, Y., Mita, K., Inoue, K., Nagahama, Y., and Yamashita, M. 2001. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 276: 20945–20953. [DOI] [PubMed] [Google Scholar]
- Nakahata, S., Kotani, T., Mita, K., Kawasaki, T., Katsu, Y., Nagahama, Y., and Yamashita, M. 2003. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 120: 865–880. [DOI] [PubMed] [Google Scholar]
- Olivas, W.M. and Parker, R. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19: 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda, J. and Wharton, R.P. 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes & Dev. 13: 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Drosophila brain tumor is a translational repressor. Genes & Dev. 15: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, G.M., da Silva, A.M., and Kuspa, A. 1999. Starvation promotes Dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development 126: 3263–3274. [DOI] [PubMed] [Google Scholar]
- Tadauchi, T., Matsumoto, K., Herskowitz, I., and Irie, K. 2001. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Zamore, P.D., and Tanaka Hall, T.M. 2001. Crystal structure of a Pumilio homology domain. Mol. Cell 7: 855–865. [DOI] [PubMed] [Google Scholar]
- Wang, X., McLachlan, J., Zamore, P.D., and Tanaka Hall, T.M. 2002. Modular recognition of RNA by a human Pumilio-homology domain. Cell 110: 501–512. [DOI] [PubMed] [Google Scholar]
- Wharton, R.P., Sonoda, J., Lee, T., Patterson, M., and Murata, Y. 1998. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1: 863–872. [DOI] [PubMed] [Google Scholar]
- White, E.K., Moore-Jarrett, T., and Ruley, H.E. 2001. PUM2, a novel murine PUF protein, and its consensus RNA-binding site. RNA 7: 1855–1866. [PMC free article] [PubMed] [Google Scholar]
- Wickens, M., Bernstein, D.S., Kimble, J., and Parker, R. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18: 150–157. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Williamson, J.R., and Lehmann, R. 1997. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3: 1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Bartel, D.P., Lehmann, R., and Williamson, J.R. 1999. The PUMILIO–RNA interaction: A single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry 38: 596–604. [DOI] [PubMed] [Google Scholar]
- Zhang, B., Gallegos, M., Puoti, A., Durkin, E., Fields, S., Kimble, J., and Wickens, M. 1997. A conserved RNA binding protein that regulates patterning of sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 1989. Computer prediction of RNA structure. Methods Enzymol. 180: 262–288. [DOI] [PubMed] [Google Scholar]