Abstract
The multisubunit eukaryotic initiation factor (eIF) 3 plays various roles in translation initiation that all involve interaction with 40S ribosomal subunits. eIF3 can be purified in two forms: with or without the loosely associated eIF3j subunit (eIF3j+ and eIF3j−, respectively). Although unlike eIF3j+, eIF3j− does not bind 40S subunits stably enough to withstand sucrose density gradient centrifugation, we found that in addition to the known stabilization of the eIF3/40S subunit interaction by the eIF2•GTP•Met-tRNAiMet ternary complex, eIF3j−/40S subunit complexes were also stabilized by single-stranded RNA or DNA cofactors that were at least 25 nt long and could be flanked by stable hairpins. Of all homopolymers, oligo(rU), oligo(dT), and oligo(dC) stimulated the eIF3/40S subunit interaction, whereas oligo(rA), oligo(rG), oligo(rC), oligo(dA), and oligo(dG) did not. Oligo(U) or oligo(dT) sequences interspersed by other bases also promoted this interaction. The ability of oligonucleotides to stimulate eIF3/40S subunit association correlated with their ability to bind to the 40S subunit, most likely to its mRNA-binding cleft. Although eIF3j+ could bind directly to 40S subunits, neither eIF3j− nor eIF3j+ alone was able to dissociate 80S ribosomes or protect 40S and 60S subunits from reassociation. Significantly, the dissociation/anti-association activities of both forms of eIF3 became apparent in the presence of either eIF2-ternary complexes or any oligonucleotide cofactor that promoted eIF3/40S subunit interaction. Ribosomal dissociation and anti-association activities of eIF3 were strongly enhanced by eIF1. The potential biological role of stimulation of eIF3/40S subunit interaction by an RNA cofactor in the absence of eIF2-ternary complex is discussed.
Keywords: eukaryotic initiation factor 1, eukaryotic initiation factor 3, mRNA, ribosome dissociation, translation
INTRODUCTION
Initiation of translation starts from separated 40S and 60S ribosomal subunits and involves at least 11 initiation factors. Initiator Met-tRNAiMet is recruited to the free 40S subunit as a ternary complex with eukaryotic initiation factor (eIF) 2 and GTP (Schreier and Staehelin 1973). eIF2-ternary complexes can bind to 40S subunits directly, but this interaction is strongly enhanced by eIF3, eIF1A, and eIF1 (Benne and Hershey 1978; Peterson et al. 1979; Chaudhuri et al. 1997, 1999; Majumdar et al. 2003). The resulting 43S preinitiation complex containing the eIF2-ternary complex, eIF3, eIF1, and eIF1A is recruited to the 5′-terminal cap-proximal region of mRNA, the secondary structure of which is unwound by eIF4F, eIF4A, and eIF4B (Gingras et al. 1999). eIF3 binds directly to mRNA and to the eIF4G subunit of eIF4F, and these interactions are likely to be important for binding of the 43S complex to the 5′-terminal region of mRNA (Setyono et al. 1984; Westermann and Nygard 1984; Carberry and Goss 1991; Imataka and Sonenberg 1997). After binding to an mRNA, the 43S complex scans downstream to the first AUG triplet in a favorable context, stops there and forms a 48S initiation complex with an established codon–anticodon interaction (Kozak 1989). At this stage, eIF5 stimulates hydrolysis of eIF2-bound GTP, eIF2 · GDP is lost, and Met-tRNAiMet is left in the peptidyl (P) site of the 40S subunit (Das and Maitra 2001; Unbehaun et al. 2004). The final stage in initiation is the displacement of other factors from the 40S subunit and its joining with a 60S subunit, which requires the initiation factor eIF5B (Pestova et al. 2000; Unbehaun et al. 2004). Thus, initiation requires a pool of separated 40S and 60S subunits, and after each cycle of translation, 80S ribosomes must therefore be released from mRNA as separated 40S and 60S subunits that must be protected immediately from reassociation, because free 40S and 60S subunits can reassociate at the physiological salt concentrations of the cytoplasm (Falvey and Staehelin 1970; Henshaw et al. 1973). Although the mechanism of post-termination recycling of ribosomes in eukaryotes is not understood (Janosi et al. 1996; Kisselev and Buckingham 2000), eIF3 and, to some extent, eIF1A have been implicated in preventing reassociation of free 40S and 60S subunits and in dissociation of empty 80S ribosomes (Thompson et al. 1977; Trachsel and Staehelin 1979; Thomas et al. 1980a). Native 40S subunits in the cytoplasm are stably associated with eIF3 (Freienstein and Blobel 1975) and purified eIF3 can bind directly to 40S subunits in the absence of other initiation factors (Benne and Hershey 1976; Peterson et al. 1979; Trachsel and Staehelin 1979; Nygard and Westermann 1982). The ability of eIF3 to bind to 40S subunits in the absence of other initiation factors depends strictly on the presence of its loosely associated eIF3j subunit (Fraser et al. 2004).
Although eIF3 plays significant roles at multiple stages in initiation from anti-association of ribosomal subunits to 48S complex formation, the molecular basis for its many activities is poorly understood. eIF3 is the largest initiation factor with a mass of >650 kDa, which, as we found recently, contains 13 rather than 12 nonidentical subunits in mammals (Mayeur et al. 2003; Unbehaun et al. 2004; Fig. 1A). The 13th subunit of eIF3 was identified as GA17, a protein of unknown function that contains a PCI domain like those found in eIF3a, eIF3c, and eIF3e subunits. eIF3 binds to many other initiation factors, including eIFs 1, 1A, 2, 4B, 4G, and 5 (Methot et al. 1996; Bandyopadhyay and Maitra 1999; Fletcher et al. 1999; Asano et al. 2000; Válasek et al. 2002; Olsen et al. 2003) and participates in organization of higher order multifactor complexes on the ribosome. However, since all activities of eIF3 involve its interaction with the 40S subunit, it is the single most important of eIF3’s partners.
FIGURE 1.
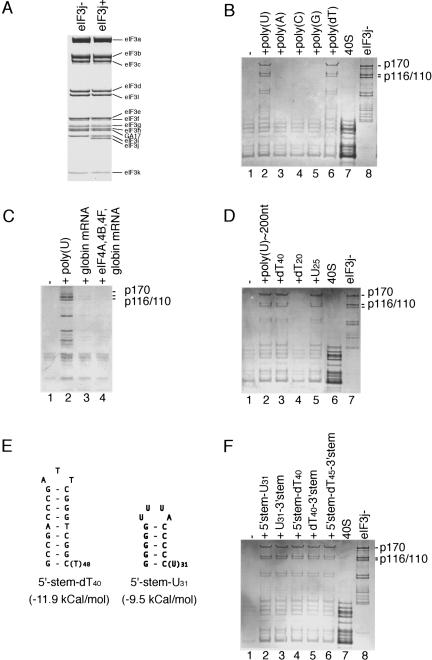
Subunit composition of HeLa eIF3j+ and eIF3j− analyzed by electrophoresis on 4%–12% Bis-Tris NuPAGE gel using MOPS buffer system (Invitrogen), followed by Coomassie staining (A). Presence of eIF3 in the peak fraction corresponding to 40S ribosomal subunits after sucrose density gradient centrifugation, analyzed by electrophoresis on (B,D,F) 4%–12% Bis-Tris NuPAGE gel using MES buffer system (Invitrogen) or (C) SDS-11% polyacrylamide, followed by Coomassie staining. (B,F) 40S subunits (lane 7) and eIF3j− (lane 8) and (D) 40S subunits (lane 6) and eIF3j− (lane 7) were incubated in the absence (lane 1) or in the presence of poly- or oligonucleotides, as indicated, and separated by centrifugation in 10%–30% linear sucrose density gradients. (C) 40S subunits were incubated with eIF3j− alone (lane 1), eIF3j−and poly(U) (lane 2), eIF3j− and globin mRNA (lane 3) or eIF3j−, globin mRNA, eIFs 4A, 4B, and 4F (lane 4) and separated by sucrose density gradient centrifugation. eIF3 subunits are labeled to the right of panels A–D and F. (E) Sequences and structures of 5′ stem-U31 and 5′stem-dT40 oligonucleotides.
Here we report that although in contrast to eIF3j+, eIF3j− does not bind 40S subunits stably enough to withstand sucrose density gradient centrifugation, eIF3j−/40S subunit interaction can be stabilized not only by eIF2-ternary complexes, but also by a single-stranded U- or dT-rich oligonucleotide cofactor, which most likely binds to the mRNA-binding cleft of the 40S subunit. We also found that contrary to common assumption, eIF3 alone (either with or without the eIF3j subunit) or even in combination with eIF1 and eIF1A does not possess ribosomal dissociation/ anti-association activities and that these activities of eIF3 require the presence of either the eIF2 ternary complex, or a compatible single-stranded oligonucleotide cofactor, in which case they are strongly enhanced by eIF1. We propose that the ability of eIF3 to associate stably with the 40S subunit and to protect it from association with the 60S subunit in the presence of RNA cofactor and the absence of other initiation factors, including eIF2 ternary complex, influences the mechanism of initiation factor release during ribosomal subunit joining and may even implicate eIF3 in post-termination recycling of ribosomes.
RESULTS
Stimulation of stable binding of eIF3j− to 40S subunits by oligonucleotide cofactors
Consistent with a recent report (Fraser et al. 2004), purified eIF3j− (Fig. 1A) and 40S subunits that were fully active in 48S complex formation (data not shown) did not associate stably enough to withstand sucrose density gradient centrifugation. Thus eIF3j− was not present in the peak corresponding by optical density to 40S subunits (Fig. 1B, lane 1). However, eIF3j− bound to the 40S subunit efficiently and stably enough to withstand sucrose density gradient centrifugation in the presence of various RNA and DNA cofactors (Table 1). Stable eIF3j−/40S subunit complexes formed in the presence of poly(U) of heterogeneous length, but not in the presence of poly(A), poly(C), or poly(G) (Fig. 1B, lanes 2–5). Formation of stable eIF3j−/40S subunit complexes was not promoted by mRNAs that use various initiation mechanisms, including β-globin mRNA (Pestova et al. 1998a), a chimeric (CAA)n-GUS mRNA comprising a 68-nt-long A-rich 5′ UTR and the β-glucuronidase coding region (Pestova and Kolupaeva 2002), and a 840-nt-long mRNA containing the highly structured encephalomyocarditis virus (EMCV) internal ribosomal entry site (Borovjagin et al. 1991; Fig. 1C, lane 3; Table 1). Inclusion in a reaction mixture with native globin mRNA of eIFs 4F, 4A, and 4B, which unwind the cap-proximal region of mRNA to facilitate binding of 43S complexes in a process that may involve interaction of eIF3 with eIF4F, did not promote stable binding of eIF3j− to 40S subunits (Fig. 1C, lane 4). The activity of the polynucleotide cofactor did not require the 2′-OH group of the ribose moiety because poly(dT) also promoted formation of stable eIF3j−/40S subunit complexes (Fig. 1B, lane 6).
TABLE 1.
Stimulation of formation of binary eIF3j−/40S subunit complexes by poly- and oligonucleotides
| Polynucleotide | Binary complex formation |
| poly(U) | +++ |
| poly(A) | − |
| poly(C) | − |
| poly(G) | − |
| poly(dT) | +++ |
| rU25 | ++ |
| dT40 | +++ |
| dT20 | −/+ |
| rU31-stem | +++ |
| stem-rU31 | +++ |
| stem-dT40 | +++ |
| dT40-stem | +++ |
| stem-dT45-stem | +++ |
| dC35 | +++ |
| d(CT)25 | +++ |
| d(CTTT)13 | +++ |
| dC20T10C20 | +++ |
| dC20T2C20 | +++ |
| dT10C20T10 | +++ |
| dA35 | − |
| d(AT)25 | − |
| d(ATT)17 | −/+ |
| d(ATTT)13 | +++ |
| dG35 | − |
| d(GT)25 | +++ |
| d(GTT)17 | +++ |
| d(GTTT)13 | +++ |
| d(CAA)12 | − |
| dT10(CAA)8 | −/+ |
| dT5(CAA)10 | − |
| d(CAA)5T5(CAA)5 | − |
| d(CAA)4T15(CAA)4 | +/− |
| rU35 | +++ |
| rC35 | − |
| r(CCCU)9 | − |
| r(CU)17 | ++ |
| r(CUUU)9 | +++ |
| rG35 | − |
| r(GU)17 | ++ |
| rA35 | − |
| Met-tRNAiMet | − |
| eIF2-GTP-Met-tRNAiMet | +++ |
| β-globin mRNA | − |
| β-globin mRNA, 4A, 4B, 4F | − |
| (CAA)n-GUS mRNA | − |
| EMCV nt 315–1155 | − |
To determine the size of the polynucleotide cofactor that was sufficient to promote this interaction, poly(U) was fractionated by gel filtration on Superdex-200. All fractions including the smallest (poly(U) of ~200 nt or less) were active (Fig. 1D, lane 2). The DNA oligonucleotide (dT)40, but not (dT)20, also supported binary complex formation (Fig. 1D, lanes 3,4). The length of a synthetic U25 oligonucleotide was sufficient to promote eIF3j−/40S subunit binding (Fig. 1D, lane 5), although stimulation was lower than by poly(U) or (dT)40. RNA oligonucleotides containing the sequence U31 and a stable stem (−9.5 kcal/mol) (Fig. 1E) at either 5′ or 3′ terminus also promoted eIF3/40S subunit binding (Fig. 1F, lanes 2,3), as did DNA oligonucleotides containing the sequence (dT)40 flanked by stable hairpins (−11.9 kcal/mol) (Fig. 1E) at either terminus (Fig. 1F, lanes 4,5), or the sequence (dT)45 flanked by stable hairpins (−11.9 kcal/mol) at both termini (Fig. 1F, lane 6). The oligonucleotide cofactor therefore need not have unstructured 5′ or 3′ ends to promote association of eIF3j− and 40S subunits.
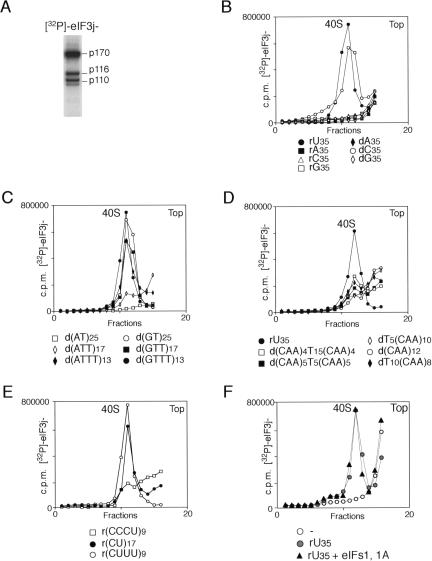
To investigate further the nature of the cofactor required to promote eIF3j−/40S subunit binding, we assayed a panel of 34–50-nt-long RNA and DNA oligonucleotides. These data are presented in Figure 2 and summarized in Table 1. To obtain quantitative data, we used [32P]-labeled eIF3j− that had been phosphorylated on p170, p116, and p110 subunits using the catalytic subunit of cAMP-dependent protein kinase (Unbehaun et al. 2004; Fig. 2A). Because of the limitations of the sucrose density gradient centrifugation method, which can be applied most successfully and reliably to investigation of stable biological complexes, but can give somewhat misleading results in the case of weak complexes that can dissociate during the actual gradient centrifugation, we do not present a quantitative comparison of the activities of weakly active oligonucleotides here.
FIGURE 2.
Activity of RNA and DNA oligonucleotides in promoting association of [32P]-labeled eIF3j− and 40S subunits. (A) [32P]-phosphorylation of eIF3a (p170), eIF3b (p116), and eIF3c (p110) subunits of eIF3j− by the catalytic subunit of cAMP-dependent protein kinase. [32P]-phosphorylated eIF3j− was resolved by gel electrophoresis on 4%–12% Bis-Tris NuPAGE gel using MES buffer system and visualized by autoradiography. (B–F) Binding of [32P]-labeled eIF3j− to 40S subunits in the presence of oligonucleotides and eIF1 and eIF1A, as indicated. Ribosomal complexes were separated by centrifugation in 10%–30% linear sucrose gradients, and aliquots of gradient fractions were analyzed by scintillation counting. The position of 40S subunits determined by optical density is indicated. Sedimentation was from right to left. Upper fractions from the gradient have been omitted for clarity.
Like the ribooligopurines rA35 and rG35, the deoxyribooligopurines dA35 and dG35 did not stimulate eIF3j−/40S subunit binding (Fig. 2B). However, in contrast to rC35, dC35 promoted formation of stable eIF3j−/40S subunit complexes (Fig. 2B). Consistent with the ability of dT40 and dC35 to promote eIF3j−/40S subunit binding, all DNA oligonucleotides with mixed dT/dC compositions (Table 1) also promoted this process, as did the DNA oligonucleotides d(GT)25, d(GTT)17, and d(GTTT)13 (Fig. 2C). d(AT)25 did not have stimulatory activity, possibly due to its ability to form double-stranded DNAs, whereas the stimulatory activity of oligo(dT) was not abrogated by interrupting its sequence with adenine residues in d(ATTT)13 (Fig. 2C). Because d(CAA)12 did not promote stable binding of eIF3j− to 40S subunits (Fig. 2D), we used the (CAA)n sequence in experiments to determine whether extension of oligo(dT) sequences that were shorter than the required 25 nt by sequences that by themselves did not promote eIF3j−/40S subunit binding could reduce the required number of contiguous (dT) residues. However, extension with (CAA)n sequences did not greatly increase the stimulatory activity of short oligo(dT) sequences: dT5(CAA)10 and d(CAA)5T5(CAA)5 were inactive and dT10(CAA)8 and d(CAA)4T15(CAA)4 had very low activities (Fig. 2D). Like its DNA counterpart, r(GU)17 promoted eIF3j−/40S subunit binding (Table 1). Since rC35 did not promote eIF3j−/40S subunit association, we also investigated the ability of RNA oligonucleotides comprising different rU/rC combinations to stimulate this process. The stimulatory activity of oligo(U) was not impaired by interrupting its sequence with cytidine residues in r(CUUU)9 (Fig. 2E). The oligonucleo tide r(CU)17 was ~85% as active as r(CUUU)9, and r(CCCU)9 did not promote eIF3j−/40S subunit binding (Fig. 2E). We conclude that to be able to promote stable binding of eIF3j−to 40S subunits, the oligonucleotide cofactor must contain a single-stranded U-rich or dT-rich stretch at least 25 nt long that can be flanked by stable hairpins on both sides. Neither eIF1 nor eIF1A, either alone or in combination, enhanced eIF3j−/40S subunit binding promoted by oligonucleotide cofactors (Fig. 2F; data not shown).
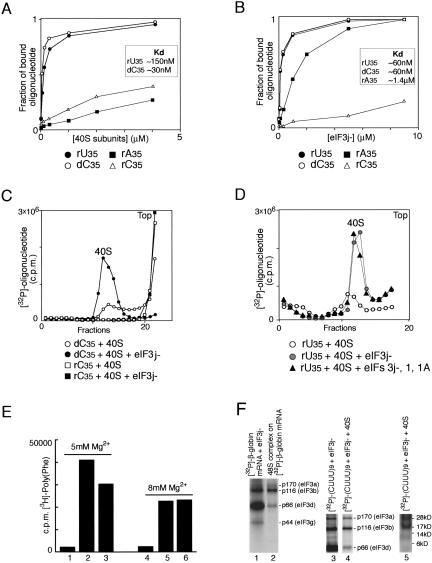
We considered three possible mechanisms by which oligonucleotide cofactors might promote the eIF3j−/40S subunit interaction. eIF3 could first bind RNA, change conformation, and then bind to the 40S subunit, or the 40S subunit could bind RNA first and only then bind eIF3, or eIF3 could first bind the 40S subunit, creating a composite RNA binding site, and subsequent binding of RNA could stabilize the whole complex. Therefore to assess the importance of binding of oligonucleotide cofactor to either 40S subunits or to eIF3 for its ability to promote the eIF3/40S subunit interaction, we investigated binding of 5′-[32P]-phosphorylated U35, rC35, dC35, and rA35 (some of which stimulated eIF3j−/40S subunit binding and some of which did not) to eIF3j− and to 40S subunits, using a filter-binding assay (Fig. 3A,B). U35 and dC35, which stimulated the eIF3j−/40S subunit interaction, bound to both eIF3j− and 40S subunits. U35 bound to 40S subunits and to eIF3j− with dissociation constants (Kd) of ~150 nM and ~60 nM, respectively, and dC35 bound to eIF3j− and to 40S subunits with Kd of ~30 nM and ~60 nM, respectively, using the current eIF3j− and 40S subunit preparations. Both eIF3j− and 40S subunits bound very weakly to rC35 (which did not stimulate eIF3j−/ 40S subunit binding). Although rA35 (which did not stimulate eIF3/40S subunit interaction) bound to eIF3j− with Kd ~1.4 μM, it had a very low affinity for 40S subunits. In similar experiments, the simultaneous presence of eIF3j− and 40S subunits in reaction mixtures had a strong synergistic affect on binding of U35 and dC35 irrespective of the order of addition of components (data not shown). Consistent with this, 10 times more U35 and five times more dC35 were associated with 40S subunits after sucrose density gradient centrifugation in the presence of eIF3j− than in its absence (Fig. 3C,D). rC35 did not occur in the peak corresponding to 40S subunits after sucrose density gradient centrifugation (Fig. 3C). eIF1 and eIF1A did not influence binding of either U35 or dC35 to 40S subunits in the presence of eIF3j− (Fig. 3D; data not shown). Although by testing the ability of different oligonucleotides to bind individually to either eIF3 or to 40S subunits we were not able to identify an oligonucleotide that could bind to 40S subunits but could not bind to eIF3 and as a result was also unable to promote eIF3/RNA/40S complex formation, and we therefore could not make a firm conclusion about the importance of direct binding of oligonucleotide cofactor to eIF3, we found that rA35 was able to bind to eIF3 but not to 40S subunits, and as a result did not promote formation of eIF3/RNA/40S subunit complex (Fig. 3A,B). This therefore allows us to conclude that binding of an oligonucleotide to the 40S subunit is essential for formation of the eIF3/RNA/ 40S subunit ternary complex.
FIGURE 3.
Binding of oligonucleotides to eIF3j− and to 40S subunits. (A,B) Affinity measurements made in filter-binding experiments using (A) 40S subunits and (B) eIF3j− with [32P]-labeled oligonucleotides as indicated. The fraction of bound oligonucleotide is the ratio of [32P]-labeled oligonucleotide retained on the filter to the input. Dissociation constants are shown in the inset boxes. (C) Binding of [32P]-labeled oligonucleotide (dC35 or rC35) to 40S subunits in the presence and absence of eIF3j−. (D) Influence of eIF1 and eIF1A on binding of [32P]-labeled rU35 to the 40S subunit in the presence of eIF3j−. Ribosomal complexes were separated by centrifugation in 10%–30% linear sucrose gradients. Sedimentation was from right to left. The position of 40S subunits determined by optical density is indicated. Upper fractions from the gradient have been omitted for clarity. (E) Template-dependent [3H]-polyphenylalanine synthesis by ribosomes assembled by incubation at 5 mM Mg2+ (lanes 1–3) or 8 mM Mg2+ (lanes 4–6) of 40S and 60S subunits without poly(U) (lanes 1,4), 60S subunits with preassembled 40S subunit/poly(U) complexes (lanes 2,5) or poly(U) with preassembled 80S ribosomes (lanes 3,6). All reaction mixtures contained [3H]-Phe-tRNAPhe, EF1A, and EF2. [3H]-polyphenylalanine was counted after TCA precipitation of reaction mixtures on nitro-cellulose filters. (F) UV-crosslinking of [32P]-β-globin mRNA (lanes 1,2) and [32P]-(CUUU)9 (lanes 3–5) to eIF3j− (lanes 1–4) and 40S subunits (lane 5) in binary eIF3j−/RNA complexes (lanes 1,3), 48S complexes (lane 2), and eIF3j−/(CUUU)9/40S subunit complexes (lanes 4,5). Polypeptides resolved by gel electrophoresis were visualized by autoradiography. eIF3a (p170), eIF3b (p116), eIF3d (p66), and eIF3g (p44) are indicated to the right of lanes 2,4. The positions of molecular weight markers are shown to the right of the radiolabeled ribosomal proteins in lane 5.
If an oligonucleotide must be bound to a 40S subunit to be able to stimulate eIF3j−/40S subunit association, this poses the obvious question as to the site on the 40S subunit to which it binds. The only homopolymeric RNA that stimulated eIF3j−/40S subunit association (i.e., poly(U)) is identical to the only RNA that can bind to the mRNA-binding cleft of 40S subunits in the absence of initiation factors (Williamson 1969; Roberts and Coleman 1971) and can be subsequently translated into polyphenylanine in the presence of only 60S subunits, Phe-tRNAPhe and elongation factors (Kemper and Merrick 1979). To confirm that poly(U) could bind productively in the mRNA-binding cleft of our 40S subunits, we assayed polyphenylalanine synthesis in such reactions both when 40S subunits were preincubated with poly(U) to form binary complexes before addition of 60S subunits (Fig. 3E, lanes 2,5) and when 40S and 60S subunits were preincubated to form 80S ribosomes prior to addition of poly(U) (Fig. 3E, lanes 3,6). At high (8 mM) Mg2+ concentration, the levels of poly(Phe) synthesis were similar under both conditions. Binding of poly(U) to 80S ribosomes is optimal at 8 mM Mg2+ (Williamson 1969). At a lower Mg2+ concentration, poly(Phe) synthesis was slightly greater if poly(U) was first preincubated with 40S subunits.
To investigate the interaction of the oligonucleotide cofactor with the 40S subunit and eIF3j− in the eIF3j−/40S/ cofactor complex in UV cross-linking experiments, we used [α-32P]UTP-labeled (CUUU)9 RNA transcripts that promoted eIF3j−/40S subunit binding (Fig. 2E). Assembled [eIF3j−/40S/(CUUU)9] complexes were purified by sucrose density gradient centrifugation and subjected to UV irradiation. eIF3 can be UV cross-linked to β-globin and other mRNAs in binary complexes (Fig. 3F, lane 1; Sonenberg et al. 1979; Setyono et al. 1984; Westermann and Nygard 1984). It can also be UV cross-linked to [32P]-labeled β-globin mRNA in 48S complexes, but the resulting pattern of UV cross-linking of eIF3j− differed from that observed for eIF3j− in binary complexes. Thus, eIF3g (p44) and eIF3d (p66) were much more strongly cross-linked to β-globin mRNA in eIF3j−/ RNA binary complexes than in 48S complexes (Fig. 3F, lanes 1,2). Cross-linking of eIF3j− to (CUUU)9 in [eIF3j−/40S/(CUUU)9] complexes (Fig. 3F, lane 4) was very similar to cross-linking of eIF3j− to β-globin mRNA in 48S complexes (Fig. 3F, lane 2) but differed from cross-linking of eIF3j− to (CUUU)9 in eIF3j−/(CUUU)9 binary complexes (Fig. 3F, lane 3), which in turn was similar to cross-linking of eIF3j− in eIF3j−/β-globin mRNA binary complexes (Fig. 4F, lane 1). The pattern of UV cross-linked ribosomal proteins in [eIF3j−/40S/(CUUU)9] complexes, including a prominent ~17-kDa band (Fig. 3E, lane 5), was similar to the pattern of labeled ribosomal proteins that we observed after UV cross-linking to mRNA containing 4-thio-uridines at different defined positions around the AUG codon in 48S initiation complexes (V.G. Kolupaeva, A. Pisarev, C. Hellen, T. Pestova, in prep.). Confirmation that these patterns are identical will require identification of the cross-linked ribosomal proteins. However, we assume that the oligonucleotide cofactor interacts with eIF3j− and 40S subunits in eIF3j−/40S/cofactor complexes in a manner consistent with it binding to the mRNA-binding cleft of the 40S subunit. The cross-linking pattern of eIF3j+ in binary eIF3/RNA and in 48S complexes did not differ from the cross-linking patterns of eIF3j− (data not shown).
FIGURE 4.
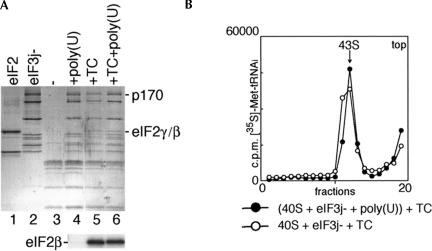
Binding of eIF2-ternary complex (TC) and eIF3j− to 40S subunits in the presence and absence of poly(U) RNA. (A) Detection of eIF2 and eIF3j− in ribosomal complexes isolated from sucrose density gradients. eIF2 (lane 1), eIF3j− (lane 2), 40S subunits (lane 3), 40S/eIF3j− binary complexes formed in the presence of poly(U) (lane 4), 43S ribosomal complexes formed from purified TC and preassembled eIF3j−/poly(U)/40S subunit complexes (lane 6), and 43S complexes assembled using TC, eIF3j− and 40S subunits (lane 5) were analyzed by gel electrophoresis followed by Coomassie staining (lanes 1–6 of the upper panel) or Western blotting with eIF2βantibodies (lanes 4–6 of the lower panel). eIF3a (p170 subunit) and eIF2β,γ are labeled to the right of the upper panel; eIF2β is labeled to the left of the lower panel. (B) Incorporation of aminoacylated [35S]Met-tRNAiMet into 43S complexes, assembled either from TC and preassembled eIF3j−/poly(U)/40S subunit complexes or from 40S subunits, eIF3j−, and TC. Ribosomal complexes were separated by centrifugation in 10%–30% linear sucrose gradients. Sedimentation was from right to left. The position of 43S complexes is indicated. Upper fractions from the gradient have been omitted for clarity.
Consistent with previous reports (Peterson et al. 1979; Trachsel and Staehelin 1979; Erni and Staehelin 1983), stable binding of eIF3j− to 40S subunits was also stimulated by eIF2•GTP•Met-tRNAiMet ternary complexes in the absence of oligonucleotide cofactor (Fig. 4A, lane 5). The stimulation of eIF3j−’s stable binding to 40S subunits by oligonucleotides and by eIF2-ternary complexes raises the question of whether the interactions between eIF3j− and 40S subunits are similar in eIF3j−/40S subunit/eIF2•GTP•Met-tRNAiMet complexes and in eIF3j−/40S subunit complexes assembled in the presence of polynucleotide cofactor. In assays done to address this question, eIF3j−/40S subunit complexes preassembled in the presence of poly(U) bound to eIF2-ternary complexes as efficiently as 40S subunits incubated simultaneously with eIF2-ternary complexes and eIF3j− in the absence of poly(U) (Fig. 4A, lanes 5,6; Fig. 4B). Binary eIF3j−/ 40S subunit complexes assembled in the presence of a polynucleotide cofactor were therefore able to interact normally with another key component of the translation apparatus.
Factor and cofactor requirements for dissociation of 80S ribosomes by eIF3
Dissociation of 80S ribosomes into 40S and 60S subunits is a prerequisite for translation initiation, and so far eIF3 and, to some extent, eIF1A have been implicated in this process (Thompson et al. 1977; Trachsel and Staehelin 1979; Thomas et al. 1980a). In light of the influence of the oligonucleotide cofactor on eIF3’s binding to 40S subunits, we also investigated its effect on eIF3’s ribosome anti-association and dissociation activities (summarized in Table 2). To assay dissociation, 80S ribosomes were preassembled from purified 40S and 60S subunits and incubated with different combinations of factors and cofactors. To assay anti-association, 40S subunits were first incubated with different combinations of factors and cofactors, then incubated with 60S subunits and analyzed by sucrose density gradient centrifugation. Consistent with its inability to bind stably to 40S subunits in the absence of polynucleotide cofactor or eIF2-ternary complex, eIF3j− alone had no ribosomal anti-association or dissociation activity (Fig. 5A; data not shown). Significantly, although eIF3j+ was able to bind to 40S subunits, it also could not dissociate 80S ribosomes or protect 40S subunits from reassociation with 60S subunits either alone, or in the presence of eIFs 1 and 1A (Fig. 5B; data not shown). Even though we recently showed that incubation of 40S subunits with eIF3j+ leads to appearance of 60–70S subunit particles, which most likely represent 40S dimers (Unbehaun et al. 2004), and therefore the small shoulders, which correspond to material migrating with a coefficient of sedimentation of ~60S in Figure 5B in reaction mixtures that contained 40S subunits and eIF3j+ in the presence or absence of eIFs 1 and 1A theoretically could contain 60S subunits and 40S dimers obtained as a result of the dissociating activity of eIF3j+, the amounts of such hypothetical 40S and 60S subunits are too small for a ribosome dissociating activity of eIF3j+ to be considered. However, in the presence of either eIF2-ternary complex, poly(U), or other oligonucleotides that promoted eIF3’s binding to 40S subunits (Table 1), both eIF3j− and eIF3j+ protected 40S subunits from association with free 60S subunits and promoted dissociation of 80S ribosomes (Fig. 5A,B,C; data not shown). The subunit anti-association activity of eIF3 in the presence of poly(U) (data not shown) was greater than its ribosomal dissociation activity. If the higher level of poly(Phe) synthesis in the case when poly(U) was preincubated with 40S subunits (Fig. 3E) reflects the difference in the abilities of 40S subunits and 80S ribosomes to interact with poly(U), that might explain why the subunit anti-association activity of eIF3 observed in the presence of poly(U) was greater than its ribosomal dissociation activity. eIF2-ternary complexes or oligonucleotide cofactors had no anti-association or dissociation activities in the absence of eIF3 (data not shown).
TABLE 2.
Cofactor requirements for ribosomal dissociation/anti-association by eIF3j− and eIF3j+
| Reaction components (with 40S + 60S subunits) | Ribosomal dissociation/anti-association |
| eIF3j− | − |
| eIF3j− + poly(U) | ++ |
| poly(U) | − |
| eIF3j− + poly(U) + eIF1A | +++ |
| eIF3j− + poly(U) + eIF1 | ++++ |
| eIF3j− + poly(U) + eIF1A + eIF1 | ++++ |
| eIF1A | − |
| eIF1 | − |
| eIF1 + poly(U) | − |
| eIF1A + poly(U) | − |
| eIF1 + eIF1A + poly(U) | − |
| eIF3j− + eIF2-ternary complex | +++ |
| eIF3j− + eIF1A | − |
| eIF3J− + eIF1 | − |
| eIF3j+ | − |
| eIF3j+ + poly(U) | ++ |
| eIF3j+ + poly(U) + eIF1A | +++ |
| eIF3j+ + poly(U) + eIF1 | ++++ |
| eIF3j+ + poly(U) + eIF1A + eIF1 | ++++ |
| eIF3j+ + eIF1 | − |
| eIF3j+ + eIF1A + eIF1 | − |
| eIF3j+ + eIF2-ternary complex | +++ |
FIGURE 5.
Ribosomal dissociation activity of eIF3. Dissociation activity of eIF3j− in the presence of (A) poly(U), (C) eIF2-ternary complex (TC) and (D) poly(U), eIF1, and eIF1A. (B) Dissociation activity of eIF3j+ in the presence of poly(U), eIF1, and eIF1A. (E,F) Resistance of 80S ribosomes to dissociation by (E) eIF1 or eIF1A alone and (F) eIF3c (p110 subunit) alone, with eIF1 or with eIF1 and poly(U). The optical density of ribosomal profiles was measured after centrifugation through 10%–30% linear sucrose gradients. Sedimentation was from right to left. Upper fractions from gradients have been omitted for clarity. (G) Dissociation by eIF3j− (in the presence and absence of poly(U)) of 80S complexes assembled on [32P]-CAAn-GUS mRNA and purified by sucrose density gradient centrifugation. After incubation with eIF3j− and poly(U) as indicated, ribosomal complexes were again centrifuged through 10%–30% linear sucrose gradients. Sedimentation was from right to left. Upper fractions from the gradient have been omitted for clarity.
In a parallel series of experiments, oligonucleotide cofactor-dependent ribosomal dissociation and anti-association activities of eIF3j− and eIF3j+ were enhanced by eIF1A and, to a much greater extent, by eIF1 (Fig. 5D; data not shown). Neither eIF1 nor eIF1A had anti-association activity individually or together either with or without poly(U) (Fig. 5E; Table 2; data not shown). eIF1 associates with eIF3 by binding to its eIF3c (p110) subunit (Fletcher et al. 1999). In the presence of poly(U) RNA, pure recombinant eIF3c did not dissociate empty 80S ribosomes by itself or in combination with eIF1 (Fig. 5F). eIF1 therefore acts as an accessory factor that enhances the activity of eIF3 in ribosomal dissociation and this activity requires components of eIF3 in addition to eIF3c.
A consequence of the assumption that RNA must be bound in the mRNA-binding cleft of the 40S subunit to be able to stabilize eIF3/40S subunit association and the fact that in the presence of oligonucleotide cofactor, eIF3 possesses ribosomal dissociation/antiassociation activities is that eIF3 might also be able to dissociate 80S ribosomes assembled on mRNA by the normal translation process. Dissociation of 80S ribosomes assembled on mRNA in a conventional factor-mediated manner might be complicated by the presence of initiator tRNA in the ribosomal P site. However, we found that sucrose density gradient centrifugation led to loss of 30%–35% of initiator tRNA from 80S ribosomes whereas mRNA remained stably bound (data not shown). Therefore, 80S ribosomes purified in this way constitute a mixed population of which a relatively high proportion contains only mRNA. To investigate the dissociating activity of eIF3j−, 80S ribosomes were assembled on [32P]ATP-labeled (CAA)n-GUS mRNA (see Materials and Methods) in the presence of eIFs 1, 1A, 2, 3, 4A, 4B, 4F, 5, and 5B and isolated by sucrose density gradient centrifugation. The integrity of 80S complexes was monitored by Cerenkov counting after they had been incubated with eIF3j− with or without excess poly(U) RNA and subjected to a second round of sucrose density gradient centrifugation. Incubation with eIF3j− led to dissociation of ~35% of 80S ribosomes, yielding 60S subunits, and 40S subunits that were bound to (CAA)n-GUS mRNA (Fig. 5G) and eIF3 (data not shown). However, in the presence of excess poly(U), eIF3j− failed to dissociate 80S ribosomes assembled on this mRNA (Fig. 5G). Similar results were obtained for eIF3j+ (data not shown). We suggest that eIF3’s RNA-binding capacity is required for it to bind to these 80S ribosomes in order to dissociate them and that in the conditions of this experiment, this capacity was saturated by the excess poly(U). Further experiments will be needed to reveal whether eIF3 can dissociate intact 80S complexes or dissociates only those ribosomes assembled on mRNA that have lost initiator tRNA.
Factor-dependent changes in the mobility of ribosomal complexes in sucrose density gradients
eIF3j−/40S subunit complexes formed in the presence of poly(U) or other oligonucleotide cofactors unexpectedly migrated more slowly than free 40S subunits on centrifugation through sucrose density gradients. Thus Coomassie staining of polypeptides from different fractions of a sucrose density gradient containing eIF3j−/40S subunit complexes assembled in the presence of poly(U) with excess 40S subunits showed that the eIF3j−/40S subunit complex corresponded to the more slowly migrating shoulder of the peak (Fig. 6A). The reduced mobility of binary eIF3j−/40S subunit complexes relative to free 40S subunits might be due to conformational changes in the 40S subunit or to changes in the shape of the complex to a less globular form caused by binding of eIF3 that increased its frictional coefficient. The presence of eIF3j in eIF3 preparation did not influence the mobility of eIF3/poly(U)/40S complexes (data not shown). Moreover, as we have shown elsewhere (Unbehaun et al. 2004), the eIF3j subunit is released from eIF3 on assembly of ribosomal complexes that contain RNA in the mRNA-binding cleft of the 40S subunit. This means that the final eIF3/poly(U)/40S subunit complexes or 48S complexes would not contain the eIF3j− subunit and would therefore have the same composition irrespective of whether or not eIF3j was present in eIF3 before formation of these ribosomal complexes.
FIGURE 6.
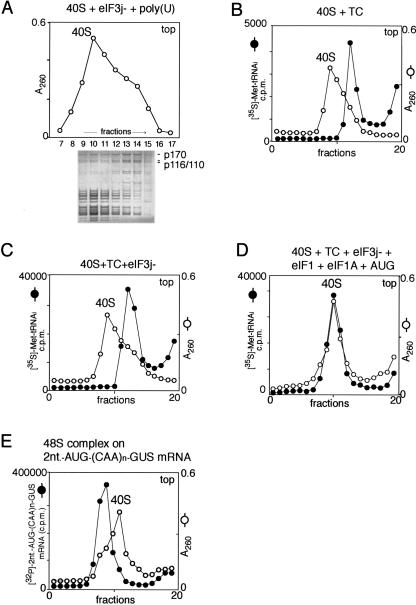
Mobility of ribosomal complexes during centrifugation through 10%–30% linear sucrose density gradients. (A) Fractionation by sucrose density gradient centrifugation of eIF3j−/poly(U)/40S subunit complexes formed with excess 40S subunits. The positions of 40S subunits and their complexes with eIF3j− were determined by optical density. Polypeptides in different fractions were resolved by electrophoresis followed by Coomassie staining. eIF3a (p170) and eIF3b/3c (p116/p110) are labeled to the right of the lower panel. (B,C) Mobility of preinitiation 43S complexes (consisting of 40S subunits and eIF2-ternary complex [TC]) with (C) or without (B) eIF3, compared to free 40S subunits. (D) Mobility of 43S preinitiation complexes (consisting of 40S subunits, eIF1, eIF1A, TC, eIF3, and AUG triplets) compared to free 40S subunits. The positions of 40S subunits and their complexes with TC, eIF3, eIF1, and eIF1A were determined by optical density and by scintillation counting of [35S]Met-tRNAiMet (B,C,D). (E) Mobility of 48S initiation complexes assembled on [32P]-2-nt-AUG-(CAA)n-GUS mRNA compared to free 40S subunits. The positions of 40S subunits and 48S complexes were determined by optical density and by Cherenkov counting of [32P]-2-nt-AUG-(CAA)n-GUS mRNA. Sedimentation was from right to left. Upper fractions from the gradient have been omitted for clarity.
The unexpected change in mobility of eIF3/40S subunit complexes prompted us to investigate the influence of other translation components on sedimentation of 40S subunits in 43S preinitiation and 48S initiation complexes during sucrose density gradient centrifugation (Fig. 6; Table 3). Since we did not detect any difference in the mobility of ribosomal complexes formed with either eIF3j− or with eIF3j+, we shall refer to this initiation factor simply as eIF3 throughout the rest of this section and also on panels B–E of Figure 6 and in Table 3. We first analyzed the mobility of complexes comprising 40S subunits and eIF2•GTP•Met-tRNAiMet. The position of ribosomal complexes containing eIF2-ternary complexes was monitored by the presence of [35S]Met-tRNAiMet; the position of free 40S subunits was monitored by measuring optical density. To distinguish between complexes that contained eIF2-ternary complexes and free 40S subunits, the latter were present in excess over other components in reaction mixtures. These complexes also migrated more slowly than free 40S subunits (Fig. 6B). Addition of eIF1, eIF1A, or eIF1 with eIF1A did not alter the mobility of ribosomal complexes assembled in reaction mixtures containing only eIF2-ternary complexes and 40S subunits (Table 3). Ribosomal complexes assembled with eIF2-ternary complex and eIF3 also migrated more slowly than 40S subunits (Fig. 6C) and their mobility was not altered further by inclusion of eIF1A or AUG triplets in any reaction mixture (Table 3). However, inclusion of eIF1 with or without AUG triplets in addition to eIF1A, eIF2-ternary complexes, and eIF3 yielded ribosomal complexes that migrated with a mobility similar to that of free 40S subunits (Fig. 6D; Table 3). The observed change in mobility can therefore be attributed to binding of eIF1 to these preinitiation complexes.
TABLE 3.
The mobility of ribosomal complexes compared to that of 40S subunits
| Constituents of ribosomal complexes | Mobility of ribosomal complexesa |
| 40S, eIF3, poly(U) | < |
| 40S, eIF3, poly(U), eIF1, eIF1A | < |
| 40S, eIF2·GTP· Met-tRNAiMet | < |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF3 | < |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF3, AUG | < |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1A | < |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1, eIF1A | < |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1, eIF1A, AUG | < |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1A, eIF3 | < |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1A, eIF3, AUG | < |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1 | < |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1, eIF3 | = |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1, eIF3, AUG | = |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1, eIF1A, eIF3 | = |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1, eIF1A, eIF3, AUG | = |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1, eIF1A, eIF3, mRNA | > |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1A, eIF3, eIF4A, eIF4B, eIF4F, mRNA | > |
| 40S, eIF2·GTP·Met-tRNAiMet, eIF1, eIF1A, eIF3, eIF4A, eIF4B, eIF4F, mRNA | > |
aMobility in sucrose density gradients of ribosomal complexes is indicated as being less than (<), greater than (>), or equal to (=) that of free 40S ribosomal subunits.
The mobility of 48S initiation complexes assembled in the presence of different sets of factors was investigated using [32P]UTP-labeled 2-nt-AUG-(CAA)n-GUS mRNA (see Materials and Methods), which is a derivative of (CAA)n-GUS mRNA with an additional AUG triplet 2 nt from its 5′ end. Its unstructured 5′ UTR allows 48S complexes to assemble on the AUG initiation codon of the GUS open reading frame in the presence of eIF1 and in the absence of eIFs 4A, 4B, and 4F, and the additional AUG triplet near the 5′ end allows very efficient 48S complex formation on it in the absence of eIF1 (Pestova and Kolupaeva 2002). 48S complexes were therefore formed in the presence of eIF2-ternary complex and (a) eIF1, 1A, and 3, (b) eIF1A, 3, 4A, 4B, and 4F, or (c) eIF1, 1A, 3, 4A, 4B, and 4F. These ribosomal complexes containing mRNA all had a greater mobility than free 40S subunits on sucrose density gradient centrifugation (Fig. 6E; Table 3). Although it is possible that the change in mobility of 48S complexes compared to 43S complexes could be due to the presence of mRNA with its high density, these data may also reflect another sequential conformational change in ribosomal complexes following binding of mRNA and establishment of the codon–anticodon interaction.
Factor-dependent stabilization of binding eIF2-ternary complexes to 40S subunits
We also investigated the influence of AUG triplets and different factors on binding of eIF2•GTP•Met-tRNAiMet ternary complexes to 40S subunits using sucrose density gradient centrifugation (Fig. 7). Again, we did not detect any difference in the activities of eIF3j− and eIF3j+ forms of eIF3 in these experiments and will therefore refer to this factor simply as eIF3 throughout this section. Consistent with previous reports (Trachsel et al. 1977; Benne and Hershey 1978; Chaudhuri et al. 1999; Majumdar et al. 2003), in the absence of AUG triplets, eIFs 1A and 3 both stabilized binding of eIF2 ternary complexes to 40S subunits (Fig. 7A). Consistent with some previous reports (Trachsel et al. 1977; Benne and Hershey 1978) but in contrast to data reported by Maitra and colleagues (Chaudhuri et al. 1999; Majumdar et al. 2003), the influence of eIF3 was significantly greater than that of eIF1A on a molar basis. It has been reported that eIF1 also stabilized binding of eIF2•GTP•Met-tRNAiMet to 40S subunits (Thomas et al. 1980b) and we therefore systematically investigated eIF1’s influence on this interaction. eIF1 alone stabilized 40S subunit/eIF2•GTP•Met-tRNAiMet complexes to a greater extent than eIF1A alone (but less than eIF3 alone) and addition of eIF1A to reaction mixtures containing eIF1 had a relatively small stimulatory effect (Fig. 7A), contrary to a recent report (Majumdar et al. 2003). Individually, eIFs 1 and 1A only slightly augmented the stabilizing effect of eIF3 (Fig. 7B). Binding of eIF2-ternary complexes to 40S subunits was most efficient in the presence of eIFs 1, 1A, and 3 (Fig. 7B) as recently reported (Majumdar et al. 2003). The differences between our results and data from Maitra’s laboratory (Majumdar et al. 2003) may in part be due to differences in conditions of sucrose density gradient centrifugation, which was done at 4 mM Mg2+ here rather than at 1 mM Mg2+, and the use of ribosomal subunits from different sources. Consistent with a previous report (Chaudhuri et al. 1999), AUG triplets stimulated binding of eIF2-ternary complexes to 40S subunits about twofold in reaction mixtures that otherwise contained only eIF1A (Fig. 7C). AUG triplets stimulated 43S complex formation by only ~20%–25% in the presence of eIF3 alone (Fig. 7D) or eIF3 together with eIF1A (Fig. 7E), by only 5%–10% in the presence of eIFs 3 and 1 (Fig. 7F), and not at all in the presence of eIFs 1, 1A, and 3 (Fig. 7G).
FIGURE 7.
(A–G) Stimulation of the binding of the eIF2•GTP•Met-tRNAiMet ternary complex (TC) to 40S subunits by other initiation factors and AUG triplets, as indicated. Ribosomal complexes were fractionated by centrifugation in 10%–30% linear sucrose gradients and analyzed by scintillation counting of [35S]Met-tRNAiMet. Sedimentation was from right to left. Upper fractions from each gradient have been omitted for clarity.
DISCUSSION
The multiple roles played by eIF3 in initiation include ribosomal dissociation and anti-association, stabilizing binding of the eIF2•GTP•Met-tRNAiMet ternary complex to 40S subunits to form 43S complexes, mediating 5′ end-dependent attachment of 43S complexes to mRNA, and subsequent formation of 48S initiation complexes. These activities all involve binding of eIF3 to the 40S subunit. Although eIF3j−, which lacks the eIF3j subunit, does not bind 40S subunits stably enough to withstand sucrose density gradient centrifugation (Fraser et al. 2004), stable binding of eIF3j− to 40S subunits can be promoted not only by eIF2-ternary complexes, but also by an rU or dT-rich unstructured oligonucleotide cofactor longer than 25 nt that did not require either the 2′-OH group of the ribose moiety or unstructured ends to be active. Of homopolymers, oligo(rU), oligo(dT), and oligo(dC) stimulated this interaction, whereas oligo(rA), oligo(rG), oligo(rC), oligo(dA), and oligo(dG) did not. The ability of these cofactors to stimulate eIF3/40S subunit interaction correlated with their ability to bind to 40S subunits. The observed binding of 40S subunits to oligo(U) and oligo(dT) and their inability to bind oligo(rA) is consistent with previous reports (Williamson 1969; Roberts and Coleman 1971; Iwasaki 1982; Potapov et al. 1992). Efficient binding of poly(dC) to Escherichia coli 30S ribosomal subunits has also been reported (Salas and Bollum 1969). The distinct common feature of poly(rU), poly(dT), and poly(dC) is a lack of ordered local structure (Adler et al. 1967; Evans and Sarma 1976; Cantor and Schimmel 1980). Despite the obvious similarities between poly(rC) and poly(dC) (except for the 2′-OH group present in poly(rC)), at neutral pH, unlike poly(dC), poly(rC) forms a narrow single-stranded helix that is stabilized by stacking interactions (Fasman et al. 1964; Adler et al. 1967; Arnott et al. 1976; Garriga et al. 1992). At neutral pH poly(rA) also forms single-stranded helical segments with a larger diameter than the poly(rC) helix that are interrupted by unstructured unstacked regions (Holcomb and Tinoco 1965; Saenger et al. 1975). Although the nature of nucleotide base (pyrimidine vs. purine) might play a discriminatory role in binding of oligonucleotide to 40S subunits, it is likely that the structure of the oligonucleotide is a more important determinant. Thus, oligo(U) or oligo(dT) sequences interrupted by other bases were also active in promoting eIF3j−/40S subunit binding. It is worth noting that although poly(rA) alone cannot bind to 40S subunits, it can do so in the presence of tRNALys (Iwasaki 1982), but unlike for poly(U) RNA, binding of poly(rA) to 40S subunits in this instance was strongly temperature dependent, suggesting that the poly(rA)/40S subunit interaction may be inhibited by stacking interactions in poly(rA) at neutral pH and low temperature. The sequence and size requirements of the cofactor, which correspond to the known RNA-binding characteristics of the mRNA-binding channel of eukaryotic ribosomes (Williamson 1969; Kozak 1977) and data on cross-linking of the cofactor with ribosomal proteins both point to a model in which the cofactor has to bind to the 40S subunit mRNA-binding cleft to be able to stimulate eIF3/40S subunit association. In this case, the stabilizing effect of the oligonucleotide cofactor could be due to conformational changes in the 40S subunit and/or to direct binding of eIF3 to 40S subunit-associated oligonucleotide, which was observed in UV cross-linking experiments. Although biophysical data on the location of eIF3 bound to the 40S subunit are contradictory (Emanuilov et al. 1978; Lütsch et al. 1986; Srivastava et al. 1992), biochemical data (Válasek et al. 2003) support a model in which eIF3 binds to the solvent side of the 40S subunit, where the eIF3’s and 40S subunit’s mRNA-binding surfaces could either dock together to form a mRNA-binding tunnel or eIF3 by itself could extend the mRNA-binding channel of the 40S subunit outside the interface surface.
The oligonucleotide cofactor also had a major influence on the ribosomal dissociation and anti-association activities of eIF3. Thus, although eIF3j+ was able to bind to the 40S subunit, it alone, like eIF3j−, was not able to dissociate 80S ribosomes or protect 40S subunits from reassociation with 60S subunits. However, in the presence of oligonucleotide cofactor both forms of eIF3 possessed ribosomal dissociation and anti-association activities even in the absence of eIF2-ternary complex.
The observations that the interaction of eIF3 with the 40S subunit can be promoted by RNA in the absence of eIF2-ternary complex raises the question of its biological significance. At the first stage in initiation, eIF3 and eIF2-ternary complex mutually promote each other’s stable binding to the 40S subunit in a process that neither involves nor requires mRNA. Consistently, we found that native capped β-globin mRNA could not promote stable association of eIF3 with the 40S subunit even in the presence of eIFs 4A, 4B, and 4F. The enhancement by oligonucleotide cofactor of eIF3j−/40S subunit binding may reflect the simultaneous interaction of eIF3 with mRNA and 40S subunits that occurs at other stages in the initiation process, including scanning along the 5′ UTR and ribosomal arrest at the initiation codon. It is worth noting that, as we showed elsewhere (Unbehaun et al. 2004), eIF3 is no longer associated with its eIF3j subunit in ribosomal complexes containing mRNA (e.g., 48S complexes). eIF2 and eIF3 remain associated with 40S subunits up to the stage of 48S complex formation until eIF5 induces hydrolysis of eIF2-bound GTP and release of eIF2-GDP. What happens to eIF3 after release of eIF2-GDP? We recently found that eIF3 is released from 40S subunits upon hydrolysis of eIF2-bound GTP in 48S complexes assembled on AUG triplets, consistent with previous reports (Das and Maitra 2001), but that it remains bound to 40S subunits even after release of eIF2-GDP from 48S complexes assembled on mRNA (Unbehaun et al. 2004). Such intermediate ribosomal initiation complexes, which contain mRNA, initiator tRNA, and other initiation factors except eIF2, were not able to join with 60S subunits in the absence of eIF5B. Although it is possible that, as suggested previously (Roll-Mecak et al. 2001), the inability of such complexes to join with 60S subunits spontaneously is because of the position of initiator tRNA, which should be adjusted by eIF5B for ribosomal joining to occur, the fact that eIF3 protects 40S subunits from joining even in eIF3/RNA/40S subunit complexes (which do not contain initiator tRNA) suggests that the stable association of eIF3 with the 40S subunit promoted by mRNA after release of eIF2 could be at least partially responsible for the requirement of eIF5B for subunit joining. Additionally, our recent data indicate that not only does mRNA stabilize binding of eIF3 to the 40S subunit but also that eIF3, in turn, stabilizes binding of mRNA to the 40S subunit after the release of eIF2-GDP until the actual subunit joining event (Unbehaun et al. 2004). It has also been suggested that eIF3 may stay bound to the 40S subunit during the first few elongation cycles and even remain associated with post-termination ribosomes, promoting reinitiation after translation of a short open reading frame (Park et al. 2001; Poyry et al. 2004). It is possible that such temporary association of eIF3 with elongating 80S ribosomes is promoted by the presence of mRNA. The strong influence of the oligonucleotide cofactor on ribosomal dissociation and the fact that eIF3 could dissociate 80S ribosomes assembled on mRNA both suggest another stage in the translation process that may require this interaction. We speculate that eIF3 might play an important role in dissociating 80S ribosomes bound to mRNA after termination of translation. This process is essentially uncharacterized in eukaryotes, but the activities of eIF3 in dissociating mRNA-associated 80S ribosomes described here and of eIF1 in dissociating aberrant initiation complexes containing only the 40S subunit (Pestova et al. 1998a; Pestova and Kolupaeva 2002) could act sequentially to dissociate post-termination ribosomes that remain bound to mRNA after peptide release. The involvement of prokaryotic IF3 in post-termination recycling of prokaryotic ribosomes indicates that initiation factors can act at stages in translation other than initiation (Karimi et al. 1999). We also believe that the stabilization of the eIF3/40S subunit interaction by oligonucleotide cofactor that we have identified may have important practical implications in structural studies of the eIF3/40S subunit interaction.
Although individually neither eIF1 nor eIF1A had ribosome dissociation activity, both factors, and particularly eIF1, enhanced the dissociation activity of eIF3 in the presence of oligonucleotide cofactor. This activity of eIF1 has not previously been reported. The observation that eIF1 and eIF1A individually or together do not have ribosome dissociation activities suggests that they cannot interact productively with 80S ribosomes whereas eIF3 is able to do so, and that the modes of action of these factors in dissociation differ fundamentally. These results are consistent with a model in which eIF3 at least initially interacts with a site on the 40S subunit that does not include its intersubunit surface, whereas eIF1 and eIF1A bind to this surface, which is accessible only in the absence of 60S subunits. Consistent with this, we recently found that eIF1 binds to the interface surface of the platform in the vicinity of the ribosomal P site (Lomakin et al. 2003). In this case, binding of eIF1 to the 40S subunit would block access of the 60S subunit to elements of 18S rRNA that participate in formation of the intersubunit bridges B2b and B2d (Spahn et al. 2001). It is also likely that eIF1A binds to the ribosomal A site on the 40S subunit just as its prokaryotic homolog IF1 binds to this site on the 30S subunit (Carter et al. 2001; for a discussion, see Pestova and Hellen 2001a). We did not detect any stabilizing/enhancing influence of eIF1 and eIF1A on the interaction of eIF3 or oligonucleotide cofactors with the 40S subunit and therefore suggest that eIF1 and eIF1A could cooperate with eIF3 in preventing subunit reassociation not by influencing the eIF3/40S subunit interaction, but instead by either inducing conformational changes in the 40S subunit that impair potential interactions with the 60S subunit or by steric hindrance, occluding sites on the 40S subunit that form intersubunit bridges with the 60S subunit in 80S ribosomes. eIF2-ternary complex also binds to the interface side of the 40S subunit (Bommer et al. 1991) and could therefore enhance eIF3’s activity in preventing subunit re-association in a similar manner. Although it has been suggested that eIF3 is attached to the back lobes of the 40S subunit, oriented away from the intersubunit interface, which would necessarily imply that eIF3’s dissociation/anti-association activity is due to induced conformational change rather than to steric hindrance (Srivastava et al. 1992; Válasek et al. 2003), this contradicts other studies (Emanuilov et al. 1978; Lütsch et al. 1986). The mechanism by which eIF3 dissociates ribosomes therefore remains an open question.
In addition to enhancing eIF3’s activity in ribosomal dissociation, eIF1 promoted association of eIF2-ternary complex with the 40S subunit in the absence of eIF3. The stimulatory activity of eIF1 was greater than that of eIF1A but weaker than that of eIF3. These data are not in complete agreement with data from Maitra and colleagues (Chaudhuri et al. 1999; Majumdar et al. 2003); possible reasons for this discrepancy were noted in Results. We also found that eIF1 altered the sedimentation of ribosomal complexes in sucrose density gradients, most likely reflecting changes in the frictional coefficient of complexes due to induced conformational changes. The mechanism by which eIF1 stabilizes binding of the eIF2 ternary complex to 40S subunits is not known but is likely indirect, because eIF1 is not known to interact directly with this complex. The stabilization of the attachment of the ternary complex to 40S subunits and the induced changes in the sedimentation of ribosomal complexes may both be manifestations of the same eIF1-induced conformational switch in the 40S subunit.
MATERIALS AND METHODS
Reagents
DNA restriction endonucleases, T7 RNA polymerase, and T4 polynucleotide kinase were from Roche. The cAMP-dependent protein kinase catalytic subunit was from New England Biolabs. Native globin mRNA and DNA oligonucleotides were from Invitrogen. AUG triplets and RNA oligonucleotides were from Integrated DNA Technologies and Dharmacon. Native rabbit total tRNA was from EMD Biosciences. Rabbit reticulocyte lysate was from Green Hectares. GTP, ATP and RNase inhibitor were from Amersham Biosciences. Poly(U), poly(A), poly(C), poly(G), poly(dT), poly(dA), and native yeast tRNAPhe were from Sigma. [35S]methionine (44 TBq/mmol), [α-32P]UTP (111 Tbq/mmol), [α-32P]CTP (111 TBq/mmol), [α-32P]ATP (111 TBq/mmol), and [γ-32P]ATP (259 TBq/mmol) were from ICN MP Biomedicals. L-[2,3,4,5,6-3H]Phe (4.63 TBq/mmol) was from Amersham Biosciences. Antibodies were against eIF2β (Santa Cruz Biotechnology).
Plasmids and primers
Vectors for expression of recombinant initiation factors have been described (Pestova et al. 1996a, 1998a, 2000). pET-eIF3c for expression of recombinant human eIF3c (p110) subunit was made by inserting the EcoRI–EcoRI fragment from pGEXp110 (Asano et al. 1997) into the EcoRI site of pET28a (Novagen). Plasmid pCUUU for T7-polymerase transcription of (CUUU)9 RNA was made by inserting complementary oligonucleotides 5′-ATCTAAGCTTAATACGACTCACTATAG(CTTT)9GGATCCTAT-3′ and 5′-ATAGGATCC(AAAG)9CTATAGGTCGTATTAAGCTTAGAT 3′ between HindIII and BamHI sites in pUC18. For transcription, pCUUU was linearized with BamHI. Plasmids (CAA)n-GUS (Wilson et al. 1990), 2-nt-AUG-(CAA)n-GUS (Pestova and Kolupaeva 2002), pTE17 containing EMCV nt 315–1155 (Borovjagin et al. 1991) and pBS-(β-globin) (Hellen et al. 1993) were linearized and transcribed as described. (CAA)n-GUS mRNA comprises a 5′ UTR with the sequence 5′-GCAAGAA-(CAA)19-CACCAUGG (in which the initiation codon is underlined) and the β-glucuronidase (GUS) coding sequence. 2-nt-AUG-(CAA)n-GUS mRNA has a 5′ UTR with the sequence 5′GG-AUG-A(CAA)15-CCAUGG (in which the 5′-proximal initiation codon and the GUS initiation codon are underlined) and the GUS coding region.
Binary eIF3/40S complex formation was investigated using oligonucleotides that contained stable stems (complementary sequences are underlined):
stem-U31: 5′-GGGGGUUUACCCCC(U)31; U31-stem: 5′-(U)31GG GGGUUUACCCCC; stem-dT40: 5′-GCCGACCCGATTCGGGTCGGC(T)40; dT40-stem 5′-(dT)40GCCGACCCGATTCGGGTCGGC; andstem-dT45-stem: 5′GCCGACCCGATTCGGGTCGGC(T)45 GCCGACCCGATTCGGGTCGGC,
as well as other oligonucleotides listed in Table 1. The stability of stems was calculated using Mfold (Mathews et al. 1999).
Purification of factors and ribosomal subunits
Aminoacyl-tRNA synthetases from E. coli strain MRE 600, ribosomal 40S and 60S subunits, and native and recombinant translation initiation and elongation factors (except eIF3) were purified as described (Pestova et al. 1996a, 1998a,b 2000; Pestova and Hellen 2003). Rabbit aminoacyl-tRNA synthetases (for aminoacylation of tRNAPhe) were purified from RRL as described (Stanley 1974). Native total tRNA and tRNAPhe were aminoacylated as described (Pestova et al. 1996a; Pestova and Hellen 2003). eIF2•GTP•Met-tRNAiMet ternary complexes were prepared and purified as described (Pestova and Hellen 2001b).
eIF3j− was purified from a 0%–40% (NH4)2SO4 precipitation fraction of the 0.5 M KCl wash of HeLa ribosomes by chromatography on DEAE and phosphocellulose (Pestova et al. 1996a). The fraction containing eIF3 was centrifuged through a 10%–30% sucrose density gradient in buffer containing 20 mM Tris (pH 7.5), 0.1 mM EDTA, 2 mM DTT, and 0.4 M KCl in a SW41 rotor for 22 h at 40,000 rpm. Fractions containing eIF3 were further purified on Mono Q (Pharmacia). eIF3 purified by this procedure (i.e., eIF3j−) contained all subunits except eIF3j, which dissociated during sucrose density gradient centrifugation. eIF3j+ was obtained when the centrifugation step was omitted. The subunit composition of eIF3 was analyzed by electrophoresis in 4%–12% NuPAGE Bis-Tris gel in MOPS buffer system (Invitrogen). The catalytic subunit of cAMP-dependent protein kinase and [γ-32P]ATP (259 TBq/mmol) were used to phosphorylate eIF3j− according to the manufacturer’s instructions.
Assembly and analysis of ribosomal complexes
Assembly of binary 40S subunit/eIF3j− complexes in the presence of oligonucleotide cofactors
Binary 40S/eIF3j− complexes were assembled by incubating 45 pmol 40S subunits and 60 pmol of unlabeled or [32P]-labeled (2 × 105 cpm/pmol) eIF3j− for 10 min at 37°C in 200 μL buffer A (20 mM Tris-HCl at pH 7.5, 100 mM KCl, 2.5 mM MgCl2, 2 mM DTT) in the presence of 10 μg poly(U), poly(dT), poly(A), poly(C), or poly(G), 150 pmol of various RNA and DNA oligonucleotides (see Table 1), or 80 pmol native globin mRNA, (CAA)n-GUS mRNA or a 840-nt-long EMCV mRNA. One reaction mixture with native globin mRNA also contained 60 pmol eIF4F, 100 pmol eIF4A, 100 pmol eIF4B, and 1 mM ATP. Complexes were isolated by centrifugation in a Beckman SW55 rotor for 2 h at 4°C and 42,000 rpm in 10%–30% linear sucrose density gradients in buffer B (20 mM Tris-HCl at pH 7.5, 100 mM KCl, 4 mM MgCl2, 2 mM DTT). Aliquots of gradient fractions that corresponded by optical density to 40S subunits were analyzed by SDS-polyacrylamide gel electrophoresis and Coomassie staining, as described (Pestova et al. 2000) or by scintillation counting. Poly(U) RNA was fractionated by FPLC gel-filtration on a Superdex G-200 column in buffer containing 20 mM Tris-HCl (pH 7.5), 200 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA. For experiments with radio-labeled oligonucleotides, they were 5′-phosphorylated with [32P]-γ ATP by T4 polynucleotide kinase to a specific activity of 1.2 × 106 cpm/pmol. Eighteen picomoles of radiolabeled oligo-nucleotide were incubated with 30 pmol 40S subunits with or without 45 pmol eIF3j− in 200 μL buffer A and subjected to sucrose density gradient centrifugation as described above. The presence of oligonucleotides in fractions that corresponded to 40S subunits was monitored by scintillation counting.
Filter-binding assay
5′-[32P]-phosphorylated U35, rC35, dC35, and rA35 (spec. act. = 1.2 × 106 cpm/pmol) were incubated at 1 nM final concentration in 50 μL buffer A for 10 min at 37°C with the indicated amounts of 40S subunits or eIF3j−. After incubation samples were diluted with 500 μL ice-cold buffer A, applied to 0.2 μm nitrocellulose filters (Schleicher and Schuell) and washed three times with 3 mL ice-cold buffer A. The radioactivity retained on filters was measured by scintillation counting. All experiments were repeated at least three times.
Binding of eIF3j− to 40S subunits in the presence of eIF2-ternary complex
43S ribosomal complexes were formed by incubating 45 pmol 40S subunits, 60 pmol eIF3, and 60 pmol eIF2-ternary complex that contained [35S]Met-tRNAiMet (spec. act. 1 × 104 cpm/pmol) in 200 μL buffer A supplemented with 0.4 mM GTP for 10 min at 37°C. To assay binding of eIF2-ternary complexes to binary eIF3j−/40S complexes assembled in the presence of poly(U), 60 pmol eIF2-ternary complex were added to preassembled eIF3/40S complexes, incubated for 10 min at 37°C and analyzed by sucrose density gradient centrifugation as described above. Radioactivity in aliquots of fractions due to [35S]Met-tRNAiMet was determined by scintillation counting. Fractions that corresponded to 43S complexes were analyzed by SDS-polyacrylamide gel electrophoresis and Coomassie staining, or western blotting using antibodies against eIF2β.
Anti-association and dissociation activity of eIF3j−and eIF3j+
To investigate the dissociation activity of eIF3j− and eIF3j+, empty 80S ribosomes assembled by incubating 25 pmol 40S subunits and 25 pmol 60S subunits in 200 μL buffer A for 10 min at 37°C were incubated for 10 min at 37°C with different combinations of 40 pmol eIF3j− or eIF3j+, 60 pmol purified eIF2 ternary complex, 100 pmol eIF1, 100 pmol eIF1A, 60 pmol eIF3c, and 5 μg poly(U) RNA, as stated in the text. To investigate eIF3’s anti-association activity, 25 pmol of 40S subunits were incubated for 10 min at 37°C in 200 μL buffer A with various combinations of 40 pmol eIF3j− or eIF3j+, 60 pmol purified eIF2 ternary complex, 100 pmol eIF1, 100 pmol eIF1A, 60 pmol eIF3c, and 5μg poly(U) RNA to promote binding of eIF3 to 40S subunits and then for 10 min more with 25 pmol 60S subunits to allow 80S formation. Ribosomal complexes were analyzed by sucrose density gradient centrifugation as described above.
UV cross-linking of eIF3j− to mRNA in eIF3j−/mRNA binary complexes and 48S initiation complexes and cross-linking of RNA cofactor to eIF3j− and ribosomal proteins in binary RNA/eIF3j− and ternary eIF3j−/RNA/40S subunit complexes
To investigate UV cross-linking of eIF3j− to RNA in binary complexes, β-globin mRNA (1 × 105 cpm total) transcribed in the presence of [α-32P]UTP, [α-32P]ATP, and [α-32P]CTP, and (CUUU)9 RNA (1 × 105 cpm total) transcribed in the presence of [α-32P]UTP were incubated with 2 μg eIF3j− in 40 μL buffer A for 10 min at 37°C. UV cross-linking was done as described (Pestova et al. 1996b). For 48S complex formation, β-globin mRNA (2 × 106 cpm total) was incubated with 45 pmol 40S subunit, 60 pmol eIF3j−, 60 pmol eIF2-ternary complex, 100 pmol eIF1, 100 pmol eIF1A, 30 pmol eIF4F, 100 pmol eIF4A, and 100 pmol eIF4B in 200 μL buffer A supplemented with 0.4 mM GTP and 1 mM ATP (buffer C) for 10 min at 37°C. The resulting 48S complex was purified by sucrose density gradient centrifugation as described above. UV cross-linking of 40 μL of the peak fraction that contained 48S complex (1 × 105 c.p.m.) was done as described (Pestova et al. 1996b). For eIF3j−/(CUUU)9/40S subunit complex formation, (CUUU)9 RNA (600,000 cpm total) was incubated with 40 pmol 40S subunit and 60 pmol eIF3j− in 200 μL buffer A for 10 min at 37°C. The resulting complex was purified by sucrose density gradient centrifugation and 40 μL of a peak fraction that corresponded to 40S subunits were subjected to UV cross-linking. Irradiated samples were analyzed by SDS-polyacrylamide gel electrophoresis.
Poly(U)-dependent polyphenylalanine synthesis
Reaction mixtures (50 μL) that contained 5 pmol [3H]Phe-tRNAPhe (spec. act. 120,000 cpm/pmol), 3 μg EF1H, 2 μg EF2, 5 μg poly(U) RNA, 10 pmol 40S subunits, and 10 pmol 60S subunits in buffer (20 mM Tris-HCl at pH 7.5, 100 mM KCl, 2 mM DTT) with 0.4 mM GTP and either 5 or 8 mM MgCl2 were incubated for 30 min at 37°C. In some experiments, 40S subunits were preincubated with poly(U) RNA for 10 min at 37°C to allow 40S/ poly(U) complexes to form and in others they were preincubated with 60S subunits to form 80S ribosomes (see Fig. 3D). The amount of synthesized poly(Phe) was determined by TCA precipitation and filtration on nitrocellulose filters (Schleicher and Schuell) of 15-μL reaction mixtures (Kemper and Merrick 1979). Radioactivity was determined by scintillation counting of filters.
Dissociation of 80S/mRNA complexes by eIF3j−
[α-32P]-ATP-labeled (CAA)n-GUS mRNA (5 × 106 cpm total) was first incubated with 100 pmol 40S subunit, 150 pmol eIF3j−, 150 pmol eIF2-ternary complex, 200 pmol eIF1, 200 pmol eIF1A, 80 pmol eIF4F, 200 pmol eIF4A, and 200 pmol eIF4B in 400 μL buffer C for 10 min at 37°C to form 48S complexes, followed by addition of 100 pmol 60S subunit, 100 pmol eIF5, and 100 pmol eIF5B(587–1220) and incubation for 10 min more to form 80S ribosomes. The resulting 80S ribosomal complex was purified by sucrose density gradient centrifugation as described above. Forty microliters of the peak fraction corresponding to 80S complex were diluted fivefold in buffer A, incubated for 10 min at 37°C with 40 pmol eIF3j− with or without 4 μg poly(U) RNA, and subjected to a second round of sucrose density gradient centrifugation. Radioactivity in fractions due to [32P]ATP-labeled mRNA was determined by Cherenkov counting.
Mobility of ribosomal complexes during sucrose density gradient centrifugation
To investigate their mobility in sucrose density gradients, eIF3j−/ poly(U)/40S subunit complexes and ribosomal preinitiation complexes were assembled by incubating 50 pmol 40S subunits with different combinations of 15 pmol eIF3j− or eIF3j+, 3 μg poly(U) RNA, 15 pmol eIF2-ternary complex (in which [35S]Met-tRNAi Met had a specific activity of 1 × 105 cpm/pmol), 30 pmol eIF1A, and 30 pmol eIF1 (as indicated in the text) in buffer C for 10 min at 37°C and analyzed by sucrose density gradient centrifugation as described above. 48S initiation complexes were assembled on [32P]ATP-labeled 2-nt-AUG-(CAA)n-GUS mRNA (2 × 106 cpm total) by incubating 50 pmol 40S subunits with 15 pmol eIF3j− or eIF3j+, 15 pmol eIF2 ternary complex, and different combinations of 30 pmol eIF1A, 30 pmol eIF1, 30 pmol eIF4F, 30 pmol eIF4A, 30 pmol eIF4B in 200 μL of buffer C for 10 min at 37°C and analyzed by sucrose density gradient centrifugation as described above.
Factor-dependent binding of eIF2-ternary complexes to 40S ribosomal subunits
Ribosomal complexes were assembled by incubation of 60 pmol 40S subunits with 40 pmol eIF2•GTP•[35S]Met-tRNAiMet ternary complex (in which [35S]Met-tRNAiMet had a specific activity of 1 × 104 cpm/pmol) and different combinations of 100 pmol eIF3j− or eIF3j+, 120 pmol eIF1A, 120 pmol eIF1, and 5 nmol AUG (as stated in the text) in 200 μL buffer C for 10 min at 37°C and analyzed by sucrose density gradient centrifugation as described above.
Acknowledgments
We thank J.W.B. Hershey for the pGEXp110 plasmid. This work was supported by grant R01 GM63940 from the National Institute of General Medical Sciences.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7215305.
REFERENCES
- Adler, A., Grossmann, L., and Fasman, G.D. 1967. Single-stranded oligomers and polymers of cytidylic and 2′-deoxycytidylic acids: Comparative optical rotatory studies. Proc. Natl. Acad. Sci. 57: 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott, S., Chandrasekaran, R., and Leslie, A.G. 1976. Structure of the single-stranded polyribonucleotide polycytidylic acid. J. Mol. Biol. 106: 735–748. [DOI] [PubMed] [Google Scholar]
- Asano, K., Kinzy, T.G., Merrick, W.C., and Hershey, J.W.B. 1997. Conservation and diversity of eukaryotic translation initiation factor eIF3. J. Biol. Chem. 272: 1101–1109. [DOI] [PubMed] [Google Scholar]
- Asano, K., Clayton, J., Shalev, A., and Hinnebusch, A.G. 2000. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5 and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes & Dev. 14: 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay, A. and Maitra, U. 1999. Cloning and characterization of the p42 subunit of mammalian translation initiation factor 3 (eIF3): Demonstration that eIF3 interacts with eIF5 in mammalian cells. Nucleic Acids Res. 27: 1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne, R. and Hershey, J.W.B. 1976. Purification and characterization of initiation factor IF-E3 from rabbit reticulocytes. Proc. Natl. Acad. Sci. 73: 3005–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1978. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253: 3078–3087. [PubMed] [Google Scholar]
- Bommer, U.A., Lütsch, G., Stahl, J., and Bielka, H. 1991. Eukaryotic initiation factors eIF-2 and eIF-3: Interactions, structure and localization in ribosomal initiation complexes. Biochimie 73: 1007–1019. [DOI] [PubMed] [Google Scholar]
- Borovjagin, A.V., Ezrokhi, M.V., Rostapshov, V.M., Ugarova, T.Y., and Shatsky, I.N. 1991. RNA–protein interactions within the internal translation initiation region of encephalomyocarditis virus RNA. Nucleic Acids Res. 19: 4999–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor, C.R. and Schimmel, P.R. 1980. Biophysical chemistry, Part 1: The conformation of biological macromolecules. W.H. Freeman, San Francisco, CA.
- Carberry, S.E. and Goss, D.J. 1991. Interaction of wheat germ protein synthesis initiation factors eIF-3, eIF-(iso)4F, and eIF-4F with mRNA analogues. Biochemistry 30: 6977–6982. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Hartsch, T., Wimberly, B.T., and Ramakrishnan, V. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, J., Si, K., and Maitra, U. 1997. Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J. Biol. Chem. 272: 7883–7891. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, J., Chowdhury, D., and Maitra, U. 1999. Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40S ribosomal preinitiation complex. J. Biol. Chem. 274: 17975–17980. [DOI] [PubMed] [Google Scholar]
- Das, S. and Maitra, U. 2001. Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation. Prog. Nucleic Acid Res. Mol. Biol. 70: 207–231. [DOI] [PubMed] [Google Scholar]
- Emanuilov, I., Sabatini, D.D., Lake, J.A., and Freienstein, C. 1978. Localization of eukaryotic initiation factor 3 on native small ribosomal subunits. Proc. Natl. Acad. Sci. 75: 1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erni, B. and Staehelin, T. 1983. Initiation of mammalian protein synthesis: Dynamic properties of the assembly process in vitro. Biochim. Biophys. Acta 740: 373–378. [DOI] [PubMed] [Google Scholar]
- Evans, F.E. and Sarma, R.H. 1976. Nucleotide rigidity. Nature 263: 567–572. [DOI] [PubMed] [Google Scholar]
- Falvey, A.K. and Staehelin, T. 1970. Structure and function of mammalian ribosomes. II. Exchange of ribosomal subunits at various stages of in vitro polypeptide synthesis. J. Mol. Biol. 53: 21–34. [DOI] [PubMed] [Google Scholar]
- Fasman, G.D., Lindblow, C., and Grossmann, L. 1964. The helical conformation of polycytidylic acid: Studies on the forces involved. Biochemistry 3: 1015–1021. [DOI] [PubMed] [Google Scholar]
- Fletcher, M., Pestova, T.V., Hellen, C.U.T., and Wagner, G. 1999. Structure and interactions of the translation initiation factor eIF1. EMBO J. 18: 2631–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, C.S., Lee, J.Y., Mayeur, G.L., Bushell, M., Doudna, J.A., and Hershey, J.W.B. 2004. The j- subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J. Biol. Chem. 279: 8946–8956. [DOI] [PubMed] [Google Scholar]
- Freienstein, C. and Blobel, G. 1975. Nonribosomal proteins associated with eukaryotic native small ribosomal subunits. Proc. Natl. Acad. Sci. 72: 3392–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga, P., Garcia-Quintana, D., and Manyosa, J. 1992. Study of polynucleotide conformation by resolution-enhanced ultraviolet spectroscopy. Poly(rC) and poly(dC). Eur. J. Biochem. 210: 205–210. [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Hellen, C.U.T., Witherell, G.W., Schmid, M., Shin, S.H., Pestova, T.V., Gil, A., and Wimmer, E. 1993. A cytoplasmic 57kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. 90: 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henshaw, E.C., Guiney, D.G., and Hirsch, C.A. 1973. The ribosome cycle in mammalian protein synthesis. I. The place of monomeric ribosomes and ribosomal subunits in the cycle. J. Biol. Chem. 248: 4367–4376. [PubMed] [Google Scholar]
- Holcomb, D.N. and Tinoco Jr., I. 1965. Conformation of polyriboadenylic acid: pH and temperature dependence. Biopolymers 3: 121–133. [Google Scholar]
- Imataka, H. and Sonenberg, N. 1997. Human eukaryotic translation initiation factor-4G (eIF4G) possesses two independent binding sites for eIF4A. Mol. Cell. Biol. 17: 6940–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, K. 1982. Translation of poly(A) in eukaryotic cell-free systems. J. Biochem. 91: 1617–1627. [DOI] [PubMed] [Google Scholar]
- Janosi, L., Hara, H., Zhang, S., and Kaji, A. 1996. Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv. Biophys. 32: 121–201. [DOI] [PubMed] [Google Scholar]
- Karimi, R., Pavlov, M.Y., Buckingham, R.H., and Ehrenberg, M. 1999. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell 3: 601–609. [DOI] [PubMed] [Google Scholar]
- Kemper, W.M. and Merrick, W.C. 1979. Preparation of protein synthesis elongation factors from rabbit reticulocytes. Methods Enzymol. 60: 638–648. [DOI] [PubMed] [Google Scholar]
- Kisselev, L.L. and Buckingham, R.H. 2000. Translation termination comes of age. Trends Biochem. Sci. 25: 561–566. [DOI] [PubMed] [Google Scholar]
- Kozak, M. 1977. Nucleotide sequences of 5′-terminal ribosome-protected initiation regions from two reovirus messages. Nature 269: 390–394. [DOI] [PubMed] [Google Scholar]
- Kozak, M. 1989. The scanning model for translation: An update. J. Cell. Biol. 108: 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin, I.B., Kolupaeva, V.G., Marintchev, A., Wagner, G., and Pestova, T.V. 2003. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & Dev. 17: 2786–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütsch, G., Benndorf, R., Westermann, P., Bommer, U.-A., and Bielka, H. 1986. Structure and location of initiation factor eIF3 within native small ribosomal subunits from eukaryotes. Eur. J. Cell. Biol. 40: 257–265. [PubMed] [Google Scholar]
- Majumdar, R., Bandyopadhyay, A., and Maitra, U. 2003. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40S preinitiation complex. J. Biol. Chem. 278: 6580–6587. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Mayeur, G.L., Fraser, C.S., Peiretti, F., Block, K.L., and Hershey, J.W.B. 2003. Characterization of eIF3k. A newly discovered subunit of mammalian translation initiation factor eIF3. Eur. J. Biochem. 270: 4133–4139. [DOI] [PubMed] [Google Scholar]
- Methot, N., Song, M.S., and Sonenberg, N. 1996. A region rich in aspartic acid, arginine, tyrosine and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interactions with eIF3. Mol. Cell. Biol. 16: 5328–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygard, O. and Westermann, P. 1982. Specific interaction of one subunit of eukaryotic initiation factor eIF-3 with 18S ribosomal RNA within the binary complex, eIF-3 small ribosomal subunit, as shown by cross-linking experiments. Nucleic Acids Res. 10: 1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, D.S., Savner, E.M., Mathew, A., Zhang, F., Krishnamoorthy, T., Phan, L., and Hinnebusch, A.G. 2003. Domains of eIF1A that mediate binding to eIF2, 3IF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 22: 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.-S., Himmelbach, A., Browning, K.S., Hohn, T., and Ryabova, L.A. 2001. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106: 723–733. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V. and Hellen, C.U.T. 2001a. Functions of eukaryotic factors in initiation of translation. Cold Spring Harbor Symp. Quant. Biol. 66: 389–396. [DOI] [PubMed] [Google Scholar]
- ———. 2001b. Preparation and activity of synthetic unmodified mammalian tRNAiMet in initiation of translation in vitro. RNA 7: 1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 17: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V. and Kolupaeva, V.G. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 16: 2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U.T, and Shatsky, I.N. 1996a. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16: 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., and Hellen, C.U.T. 1996b. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16: 6870–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Borukhov, S.I., and Hellen, C.U.T. 1998a. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J., and Hellen, C.U.T. 1998b. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal initiation of translation of Hepatitis C virus and Classical swine fever virus RNAs. Genes & Dev. 12: 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Lomakin, I.B., Lee, J.H., Choi, S.K., Dever, T.E., and Hellen, C.U.T. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335. [DOI] [PubMed] [Google Scholar]
- Peterson, D.T., Merrick, W.C., and Safer, B. 1979. Binding and release of radiolabeled eukaryotic initiation factors 2 and 3 during 80S initiation complex formation. J. Biol. Chem. 254: 2509–2516. [PubMed] [Google Scholar]
- Potapov, A.P., Ovcharenko, G.V., Soldatkin, G.O., Shul’ga, N.I., Soldatkin, A.P., and El’skaya, A.V. 1992. Cell-free translation systems from different eukaryotes differ in their sensitivity to a template sugar-phosphate backbone. Biochimie 74: 435–441. [DOI] [PubMed] [Google Scholar]
- Poyry, T.A., Kaminski, A., and Jackson, R.J. 2004. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes & Dev. 18: 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, W.K. and Coleman, W.H. 1971. Polyuridylic acid binding by protein from Ehrlich ascites cell ribosomes and its inhibition by aurintricarboxylic acid. Biochemistry 10: 4304–4312. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak, A., Shin, B.S., Dever, T.E., and Burley, S.K. 2001. Engaging the ribosome: Universal IFs of translation. Trends Biochem. Sci. 26: 705–709. [DOI] [PubMed] [Google Scholar]
- Saenger, W., Riecke, J., and Suck, D. 1975. A structural model for the polyadenylic acid single helix. J. Mol. Biol. 93: 529–534. [DOI] [PubMed] [Google Scholar]
- Salas, J. and Bollum, F.J. 1969. Binding of biosynthetic polyribo- and polydeoxynucleotides to ribosomes. J. Biol. Chem. 244: 1152–1156. [PubMed] [Google Scholar]
- Schreier, M.H. and Staehelin, T. 1973. Initiation of eukaryotic protein synthesis: (Met-tRNAf-40S ribosome) initiation complex catalysed by purified initiation factors in the absence of mRNA. Nat. New Biol. 242: 35–38. [DOI] [PubMed] [Google Scholar]
- Setyono, B., van Steeg, H., and Voorma, H.O. 1984. Ultraviolet-crosslinking reveals specific affinity of eukaryotic initiation factors for Semliki Forest virus mRNA. Biochim. Biophys. Acta 782: 242–246. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N., Morgan, M.A., Testa, D., Colonno, R.J., and Shatkin, A.J. 1979. Interaction of a limited set of proteins with different mRNAs and protection of 5′-caps against pyrophosphatase digestion in initiation complexes. Nucleic Acids Res. 7: 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, C.M., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., and Frank, J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae -tRNA-ribosome and subunit–subunit interactions. Cell 107: 373–386. [DOI] [PubMed] [Google Scholar]
- Srivastava, S., Verschoor, A., and Frank, J. 1992. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit–subunit interface. J. Mol. Biol. 226: 301–304. [DOI] [PubMed] [Google Scholar]
- Stanley, W.M. 1974. Specific aminoacylation of the methionine-specific tRNAs of eukaryotes. Methods Enzymol. 29: 530–547. [DOI] [PubMed] [Google Scholar]
- Thomas, A., Goumans, H., Voorma, H.O., and Benne, R. 1980a. The mechanism of action of eukaryotic initiation factor 4C in protein synthesis. Eur. J. Biochem. 107: 39–45. [DOI] [PubMed] [Google Scholar]
- Thomas, A., Spaan, W., van Steeg, H., Voorma, H.O., and Benne, R. 1980b. Mode of action of protein synthesis initiation factor eIF-1 from rabbit reticulocytes. FEBS Lett. 116: 67–71. [DOI] [PubMed] [Google Scholar]
- Thompson, H.A., Sadnik, I., Scheinbuks, J., and Moldave, K. 1977. Studies on native ribosomal subunits from rat liver. Purification and characterization of ribosome dissociation factors. Biochemistry 16: 2221–2230. [DOI] [PubMed] [Google Scholar]
- Trachsel, H. and Staehelin, T. 1979. Initiation of mammalian protein synthesis. The multiple functions of the initiation factor eIF-3. Biochim. Biophys. Acta 565: 305–314. [DOI] [PubMed] [Google Scholar]
- Trachsel, H., Erni, B., Schreier, M.H., and Staehelin, T. 1977. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J. Mol. Biol. 116: 755–767. [DOI] [PubMed] [Google Scholar]
- Unbehaun, A., Borukhov, S.I., Hellen, C.U.T., and Pestova, T.V. 2004. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes & Dev. 18: 3078–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Válasek, L., Nielsen, K.H., and Hinnebusch, A.G. 2002. Direct eIF2–eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 21: 5886–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Válasek, L., Mathew, A.A., Shin, B.S., Nielsen, K.H., Szamecz, B., and Hinnebusch, A.G. 2003. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes & Dev. 17: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, P. and Nygard, O. 1984. Cross-linking of mRNA to initiation factor eIF-3, 24 kDa cap binding protein and ribosomal proteins S1, S3/3a, S6 and S11 within the 48S pre-initiation complex. Nucleic Acids Res. 12: 8887–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, A.R. 1969. The attachment of polyuridylic acid to reticulocyte ribosomes. Biochem. J. 111: 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T.M.A., Saunders, K., Dowson-Day, M.J., Sleat, D.E., Trachsel, H., and Mundry, K.W. 1990. Effects of the 5′ leader sequence of tobacco mosaic virus RNA, or derivatives thereof, on foreign mRNA and native viral gene expression. In Post-transcriptional control of gene expression (eds. J.E.G. McCarthy and M.F. Tuite), pp. 261–275. NATO ASI Series, v.H49. Springer Verlag, Berlin/Heidelberg, Germany.