Abstract
Pre-messenger RNA (pre-mRNA) splicing is a central step in gene expression. Lying between transcription and protein synthesis, pre-mRNA splicing removes sequences (introns) that would otherwise disrupt the coding potential of intron-containing transcripts. This process takes place in the nucleus, catalyzed by a large RNA–protein complex called the spliceosome. Prp8p, one of the largest and most highly conserved of nuclear proteins, occupies a central position in the catalytic core of the spliceosome, and has been implicated in several crucial molecular rearrangements that occur there. Recently, Prp8p has also come under the spotlight for its role in the inherited human disease, Retinitis Pigmentosa.
Prp8 is unique, having no obvious homology to other proteins; however, using bioinformatical analysis we reveal the presence of a conserved RNA recognition motif (RRM), an MPN/JAB domain and a putative nuclear localization signal (NLS). Here, we review biochemical and genetical data, mostly related to the human and yeast proteins, that describe Prp8’s central role within the spliceosome and its molecular interactions during spliceosome formation, as splicing proceeds, and in post-splicing complexes.
NOTE ON NOMENCLATURE
In this review, Prp8 and Prp8p represent the protein product of the wild-type PRP8 gene of Saccharomyces cerevisiae, while prp8–1 is an example of a mutant allele of the PRP8 gene. Human Prp8 protein is designated hPrp8, also known in the literature as PRPF8, PRPC8, p220, and 220K. In some places, to avoid confusion, yPrp8 is used to distinguish the yeast (S. cerevisiae) protein from the human form. sPrp8SPP42 (Schmidt et al. 1999) or sPrp8cwf6 (McDonald et al. 1999; Ohi et al. 2002) defines the ortholog in Schizosaccharomyces pombe. Confusingly, the cdc28 gene in S. pombe is also referred to as prp8 as a consequence of its role in both pre-mRNA splicing and cell cycle progression, and there is also a temperature-sensitive allele called prp8–1 that causes accumulation of pre-mRNA upon a shift to nonpermissive temperatures (Lundgren et al. 1996; Imamura et al. 1998). Cdc28prp8 encodes a 112-kDa DEAH-box protein. This protein is not the ortholog of the U5 snRNP protein of S. cerevisiae that is discussed here.
In discussions of pre-mRNA–protein cross-links or mutations that affect intron–exon junctions, 5′SS+2 refers to the second intronic base from the 5′ end of the intron, 5′SS-2 is the penultimate base of the 5′ exon, and 3′SS-2 is the penultimate base of the intron.
PRE-mRNA SPLICING
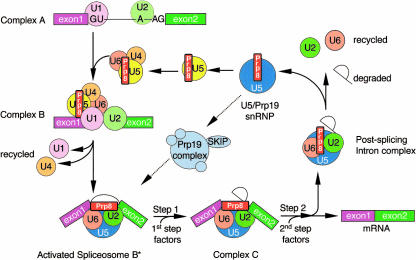
Pre-mRNA splicing involves two trans-esterification reactions within the highly dynamic spliceosome complex. A vast amount of mainly biochemical data led to a consensus view of an ordered pathway of spliceosome assembly that will be described in outline here (for further details, see Kramer 1996; Burge et al. 1999; Brow 2002). The small nuclear RNA–protein (snRNP) complexes, known as U1, U2, U4, U5, and U6 snRNPs, play key roles. U1 is the first snRNP to associate with pre-mRNA, interacting with the 5′ splice site (5′SS). The U1 snRNA becomes base-paired to the 5′ end of the intron, and a “commitment complex” forms, committing the pre-mRNA to the splicing pathway. The U2 snRNP associates with the branchpoint region of the intron to form a pre-spliceosome or “complex A” (Fig. 1 ▶). The U4 and U6 snRNAs share extensive sequence complementarity and are mainly found base-paired together in a U4/U6 di-snRNP. The U4/U6 di-snRNP interacts with U5 snRNP to form a U5•U4/U6 tri-snRNP, which then associates with the pre-spliceosome to form “complex B”. In a simplified trans-splicing system, evidence has been obtained that the early steps of spliceosome assembly can be bypassed under certain conditions; a DNA oligonucleotide complementary to the 5′ end of U1 snRNA was used to block binding of U1 to a short 5′SS RNA substrate, thereby promoting substrate interaction with U5•U4/U6 snRNP and U2/U4/U5/U6 splicing complexes (Kameoka et al. 2004).
FIGURE 1.
Schematic diagram showing the assembly and recycling of spliceosome components with respect to Prp8. The conserved 5′ and 3′ splice sites represent the U2 cis-spliceosomal GU and AG residues, respectively. Prp8 has been found in two different U5 complexes: the U5 snRNP (small, yellow) and the larger U5/Prp19 snRNP (large, blue). (Adapted with permission from Makarov et al. © 2002 AAAS [www.sciencemag.org].)
Formation of the catalytically competent spliceosome (C complex) requires an intricate series of protein and RNA rearrangements, some of which are catalyzed by RNA-dependent NTPases/RNA unwindases: Brr2, Prp2, Prp5, Prp16, Prp22, Prp28, Prp43, and Sub2 (de la Cruz et al. 1999). The concurrent unwinding of the U1 snRNA/5′SS and U4/U6 RNA helices is promoted by the U5 snRNP helicases, Prp28p and Brr2p, respectively. The U6 snRNA then base-pairs with the 5′SS and with U2 snRNA, forming part of the catalytic center of the spliceosome (Staley and Guthrie 1998, 1999). Prp2p, an RNA-dependent NTPase and putative RNA helicase, then appears to interact transiently with the spliceosome, activating it for the first trans-esterification reaction. Following ATP hydrolysis, Prp2p dissociates from the spliceosome (King and Beggs 1990; Kim and Lin 1996).
Upon completion of the first catalytic step, Prp16p, another RNA-dependent NTPase, joins the spliceosome, interacts with the 3′ splice site (3′SS) and drives further rearrangements (Wang and Guthrie 1998; S. Schneider et al. 2002a). The activities of Prp8p, Slu7p, Prp17p, and Prp18p are also required for completion of the second step (Umen and Guthrie 1995b; McPheeters et al. 2000; James et al. 2002). Two further helicases, Prp22p and Prp43p, are required for release of the spliced mRNA and excised intron, respectively (Martin et al. 2002; Schneider et al. 2003).
Following completion of the splicing reaction, spliceosomes are thought to dissociate and reassemble on other pre-mRNAs for further rounds of splicing, at least in vitro. However, preassembled penta-snRNP complexes have been isolated that, when supplemented with additional factors and pre-mRNA, catalyze splicing, suggesting that spliceosomes need not reassemble de novo on pre-mRNA (Stevens et al. 2002; Malca et al. 2003). Thus, alternative spliceosome assembly (Maroney et al. 2000; Nilsen 2003) or recycling pathways (Verdone et al. 2004) may exist.
OVERVIEW OF Prp8 PROTEIN
Prp8p is highly conserved both in its sequence and in its large size, which varies between ~230 and 280 kDa in different organisms. Prp8p is a component of the U5 snRNP (Lossky et al. 1987; Stevens et al. 2001) and U5•U4/U6 tri-snRNP (Teigelkamp et al. 1997; Gottschalk et al. 1999; Stevens and Abelson 1999). Prp8p can be photochemically cross-linked to the 5′SS, the branchpoint (BP) and the 3′SS in the pre-mRNA (Vijayraghavan et al. 1986; Wyatt et al. 1992; Konforti and Konarska 1994 Konforti and Konarska 1995; MacMillan et al. 1994; Teigelkamp et al. 1995a,b; Umen and Guthrie 1995b, Reyes et al. 1996, 1999; Maroney et al. 2000; Ismaili et al. 2001; McPheeters and Muhlenkamp 2003) and to the U5 (Dix et al. 1998) and U6 snRNAs (Vidal et al. 1999), all of which are considered to be in the catalytic center of the spliceosome. This is consistent with the hypothesis that Prp8p may function as a cofactor in RNA catalysis (Collins and Guthrie 2000). In addition to its interaction with the pre-mRNA substrate, Prp8p is found associated with spliceosomes containing the reaction intermediates (products of the first trans-esterification reaction) or the excised intron (Teigelkamp et al. 1995b), and with many large protein complexes (Gottschalk et al. 1999, 2001; Stevens and Abelson 1999; Stevens et al. 2001; Gavin et al. 2002; Hartmuth et al. 2002; Jurica et al. 2002; Makarov et al. 2002; Ohi et al. 2002; Rappsilber et al. 2002; Stevens et al. 2002; Wang et al. 2002; Zhou et al. 2002; Zhang et al. 2003; Krogan et al. 2004; Lee et al. 2004; Masciadri et al. 2004). Thus, Prp8p is present in many conformers of the spliceosome during the splicing cycle as well as being functional in three different types of spliceosomes: The classical U2 spliceosome (shown in Fig. 1 ▶), the U12 spliceosome (Luo et al. 1999), and the trans-spliceosome in which U1 snRNP is functionally replaced by the spliced leader (SL) snRNP (Maroney et al. 1996, 2000; Lucke et al. 1997; Mair et al. 2000). There is no evidence for any additional U12- or trans-splicing-specific protein domains, suggesting that Prp8p functions in a similar way in each type of spliceosome.
IDENTIFICATION OF Prp8p AS A PRE-mRNA SPLICING FACTOR
In 1967, Lee Hartwell and coworkers performed a genetic screen to isolate temperature-sensitive (ts) mutations in Saccharomyces cerevisiae (Hartwell 1967; Hartwell et al. 1970). They identified 10 complementation groups, rna2-rna11, of mutations that caused accumulation of RNA following a shift from the permissive (23°C) to the restrictive (36°C) temperature. Many of these RNA genes were later renamed PRP genes, to indicate their involvement with pre-mRNA processing (Vijayraghavan et al. 1989). At the non-permissive temperature (36°C), rna8–1 (now known as prp8–1) cells accumulated the RPL59 (ribosomal protein 59) pre-mRNA (Larkin and Woolford 1983). Similarly, extract from prp8–1 cells was defective in splicing actin pre-mRNA and could not form the 40S (active) spliceosome (Lustig et al. 1986). Splicing of actin pre-mRNA could be restored upon addition of inactivated extracts from other rna mutants or the wild-type spliceosome fraction I (Brody and Abelson 1985; Lustig et al. 1986; Lin et al. 1987). These results identified the RNA8/PRP8 gene product as an essential mRNA splicing factor that was involved early in the splicing process.
The PRP8 gene was cloned by complementation of the heat-sensitive growth defect of the prp8–1 mutation in S. cerevisiae (Jackson et al. 1988). The 7.4-kb transcript was detected at approximately one copy per yeast cell, and the product was identified as a large, 280-kDa protein. The production of antibodies against Prp8p (Fig. 2 ▶) helped to further identify its role in RNA splicing (Lossky et al. 1987; Jackson et al. 1988). Immunodepletion of Prp8p from yeast extracts with 8.1 antibodies caused loss of splicing activity (Jackson et al. 1988). Metabolic depletion of Prp8p, via a galactose-inducible, glucose-repressible promoter, inhibited splicing in vivo and caused an accumulation of complex A (pre-spliceosome) in vitro; thus U5•U4/U6 tri-snRNP binding to pre-spliceosomes was prevented (Brown and Beggs 1992). Metabolic depletion of Prp8p or heat-inactivation of prp8–1 resulted in a substantial reduction in the levels of U4, U5 and U6 snRNAs, whereas U1 and U2 were unaffected (Brown and Beggs 1992). The loss of U4, U5, and U6 under these conditions was interpreted as being a consequence of aberrantly formed tri-snRNPs being targeted for degradation. The loss of U5 snRNPs was not thought to be the cause as genetic depletion of U5 snRNA had little effect on U4 and U6 levels (Seraphin et al. 1991), nor did prp8–1 show an effect on U4/U6 assembly (Brown and Beggs 1992)
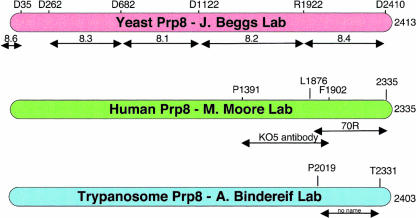
FIGURE 2.
Maps showing regions of Prp8p against which antibodies have been generated. The regions recognized are represented as arrows below the antibody names. S. cerevisiae antibodies 8.1–8.4 were raised against lacZ–protein fusions (Lossky et al. 1987; Jackson et al. 1988) whereas all the others are raised against peptides. α-8.1 only precipitates yPrp8 in U5 snRNPs, whereas α-8.4 and α-8.6 are the only ones that precipitate yeast spliceosomes and co-complexes. The 70R antibody against hPrp8 (Luo et al. 1999) is noted as being superior to KO5 for immunoprecipitation. One other antibody to tbPrp8 is known (Lucke et al. 1997).
Incubation of yeast splicing reaction with 8.4 antibodies resulted in the coimmunoprecipitation of U4, U5 and U6 snRNAs, spliceosomal complexes and the excised intron product of the reaction (Jackson et al. 1988; Whittaker et al. 1990). In contrast, 8.1 antibodies only precipitated U5 snRNP and disrupted the U5•U4/U6 tri-snRNP under the same conditions (Lossky et al. 1987). This indicates that Prp8p is present in spliceosomes during both steps of the RNA splicing reaction and in a post-splicing complex containing the excised intron, and that the 8.1 epitope region of Prp8 (amino acids 682–1122) is required for association of Prp8p with U4/U6 di-snRNP (Brown and Beggs 1992). Yeast Prp8 protein is recognized by 8.2 and 8.3 antibodies only after denaturation (Brown and Beggs 1992).
Prp8p orthologs were soon identified in a variety of higher eukaryotes. Human Prp8 was detected in HeLa cell nuclear extract by Western blotting, using anti-yeast 8.1, 8.2 and 8.4 but not 8.3 antibodies (Anderson et al. 1989). The human 20S U5 snRNP was purified and anti-8.1, 8.2 and 8.4 antibodies identified hPrp8 by Western blotting as a doublet running at ~200 kDa (Bach et al. 1989; Pinto and Steitz 1989). In addition, a HeLa 220-kDa protein that bound to Ad10 pre-mRNA in a splicing-dependent manner, cross-reacted with a mixture of anti-yeast antibodies 8.2, 8.3, and 8.4; immunoblotting with yeast anti-8.2 showed a doublet >200 kDa (Garcia-Blanco et al. 1990). Nuclear extracts from drosophila, tobacco, and pea were also shown to contain proteins that reacted to anti-yeast Prp8 antibodies (Paterson et al. 1991; Kulesza et al. 1993).
Polyclonal antibodies (70R and KO5) raised against the C-terminal 50-kDa region of hPrp8 immunoprecipitated both the U2 and U12 spliceosomes from HeLa cell nuclear extract, with 70R noted as being more efficacious (Hinz et al. 1996; Luo et al. 1999). One other antibody, against the C-terminal region of the trypanosome Prp8 protein, specifically coimmunoprecipitated U5 snRNA and U5 snRNP proteins, suggesting that basic functions of the U5 snRNP are conserved between cis- and trans-splicing systems (Lucke et al. 1997).
Prp8: EVOLUTIONARY CONSERVATION AND PROTEIN DOMAINS
Prp8 protein exhibits a high degree of conservation throughout all eukaryotes, with an overall 61% identity from yeast to humans in its amino acid sequence (Hodges et al. 1995; Luo et al. 1999). Comparison of the yeast proteome database (http://proteome.incyte.com) with the human proteome database showed that Prp8p is one of the most highly conserved nuclear proteins known. Twenty-eight full-length sequences of the PRP8 genes from 26 eukaryotic organisms are currently available (see Supplemental Table S1 at http://www.ed.ac.uk/~jeanb/) and the databases contain partial sequence information for many more. One overriding conserved feature of the Prp8 protein is its large size, most orthologs being between 2317 and 2416 amino acids long. The two smallest Prp8 proteins belong to the microsporidia Encephalitozoon cuniculi and the minute nucleomorph genome from Guillardia theta, respectively. E. cuniculi is a human parasite with a genome size of less than three megabases (Katinka et al. 2001) and, interestingly, its Prp8p, at 2172 amino acids, is 10% smaller than in most other eukaryotes. G. theta (G.t.) is a cryptomonad, an evolutionary chimaera of two eukaryotes, red algae and a non-photosynthetic host. G.t. has retained the enslaved red algal nucleus as a minute nucleomorph containing 0.55 mega-bases and, at 2057 amino acids, its Prp8p is the shortest known, although still very large (Douglas et al. 2001). In these organisms, Prp8p has been severely compacted, resulting in reduced N and C termini. On the other hand, the largest Prp8 proteins belong to the two plasmodium species; they are >2747 amino acids and have multiple, large insertions throughout their sequences. These could form potential targets for anti-malaria drugs.
Arabidopsis thaliana has two copies of PRP8, one on chromosome 1 and another on chromosome 4, that encode proteins with 92% identity. The results of random T-DNA insertions indicate that the gene on chromosome 1 is involved in embryogenesis, with linkage to the emb and sus2 phenotypes. Typically, such mutations cause developmental arrest producing embryos with an abnormal suspensor. There is no known phenotype for mutations of the chromosome 4 gene (www.arabidopsis.org; Huala et al. 2001). Rice also has two copies of PRP8, one on chromosome 5 the other on chromosome 6 (see Supplemental Table S1 at http://www.ed.ac.uk/~jeanb/), that encode proteins which are 99% identical. Duplicated genes in plants usually have different expression patterns; therefore, one gene functionally predominates over the other.
Human Prp8 is encoded by a single gene with 42 exons. As it is essential for pre-mRNA splicing, presumably Prp8p function is required in all tissues, however, its expression is notably higher in skeletal and cardiac muscle (Luo et al. 1999). Mouse Prp8 differs by only three amino acid residues from the human ortholog and, in adult mice, is detected most strongly in the testis and ovary (Takahashi et al. 2001). RNAi performed against a portion of the prp-8 mRNA in the gonads of Caenorhabditis elegans, resulted in the arrest of embryogenesis at the late-gastrulation stage (Takahashi et al. 2001). Other RNAi experiments against prp-8 in adult C. elegans also showed a high level of sterility (>90%) (Gonczy et al. 2000; MacMorris et al. 2003), a clear body morphology, and a protruding vulva (Kamath et al. 2003). These results suggest that Prp8p plays an important role in reproduction and development. Recently, hPrp8 has been associated with Retinitis Pigmentosa, a disorder that starts with loss of peripheral vision and night blindness in childhood and often leads to blindness in adults (McKie et al. 2001).
The primary amino acid sequence of Prp8p lacks any obvious protein motif; however, by comparing the many different Prp8p sequences, a number of features can be identified. Here we describe protein motifs that may be relevant to Prp8p function.
Proline-rich tracts
Proline-rich regions are found at the N termini of all fungal Prp8 proteins and the two rice sequences, but in most organisms they are absent (Fig. 3A ▶). Polyproline sequences adopt an extended helical structure with three residues per turn (Kay et al. 2000) and are generally found at the N or C termini where they are exposed and available to bind to other proteins. Recently, the yLin1/Snu40 (YHR156C) protein was shown to interact with the N terminus of yPrp8 (Bialkowska and Kurlandzka 2002). Lin1p/Snu40 is a nonessential protein that associates with Prp8p and the U5 snRNP, but not with the yeast tri- or penta-snRNP complexes, and its role may be in U5 snRNP biogenesis (Stevens et al. 2001; Gavin et al. 2002). It has a GYF domain; GYF domains are known to bind polyproline sequences (Freund et al. 1999). As Lin1p is nonessential and the Prp8p polyproline domains are not conserved, these sequences may have functions that are specific to fungi and plants. Another protein, yPrp40, was also reported to bind to this region of Prp8p (Abovich and Rosbash 1997).
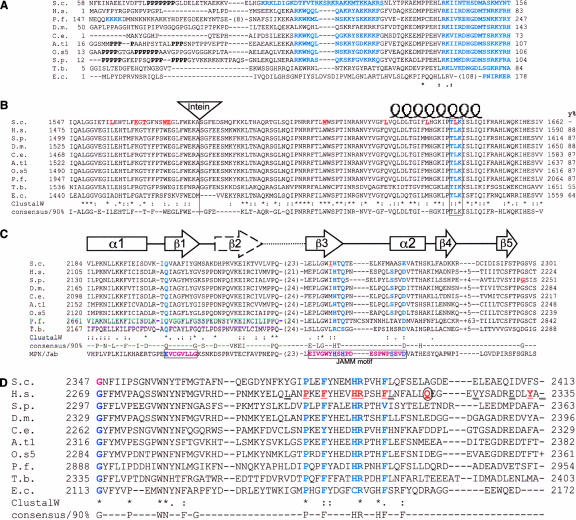
FIGURE 3.
Alignments of 10 Prp8 orthologs showing four conserved motifs. Important residues are highlighted in bold blue type and known alleles underlined in red. (S.c.) Saccharomyces cerevisiae; (H.s.) Homo sapiens; (P.f.) Plasmodium falciparum; (D.m.) Drosophila melonagaster; (C.e.) Caenorhabditis elegans; (A.t1.) Arabidopsis thaliana chromosome 1; (O.s5) Oryza sativa chromosome 5; (S.p.) Schizosaccharomyces pombe; (T.b.) Trypanosoma buceii; (E.c.) Encephalitazoon cuniculii. Numbers refer to the amino acid residues. All sequences aligned using ClustalW and consensii are derived from 25 Prp8 orthologs using the ‘consensus’ program. (A) Alignment of the N-terminal regions. The polyproline tracts can be seen in the nonconserved N termini of some species. Potential bipartite NLS sequences are highlighted in bold blue type and are separated by 15 amino acids. In the case of S. cerevisiae an overlapping nuclear localization signal (NLS) sequence is underlined. In the cases of P. falciparum and E. cuniculi, a shorter SV40 type NLS and pat7 types are highlighted, respectively. The NLS sequences always occur within the first 500 N-terminal residues as predicted by PSORTII (Nakai and Horton 1999). (B) The highly conserved Prp8 domain 3.2. The position of the intein, present in four orthologs is indicated by the triangle. A potential protein kinase C phosphorylation site (T/S-x-R/K) that is conserved in all species is boxed and highlighted. This is central to the postulated coiled-coil domain represented above the text by the coil. (y%) The level of conservation compared with the yeast sequence. (C) The MPN/JAB domain found in Prp8. The mutations yprp8–28 and sprp8spp42–1 are shown in bold red underlined text. The representative MPN/JAB domain from NCBI’s domain database is shown at the bottom of the alignment. Two important MPN motif residues identified previously are boxed in plum coloured text (Maytal-Kivity et al. 2002). The five residues in the MPN structure that coordinate the zinc are highlighted in bold blue text. The secondary structure diagrams are shown above the alignment (from Tran et al. 2003). (D) The C terminus of Prp8 showing the single mutated residues altered in human Retinitis Pigmentosa (bold, red underlined type), the nonsense mutation (bold, red, and circled) and the residues where frameshift mutations occur (doublly underlined text). The temperature-sensitive yprp8–1 mutation affects the first glycine residue on the left-hand side of the alignment (bold plum).
Nuclear localization signals (NLS)
Using the algorithm of Nakai (Nakai and Horton 1999), putative bipartite nuclear localization signals (NLS) are identifiable within 500 amino acids of the N terminus of Prp8 in all organisms. A consensus bipartite NLS for Prp8 contains two clusters of positively charged residues separated by a variable region of 10–12 amino acids. At the N terminus in the majority of species, pairs of such bipartite sequences are found, often separated by 15 amino acids. Figure 3A ▶ shows the consensus bipartite NLSs in most Prp8 proteins according to PSORT II (Nakai and Horton 1999). Those species lacking a consensus sequence appear to contain alternatives, such as the simple NLS typified by SV40 or the type known as pat7. These are also located within the first 500 amino acids of the proteins and examples are highlighted in Figure 3A ▶ for Plasmodium falciparum and E. cuniculii, respectively.
If the bipartite sequences function as nuclear localization signals, they will allow the protein to enter the nucleus assisted by the importin-α family of proteins (Izaurralde et al. 1998). The putative NLS sequence of Prp8p is sufficient to localize Prp8p to the nucleus in S. cerevisiae (K-L. Boon and J.D. Beggs, unpubl.), but whether this also directs the nuclear uptake of other associated U5 snRNP proteins is currently not known. Prp8p has also been detected in the human nucleolus (Andersen et al. 2002).
3′ splice site fidelity region 3.2
A 3′ splice site (3′SS) fidelity region between amino acids 1372 and 1660 in yeast Prp8p was originally identified by yPrp8 mutations that suppressed defects in splicing pre-mRNA 3′SS mutations (Umen and Guthrie 1996). Subsequently, a large number of additional yeast genetic loci that suppress 3′SS splicing defects have been identified (Collins and Guthrie 1999; Siatecka et al. 1999; Ben-Yehuda et al. 2000b; Dagher and Fu 2001; Query and Konarska 2004). Scrutiny of sequence alignments of the affected residues has prompted us to split this domain in two. The C-terminal half, amino acids 1547–1660 in yeast, is referred to here as region 3.2 (Fig. 3B ▶). It is exceptionally well-conserved, with 55%–87% identity over 113 amino acids (Hodges et al. 1995; Luo et al. 1999; Siatecka et al. 1999). The amino acid sequence from 1372–1546, which we refer to as region 3.1, is less highly conserved (39%–68% identity) and includes only two 3′ splice site fidelity mutations: M1399I (Umen and Guthrie 1996) and prp8–39, a mutation that is also synthetic lethal with sky1Δ (Dagher and Fu 2001).
Region 3.2 (y1547–1660) contains all but two of the 3′SS suppressor mutations and a conserved consensus site for phosphorylation by protein kinase C. In vivo, protein kinase C exhibits a preference for the phosphorylation of serine or threonine residues found on the N-side of a basic residue. This sequence in yPrp8p is embedded within a predicted coiled-coil domain that is hypothesized to interact with residues y643–669 (Kuhn and Brow 2000). Conceivably, intramolecular Prp8 interactions may be controlled by phosphorylation at this site.
In a number of fungal pathogens this conserved region of Prp8p (WERA*SGFE) (yA1578) is the insertion site (*) for an internal protein (intein). Inteins are peptides that can catalyze their own excision followed by ligation of the flanking protein regions. The pathogenic Cryptococcus species contain a 170–172-amino-acid intein (Butler et al. 2001), Ajellomyces capsulatus a 530-amino-acid intein and Aspergillus species a 605–819 residue insertion. These inteins are potential targets for drug design (Liu and Yang 2004). Inteins are usually only found in proteins that are essential for the survival of the organism. In these cases the DNA coding for the protein is not permitted to alter greatly over the evolutionary timescale; thus, intein removal from this DNA is not selected for (Pietrokovski 2001). The unusually high conservation of the DNA sequence that encodes motif 3.2 (72% identity between yeast and human) provides powerful evidence for common ancestry. This highly conserved sequence must reflect a crucial but so far unknown role in splicing. Consequently, we propose that region 3.2 may be responsible for Prp8p’s postulated role in promoting RNA-mediated catalysis between the conserved 5′SS, 3′SS, and branchpoint, (a refinement of the model of Collins and Guthrie 2000).
MPN (Mpr-1, Pad-1, N-terminal) domain
Prp8p contains a conserved MPN domain (also known as a Mov34, JAB, MPN+, PAD-1, or JAMM domain) at its C terminus (2178–2310 in yeast). MPN domains are found in the N termini of proteins with a variety of functions: proteasome regulatory subunits, eukaryotic initiation factor 3 (eIF3) subunits, the signalosome, and regulators of transcription factors. Rpn11, a proteasome component, is a typical example of the MPN superfamily. The structure of an MPN domain was recently solved (Tran et al. 2003; Ambroggio et al. 2004) and shows that five conserved residues are responsible for coordinating a Zn2+ ion. The MPN domain contains a conserved glutamate (E) and a JAMM motif that includes a conserved H-x-H-x[7]-S-xx-D sequence, where x represents nonconserved residues. Prp8’s MPN domain is slightly different, having the consensus H-x-Q-x[7]-S-xx-D and having glutamine instead of the conserved glutamate (Fig. 3C ▶). Therefore, although Prp8’s MPN domain fits into the family (Maytal-Kivity et al. 2002) and may fold in the same way as other MPN domains, as it contains only three out of the five conserved residues, it may not bind zinc. In a comparable MPN structure in cytidine deaminase, an α helix can slot into the furrow formed at the zinc-binding site (Tran et al. 2003; Ambroggio et al. 2004), and so the MPN domain in Prp8 may simply act as a protein interaction domain. Substituting alanine for four of the residues in yPrp8p that are thought to coordinate zinc produced a temperature-sensitive phenotype (P. Bellare and E. Sontheimer, pers. comm.).
In the case of two interacting MPN proteins, Rpn11p and Rpn8p, of the proteasome regulatory particle, it was found that the two MPN domains as well as C-terminal flanking sequences were required for the interaction (Fu et al. 2001). Two-hybrid analyzes have indicated an interaction of the C terminus of yPrp8p with Brr2p, and indeed, a yeast prp8–28 mutation (I2259N) that lies in the putative zinc-binding region of the MPN domain abolishes this interaction (van Nues and Beggs 2001). Brr2p does not have an MPN domain but, as previously discussed, a simple α helix may be all that is required.
Mutagenizing the conserved JAMM residues in Rpn11 (proteasome) and its related subunit Csn5 (signalosome) revealed functions in ubiquitin removal and removal of the ubiquitin-like Nedd8, respectively (Fu et al. 2001; Ambroggio et al. 2004). The spliceosomal Prp19p protein has been shown to promote the ligation of ubiquitin to proteins in an ubiquitin ligase reaction (Hatakeyama et al. 2001; Ohi et al. 2003). This raises the possibility that Prp8’s MPN domain may be involved with (de-)ubiqitination during the spliceosome cycle.
As the two smallest Prp8 proteins, of E. cuniculi and the nucleomorph from G. theta, lack this domain, the C-terminal portion of Prp8 may have a regulatory function that is specific to higher eukaryotes.
The human Retinitis Pigmentosa mutations
Autosomal dominant Retinitis Pigmentosa is a genetic disorder causing degeneration of the photoreceptors in the retina. A severe form of this disease (RP13) was found to be associated with mutations in human PRP8 (McKie et al. 2001; De Erkenez et al. 2002; van Lith-Verhoeven et al. 2002; Kondo et al. 2003; Martinez-Gimeno et al. 2003). The association of hPrp8p with a retinal-specific disorder was a surprising result, as Prp8 function is required in all cell types. All the RP13 mutations are concentrated in the last exon, encoding the very C terminus of the protein. Nine RP13 missense mutations affect seven very highly conserved amino acid residues in the region hP2301–F2334 (Fig. 3D ▶), suggesting that a conserved function may be involved. This corresponds to the region of yPrp8p that interacts with Brr2p, and is close to the prp8–1 (G2347D) mutation, which ablates the binding of C-terminal yPrp8 peptides by yBrr2p in vitro (van Nues and Beggs 2001). The C-terminal 94-amino-acid region of hPrp8p interacts with the multifunctional PAI-2 protein, the significance of which is unclear (Fan et al. 2004). Other RP13 mutations correspond to frameshifts that extend from residues h2298, 2315, 2325, 2331, and 2335 (the normal stop codon), all resulting in the introduction of nonconserved amino acids (De Erkenez et al. 2002; Martinez-Gimeno et al. 2003). In the case of the mutation of the stop codon, the protein extension may be deleterious, as the conserved sequences at the C terminus are unaffected. Extra length per se cannot be concluded to affect normal Prp8 function because one rice Prp8p paralog may contain an extra 30 amino acids at the C terminus and the addition of C-terminal epitope tags to yPrp8p does not affect cell viability (Gavin et al. 2002; Krogan et al. 2004). Three other splicing proteins, Prp3 (Chakarova et al. 2002), Prp31 (Vithana et al. 2001; Martinez-Gimeno et al. 2003), and PAP-1 (Maita et al. 2004, 2005), are also linked to Retinitis Pigmentosa. Like Prp8p, PAP-1, Prp31, and Prp3 are involved in formation of the U5•U4/U6 tri-snRNP (Anthony et al. 1997; Makarova et al. 2002).
The mammalian protein midkine (MK) is a 15-kDa protein with a heparin-binding domain, and is produced in response to constant light-induced retinal activation. Pre-injection of MK into the eyes of albino rodents prior to exposing them to constant light for up to 21 d showed that midkine prevented retinal degeneration (Unoki et al. 1994, 1995; Masuda et al. 1995). Interestingly, a midkine protein complex from mouse membrane fractions contained Prp8p, identified by mass spectrometry. Further analysis showed that when purified Prp8p was treated with RNase it still bound to the MK complex in up to 300 mM NaCl (Takahashi et al. 2001). The production of a mouse model to examine the effects of RP13 mutations on the midkine complex may determine whether this is relevant to the pathology of the disease.
A conserved RNA recognition motif (RRM)
Here, we identify for the first time, a conserved sequence in Prp8p that closely resembles an RNA recognition motif (RRM) (Fig. 4 ▶). An analysis of this sequence using a Hidden Markov Model with the HMMER package (Eddy 1998; see Supplemental Fig. S1 at http://www.ed.ac.uk/~jeanb/) against the human NCBI database gives a high confidence value that this represents a conserved RRM. All Prp8 proteins for which sequences are available contain the motif. This provides a possible RNA binding center for the 5′SS, BP, or 3′SS of pre-mRNA, or for the binding of the U5 or U6 snRNAs, all of which are known to contact with Prp8p (see below). The most conserved regions of an RRM are defined as the RNP1 and RNP2 sequences (Birney et al. 1993; Burd and Dreyfuss 1994). RNP1 usually contains an aromatic residue, either phenylalanine or tyrosine, that stacks between the RNA bases and effects RNA recognition. Recognition of RNA targets can also be modulated by a number of other factors, most notably the two loops β1–α1, β2–β3 and the amino acid residues C-terminal to the RNP2 domain (Varani and Nagai 1998; Perez-Canadillas and Varani 2001; Kielkopf et al. 2004; Stefl et al. 2005).
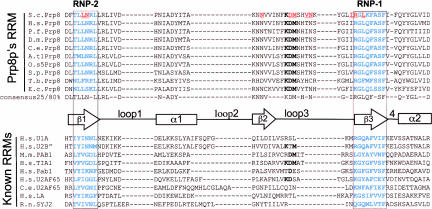
FIGURE 4.
Prp8 contains a conserved RNA recognition motif (RRM). Using a hidden Markov model with the HMMER package (Eddy 1998) against the human NCBI database, only RRM containing proteins were identified. (The model is available as Supplementary Data at http://www.ed.ac.uk/~jeanb/; Supplemental Fig. S1). The alignment between 10 different Prp8 sequences and nine known RRMs from the Interpro database is carried out using ClustalW or T-coffee in combination with Jalview (Thompson et al. 1994; Notredame et al. 2000). The conserved RNP-1 and RNP-2 motifs are boxed and highlighted in bold blue type. A number of mutations are known in yeast Prp8 that suppress brr2–1 and/or U4–cs1 mutations and these are highlighted by bold red underlined text. A unique feature of Prp8’s RRM is the conserved amino acid triplet KDM in the center of the domain. The consensus calculations are derived from 25 different Prp8p sequences from 23 species using ClustalW and the online “consensus” program. A stylized representation of the RRM secondary structure based on the U1A crystal structure is shown between the primary sequences.
Several mutations in the RRM of yPrp8p (mutated residues are underlined in Fig. 4 ▶) are known to suppress the cold-sensitive U4–cs1 and/or brr2–1 mutations, both of which affect the unwinding of U6 from U4 (Kuhn et al. 1999, 2002; Kuhn and Brow 2000). These mutations occur mainly in the β sheet regions, which are known to form the main protein surfaces that interact with RNA. A conserved unique feature about the RRM of Prp8 proteins, that distinguishes it from all others, is the amino acid triplet, KDM (Fig. 4 ▶). Three suppressors of U4–cs1 lie in this triptych at the loop region connecting β sheets 2 and 3, a region that plays an important role in determining RNA specificity. The mechanism by which Prp8’s RRM might normally affect U4/U6 unwinding may involve: (1) stabilizing an RNA structure within the U4/U6 di-snRNP; (2) masking the other U6-binding partner (i.e., the 5′SS); or (3) acting indirectly, by modulating the activity of another protein that affects U4/U6 unwinding, RRMs are also known to bind proteins (Kielkopf et al. 2004 and references therein).
Prp8p–RNA CROSS-LINKING
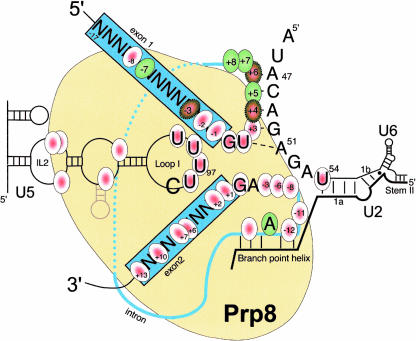
Photoactivatable base analogs in nucleic acids will cross-link efficiently to certain residues in contacting proteins (Reed and Chiara 1999). Exposure of normal bases to short wavelength UV light (254 nm) and of 4-thiouridine (SU) or other photoactivatable analogs to long wavelength UV (365 nm) induces cross-linking of close molecular contacts. Short-range cross-links form with a radius of 1.4Å for oxygen in unmodified uridine and 1.85Å for the sulphur atom in SU. The lower electronegativity of sulphur renders the uridine ring more sensitive to photo-activation and so SU efficiently forms short-range cross-links (Favre 1990; Favre et al. 1998). Other photoreagents can form longer-range cross-links (e.g., benzophenone protrudes from the RNA by 15Å) (MacMillan et al. 1994). If a photoactivatable analog is radiolabeled and incorporated at a specific site in an RNA, then proteins which contact that site can be cross-linked there and become indirectly radiolabeled. This approach has been extensively used to identify nucleic acid and protein residues interacting with the 5′SS and other RNA sequences in and around the spliceosome’s active site. Numerous cross-linking studies have placed Prp8p at or near the catalytic core, as summarized in Figure 5 ▶. To date, Prp8p is the only spliceosomal protein that has been shown to cross-link to all three regions in pre-mRNA that are required for splicing (5′SS, 3′SS, and BP), as well as to U5 snRNA and U6 snRNA.
FIGURE 5.
A summary of the RNA residues that are known to cross-link to Prp8 in the context of the spliceosome. Cross-linked residues are circled, the exons are boxed, and the intron is represented by a thin blue line. (Red circles) Short-range cross-links using uridine and 4-thiouridine; (green circles) long-range cross-links using the reagent benzophenone. Where both cross-linking agents were used at the same site the circles colored red and green with a dotted outline. Extensive Prp8 cross-links with U5 snRNA are also represented as is the U6 residue 54. Both the U5 and U6 cross-links were only demonstrated in snRNPs. All numbering relates to S. cerevisiae.
Prp8p–RNA cross-linking: The 5′ splice site
Yeast and HeLa proteins that directly bind to pre-mRNA during the splicing reaction were studied using UV cross-linking combined with immunoprecipitation to detect the cross-linked proteins. yPrp8 was found to cross-link to uniformly-labeled wild-type pre-mRNA substrates, but not to RNAs with mutations in the 5′SS or BP that prevented spliceosome assembly (Whittaker and Beggs 1991). To identify sites in the RNA where Prp8p bound, cross-linking reagents were incorporated at specific sites in pre-mRNA substrates, for example hPrp8 contacts were detected at positions -7, -3, -2, +3 but not −17 or −27 with respect to the 5′SS (Wyatt et al. 1992; Chiara et al. 1996; Reichert et al. 2002). U5 snRNA also cross-linked to the 5′SS -2 position in HeLa nuclear extract before the first step of splicing had occurred (Wyatt et al. 1992). The U5 snRNA cross-links were localized to a phylogenetically invariant 9-nucleotide sequence in loop 1. In yeast extracts, Prp8 cross-linked at the 5′SS -1, -2, and -8 positions but not at +4 in the intron before step one in a stalled (Prp2Δ) spliceosome (Teigelkamp et al. 1995a,b). This indicates that Prp8p interacts with the 5′SS and at least 8 nucleotides of the 5′ exon before step one occurs (Fig. 5 ▶).
Cross-linking of hPrp8p to the conserved 5′SS+1 (G) residue in the intron was observed before step one of splicing, in the commitment complex (with spliceosome assembly blocked by preventing U2 snRNP binding). RNase H-mediated removal of U4, U6 and U5 snRNAs showed that the hPrp8/5′SS interaction occurred when hPrp8p was part of the tri-snRNP, and that it was ATP-dependent (Maroney et al. 2000). The use of ATP-depleted extracts identified a U1 snRNP footprint around the 5′SS that spanned residues −5 to +14 (region A). Upon incubation with ATP and the addition of the tri-snRNP, the footprint extended from −15 to +14. This suggests that the 5′SS can be simultaneously occupied by the U1 snRNP and the tri-snRNP before U2 binds to the 3′SS (Maroney et al. 2000). As Prp8 is known to cross-link to the 5′SS region from at least −8 to +1 before step one, Prp8p may be a component of the U2-independent tri-snRNP footprint at the 5′SS.
Konarska and coworkers used a short RNA endecamer (11-mer) comprising the 5′SS consensus sequence in an in vitro cross-linking system. The 5′SS-endecamer binds efficiently to hPrp8p in the U5•U4/U6 tri-snRNP in a U2 snRNP-independent manner (Konforti and Konarska 1994, 1995; Reyes et al. 1996, 1999). UV cross-linking at 254 nm identified hPrp8 (p220), U1 and U6 all cross-linking to the 5′SS. The U1 and U6 cross-links were mutually exclusive, with the U1 interaction occurring prior to the U6 contact. Altering the 5′SS sequence away from the consensus U + 2 and G + 1 reduced the cross-linking to hPrp8 by 90% and 95%, respectively. Mutating the 5′ exon positions G - 1 and A - 2 reduced hPrp8p cross-linking by 66%. The hPrp8p cross-link was confirmed using 4-thiouridine at intron position +2; whereas, no cross-link was obtained at position +6. Neither U1 snRNA nor U6 snRNA cross-linked to SU + 2, but SU+6 generated a cross-link with U1 snRNA. Using benzophenone, the 5′SS positions −3 and +4 to +8 cross-linked to hPrp8p; however, cross-links closer to the 5′SS junction could not be assessed due to the benzophe-none preventing complex formation (Ismaili et al. 2001).
Using proteases and chemical agents, the position of the cross-link to the 5′SS GU dinucleotide was mapped within hPrp8p to the five residues QACLK (positions 1894–1898). This corresponds to SAAMS (positions 1966–1970) in Prp8p of S. cerevisiae, which is also part of the polypyrimidine tract recognition region (see below). Further analysis by Konarska et al. showed that a recombinant hPrp8 fragment (h1669–2034), referred to as domain IV, is sufficient to bind and cross-link to the 5′SS-endecamer (M. Konarska, pers. comm.). Chimeric constructs of domain IV containing the SAAMS sequence in place of QACKL still cross-linked to the 5′SS. This shows that the penta-peptide is not strictly required for cross-linking to the 5′SS although it is in close proximity to it. This tallies with this region of Prp8p not being conserved and suggests that other residues within domain IV contribute to binding to the 5′SS (M. Konarska, pers. comm.).
Prp8p–RNA cross-linking: The 3′ splice site
Cross-linking of yPrp8 to SU at the exonic 3′SS +1 position occurred under splicing conditions but was not detected in Prp2p-depleted spliceosomes that were stalled prior to step one. When Prp2p was added back to the Prp2p-depleted reaction, the Prp8–3′SS interaction was restored, indicating that this contact was initiated during or subsequent to step one (Teigelkamp et al. 1995a). In yeast extracts, pre-mRNA with the 3′SS CAG>CUC mutation, that replaces the evolutionarily conserved AG, cannot undergo the second step of splicing (Vijayraghavan et al. 1986). Nevertheless, the 3′SS +1 cross-link to Prp8p still occurred at the mutant site (Teigelkamp et al. 1995a). Human Prp8 cross-linked strongly at the lariat 3′SS -6 residue in spliceosomes stalled prior to step two. It also cross-linked to the other side of the 3′ splice site at the exonic +6 and +7 positions (Chiara et al. 1996, 1997). Therefore this 3′SS cross-link was established at or after step one but before step two and the interaction did not depend on a functional 3′ splice site. Thus cross-links to yPrp8p and hPrp8 span a region from the BP to 3′SS plus 13 bases into the 3′ exon (MacMillan et al. 1994; Teigelkamp et al. 1995a,b; McPheeters and Muhlenkamp 2003; Fig. 5 ▶).
The yeast prp8–101 (E1960K) mutation lies within the 5′SS cross-link area of Prp8p, but it also causes a defect in 3′SS recognition, resulting in a two- to fourfold decreased cross-linking signal at the intronic 3′SS -1G compared to wild-type yPrp8p (Umen and Guthrie 1995b). In genetic experiments, prp8–101 showed synthetic lethality with alleles of PRP16, PRP17, PRP18, and SLU7, all of which cause step two splicing defects, but not with first step splicing mutations. Prp16p, Prp17p, Prp18p, Slu7p, and Prp22p function exclusively at the second step by interacting with the 3′ splice site in a sequential manner (Umen and Guthrie 1995b; James et al. 2002). Of these second step proteins, only Prp8p, Prp16p and Slu7p cross-linked specifically with the 3′SS.
Although Prp8p and Slu7p were detectably cross-linked to the 3′SS in spliceosomes that were stalled prior to step two of splicing by immunodepleting Prp16p, maximal cross-linking required both functional Prp16p and ATP (Umen and Guthrie 1995b; McPheeters and Muhlenkamp 2003). Also, use of heat-inactivated prp16–1 protein, that is strongly defective for ATP binding and cannot release from spliceosomes, abolished the Prp8p and Slu7p 3′SS cross-links. Therefore an order of interaction was proposed in which Prp16p binds to the 3′SS first, and its ATP-dependent release may be required for contact by Prp8p and Slu7p.
In prp17 and prp18 mutant extracts that block the second step of splicing, complementation of the defects resulted in a slight increase in the cross-linking of Prp8p and Slu7p, while that of Prp16 was unchanged. In contrast, with extracts immunodepleted for Slu7p, cross-linking of Prp8p to the 3′SS was reduced, and Prp16p cross-links were enhanced (Umen and Guthrie 1995b). It is known that Prp17p acts before an ATP-requiring step (Jones et al. 1995), that Slu7p is required to hold cleaved exon 1 to the spliceosome (Chua and Reed 1999) and that Prp18p cooperates with Slu7p to help bind Prp22p to the spliceosome (James et al. 2002).
Summary of cross-linking and the “lock and load” hypothesis
Before splicing occurs, Prp8 is involved in cross-linking to residues −8 to +3 that span the 5′SS. After the first step of splicing has occurred, Prp8p and Slu7p lock the cleaved 5′ exon into the spliceosome, possibly involving the newly defined domain IV of Prp8p. At least two distinct conformational changes then occur at the 3′SS. The second step RNA helicase, Prp16p, binds to the 3′SS, hydrolyzes ATP and releases, followed by stronger binding of Prp8p and Slu7p at the 3′SS, possibly facilitated by Prp8’s 3′SS fidelity region 3.2, Prp17p and Prp18p (Umen and Guthrie 1995b).
As the binding of Prp8p and Slu7p at the 3′SS is the last defined event prior to catalysis of the second step of splicing, it seems likely that when Prp16p vacates the 3′SS, Prp8p and Slu7p, along with loop1 of U5 snRNA, hold the 3′SS in close proximity to the 5′ exon, positioning the two RNAs for catalysis. Prp8p might be considered as a protein cofactor in the catalytic reaction (Ben-Yehuda et al. 2000a; Collins and Guthrie 2000). We have already proposed that region 3.2 is the possible protein cofactor at the transition site; in effect, domain IV of Prp8p might lock the 5′SS-OH (exon) and load it onto the 3′SS-associated region 3.2.
Prp8p–RNA cross-linking: U5 and U6 snRNAs
In reconstituted yeast U5 snRNPs, the whole 5′ stem-loop system of yU5 snRNA, including the invariant loop 1 nucleotides yU96–U99 (hU40–hU43) and the internal loops IL1 and IL2 were cross-linked to yPrp8p (Dix et al. 1998). Snu114p also cross-linked to the yU5 IL1 (=hIL2) (Dix et al. 1998). In HeLa nuclear extract, coprecipitation of U5 snRNA using hPrp8 antibodies showed that the hIL2 and stem 1b were essential for maximal binding to hPrp8 (Hinz et al. 1996), and in the tri-snRNP, hPrp8p cross-linked to hU5 snRNA loop 1 ~ 50-fold stronger than in the U5 snRNP alone (Urlaub et al. 2000).
With yU6 snRNA, Prp8p cross-linked at position U54, which is in the conserved region that contributes to the catalytic core of the spliceosome (Vidal et al. 1999). This is consistent with the hU6 A51 cross-link to 5′SS +2 (Sontheimer and Steitz 1993; Kim and Abelson 1996), where Prp8 also interacts.
Both U5 snRNA loop 1 and Prp8p can be cross-linked to the pre-mRNA 5′ and 3′ splice sites in HeLa and yeast extracts, and these contacts occur at similar stages in the splicing reaction (Newman and Norman 1992; Wyatt et al. 1992; Sontheimer and Steitz 1993; Newman et al. 1995; Teigelkamp et al. 1995a; Umen and Guthrie 1995a; O’Keefe et al. 1996; Reyes et al. 1996; Dix et al. 1998; Segault et al. 1999; Alvi et al. 2001; McConnell and Steitz 2001; O’Keefe 2002). U5 snRNA was proposed to anchor and align the ends of the exons in the catalytic center of the spliceosome; however, U5 snRNA loop 1 is not essential for splicing in HeLa nuclear extract (Segault et al. 1999), and is only required for step two in yeast extracts (O’Keefe et al. 1996; O’Keefe and Newman 1998). Also, cross-linking of yPrp8p to the 3′SS occurred even in the absence of U5 loop 1 (Dix et al. 1998). It has been hypothesized that Prp8p may be the principle factor that anchors the exons in spliceosomes, acting in conjunction with U5 loop 1 (and possibly Slu7p) to position the ends of the exons for the second trans-esterification reaction (Beggs et al. 1995; Teigelkamp et al. 1995a; Dix et al. 1998; O’Keefe and Newman 1998), consistent with our lock and load hypothesis.
Prp8 PROTEIN COMPLEXES AND INTERACTIONS
Yeast two-hybrid screens have been used to identify proteins that associate directly or indirectly with Prp8p, whereas affinity purification of complexes and mass spectrometric analyzes have itemized the components of Prp8p-containing complexes (see MIPS-CYGD, BIND, and GRID in the Saccharomyces Genome Database). Splicing-related complexes that contain Prp8p include the U5 snRNP (Stevens et al. 2001), the U5•U4/U6 tri-snRNP (Gottschalk et al. 1999; Stevens and Abelson 1999; Schneider et al. 2002b), the spliceosome (Hartmuth et al. 2002; Jurica et al. 2002; Zhou et al. 2002) and a U5 snRNP/Prp19p complex (Makarov et al. 2002; Ohi et al. 2002; Stevens et al. 2002; Wang et al. 2002; see Fig. 1 ▶). hPrp8p forms a stable RNA-free core complex with several U5 snRNP proteins. Using the chaotropic salt sodium thiocyanate to preserve only strong protein–protein interactions, Achsel et al. (1998) showed that hPrp8p binds very strongly to h116K (Snu114p in yeast). h116K is a 116-kDa GTPase that is homologous to the elongation factor EF-2, a GTPase that catalyzes the rearrangement of ribosomes during the translocation step of protein synthesis. It has been suggested that h116K may permit a similar translocation event in the spliceosome (Fabrizio et al. 1997; Liu et al. 1997). Reducing the level of chaotropic agent allowed hPrp8p to be purified in association with h116K, h200K (Brr2p in yeast) and a human 40-kDa protein (Cwf17/Spf38 in S. pombe). Brr2p/h200K is an ATP-dependent RNA unwindase that has been shown to unwind the base-pairing between U4 and U6 snRNAs in vitro (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998).
A 16S yU5 snRNP has been coisolated with the U1 snRNP; however, this 16S complex lacked the majority of known Prp8-associated proteins, including Brr2p. In addition to Prp8p, it contained the conserved protein yAar2p, Snu114p and the Sm proteins (Gottschalk et al. 2001). With Brr2p being absent from the Aar2-U5 snRNP and Aar2p absent from mature tri-snRNPs, it has been suggested that this Aar2-U5 snRNP complex represents an intermediate particle in U5 snRNP biogenesis that involves U1 snRNP (Gottschalk et al. 2001). It is thought that a raft of U1 snRNP proteins acts as the docking site for the tri-snRNP on the pre-spliceosome and specifically guides Prp8p to the 5′SS region (Maroney et al. 2000; Awasthi et al. 2001), and it is known that the human U1 and U5 (loop 1) snRNAs can be cross-linked together before the first step of splicing (Ast and Weiner 1997).
Full-length yPrp40p has been shown to interact with the N terminus of yPrp8p (1–349) and it was proposed that the two WW domains in Prp40p interact with the PPxY (where x is any residue) motif in Prp8p (Abovich and Rosbash 1997). More recent NMR data on WW domains and their ligand preference show the Prp40p WW domains binding strongly to PPΨΨP sequences, where Ψ is an aliphatic residue, and only weakly to Prp8’s PPxY motifs (Macias et al. 2002; Wiesner et al. 2002). As yPrp40p can coprecipitate the human Prp8p which contains no proline-rich motifs (Abovich and Rosbash 1997), it seems unlikely that the PPxY motifs in Prp8 are responsible for its interaction with Prp40p. As Prp8p associates closely with the 5′SS alongside U1 snRNP (Maroney et al. 2000), it is not surprising that yeast two-hybrid results show interactions between U5 proteins and U1 proteins.
Although full-length Prp8p does not perform well as a bait in yeast two-hybrid screens (Uetz et al. 2000; Ito et al. 2001; van Nues and Beggs 2001), some parts of Prp8p do show interactions. The U1 snRNP protein, Prp39p, was found with an N-terminal portion of yPrp8p as bait (residues 1–263; van Nues and Beggs 2001) and ySnp1p, the yeast homolog of the human U1–70K, interacted in a two-hybrid screen with a 28-amino-acid portion of yPrp8 (1166–1193) as was confirmed by coimmunoprecipitation (Awasthi et al. 2001). Snp1p was also shown to interact with yExo84, a secretory protein that affects splicing, and Exo84 itself interacts with amino acids ~750–930 of yPrp8 and Prp40p (Kuhn and Brow 2000). In addition, Snp1p interacts with Brr2p (Fromont-Racine et al. 1997; for summary, see Fig. 6A ▶ ).
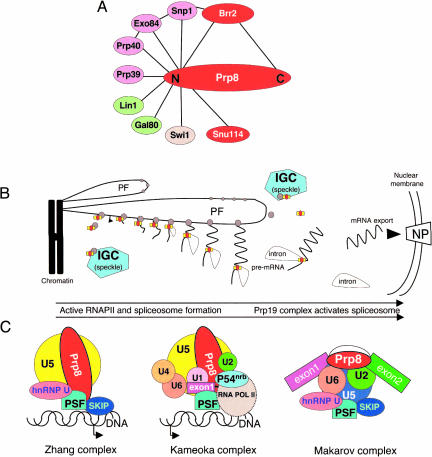
FIGURE 6.
Pictoral representations of (A) proteins shown to interact with yeast Prp8p, (B) gene expression coupled with pre-mRNA splicing, and (C) three purified Prp8-containing protein complexes involving the nuclear receptor activated promoter VDR (Zhang et al. 2003), the transcription elongation complex (Kameoka et al. 2004) and the spliceosome (Makarov et al. 2002). (Gray circle) RNA pol II; (rectangle and red oval) the spliceosome. The interchromatin granular clusters (IGC), or “speckles” as they are also known, store/assemble the RNAPII and splicing factors. Upon chromatin remodelling, the DNA becomes accessible, the RNAPII/spliceosome proteins assemble on the perichromatin fibrils (PF) and produce pre-mRNA. After removal of the intron the mRNA is exported through the nuclear pore (NP) for translation and RNAPII/spliceosome proteins are stored or reassembled in the IGCs.
Yeast two-hybrid screens also pulled out Prp8p’s binding partners, Brr2p and Snu114p. The N-terminal region of Prp8p (1–263), encompassing the proline-rich repeats and a basic segment, selected a C-terminal fragment of Brr2p (1282–1749). Notably the second-step factor Slu7p also selected this section of Brr2p. Full-length Brr2p screens identified three C-terminal Prp8p (cPrp8) fragments spanning residues 2010 to the carboxy-end of the protein. In reciprocal tests, the C-terminal region of Prp8p did not interact with fragments of Brr2p, suggesting that two or more widely separated Brr2 peptide sequences may be required for the interaction. Interestingly, the interaction with full-length Brr2p was weakened by Prp8p residues upstream of 2010 and by C-terminal truncations. Therefore this interaction is potentially sensitive to conformational changes in Prp8p. A Snu114p yeast two-hybrid screen showed an association with Prp8p, the minimal region of interaction being amino acids 420–464 in Prp8p (Dix et al. 1998; I. Dix and J.D. Beggs, unpubl.).
With the N terminus of Prp8p associated with U1 snRNP proteins as well as with Brr2p and Snu114p, it seems possible that the N terminus of Prp8p may coordinate the unwinding of the U1/5′SS interaction in connection with U4/U6 unwinding by Brr2p. This is also supported by the finding that the L280P mutation close to the N terminus in Prp8 suppresses the cold-sensitivity of prp28–1; Prp28p is required for release of U1 RNA from the 5′SS (Kuhn et al. 2002).
YEAST GENETIC INTERACTIONS
prp8 mutations that affect protein–protein interactions
Informative genetic interactions have been detected between PRP8 and BRR2. The temperature-sensitive prp8–1 allele (G2347D) was shown to be detrimental to the interaction of Prp8p with Brr2p in yeast two-hybrid and coimmunoprecipitation assays (van Nues and Beggs 2001), and a reverse two-hybrid screen (seeking random mutations that alter the two-hybrid interaction) identified prp8–28 (I2259N), which lies in the MPN domain and causes reduced coimmunoprecipitation with Brr2p. In contrast, amino acid substitutions in Prp8 residues 2033–2067, including prp8–52 (Y2037H, I2051T), enhance the strength of the interaction with Brr2p (van Nues and Beggs 2001). Yeast cells containing the prp8–52 allele grow like wild-type, while the prp8–1 and prp8–28 alleles do not support growth at elevated temperatures. Thus, alleles prp8–1 and prp8–28 that cause Prp8p to have a reduced affinity for Brr2p, also cause defects in cell growth. In addition, prp8–1 causes synthetic lethality with slt22–1 (E909K), a temperature-sensitive mutation of BRR2, whereas the prp8–52 allele suppresses the temperature-sensitive phenotype of slt22–1, and the prp8–28 has no effect (van Nues and Beggs 2001).
Prp8p and U4/U6 unwinding
Pre-mRNA complexes that are committed to splicing have the U1 snRNA base-paired with the 5′SS. However U6 replaces U1 and itself base-pairs with the 5′SS around the time that the U4/U6 snRNAs are unwound. An extensive study of intra- and intergenic interactions of PRP8 led to the proposal that Prp8p may regulate this exchange of U6 for U1 at the 5′SS (Kuhn et al. 1999; 2002; Kuhn and Brow 2000). Brow and colleagues isolated a triple nucleotide substitution mutation (AAA>UUG) in yeast U4 snRNA, referred to as U4–cs1, that results in U4 binding to the normally unbound ACAGA sequence in U6 snRNA (Li and Brow 1996). This increases the length of stem 1 in the U4/U6 heterodimer, thereby hyperstabilizing it and inhibiting its unwinding at low temperatures. Adding a second ACAGA box to U6 in the presence of the U4–cs1 mutation suppressed the cold-sensitive phenotype (Li and Brow 1996); consequently, it was determined that the binding of U6’s ACAGA box to the 5′ splice site was prevented in the presence of U4–cs1, because the ACAGA sequence was masked by binding to the mutant U4, thus blocking pre-mRNA splicing at 16°C. The U4–cs1 mutation was shown to allow spliceosome assembly, but the stalled spliceosomes contained stable U4/U6 heterodimer and higher levels of U1 snRNA compared to wild type. Increasing the temperature allowed U4/U6 to be unwound and U1 released (Staley and Guthrie 1998, 1999; Kuhn et al. 1999; Murray and Jarrell 1999).
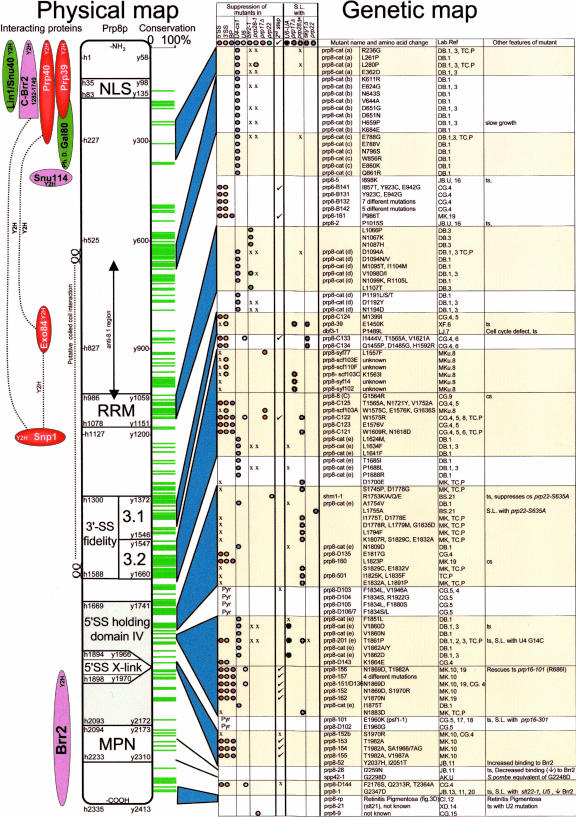
A large-scale screen for prp8 alleles that suppress the cold-sensitive phenotype of the U4–cs1 mutation identified 44 different single-site mutant alleles, known as the prp8–cat mutations, that map to five regions of PRP8, designated regions a–e (Fig. 7 ▶). When tested as heterozygous diploids, some of the prp8–cat alleles conferred semidominant or dominant suppression of the U4–cs1 growth defect, indicating that the mutant proteins were able to compete with wild-type Prp8p for incorporation into U5 snRNPs and into spliceosomes. Therefore, these mutations seem to affect only a very specific function of Prp8p that is important for spliceosome activation (Kuhn and Brow 2000).
FIGURE 7.
A stylized diagram of yPrp8 mapping the known motifs and yeast mutations. The numbers along the length of Prp8 represent the amino acid numbers for human and yeast. On the left are the yeast proteins known to interact with yPrp8 aligned approximately with the yPrp8 minimal region of interaction. (Y2H) yeast two hybrid; (Ph.D) phage display; (…) protein–protein interactions. Down the center of the stylized Prp8p are the motifs and domains discussed in the text. On the right a conservation bar code shows only residues that are 100% conserved between nineteen different species. The two plasmodium sequences were omitted because they contain excessive insertion sequences. The similarity line-up was performed with Vector NTI (Alignment; Invitrogen). In the table on the right are the allele names and their relevant genotype/amino acid alteration. A simple coloured dots scheme highlights their phenotypes. Alleles have then been aligned against the amino acid conservation map of Prp8 using blue trapezoids and yellow highlighting. (prp28Δ*) prp28Δ only in the presence of yhc-1; (S.L.) synthetic lethal; (Pyr) pyrimidine tract recognition mutation; (x) alleles tested and no phenotype noted; (ts) temperature sensitive; (cs) cold sensitive. The lab that first reported the mutations is indicated by the lab head’s initials; references numbers refer to a list posted on Jean Beggs’ Web site (http://www.ed.ac.uk/~jeanb/). P.I. initials and names are: (DB) D. Brow; (TC) T-C Chang; (JB) J. Beggs; (CG) C. Guthrie; (MK) M. Konarska; (MKu) Martin Kupiec; (XF) X. Fu; (LJ) L. Johnston; (BS) B. Schwer; (DX) D. Xu; and (CI) C. Inglehearn.
Four prp8–cat substitutions map to region a (amino acids 232–362), and one of these, L280P, also suppresses the cold-sensitive prp28–1 (G297E). Prp28p is the RNA unwindase that is believed to be responsible for displacing U1 from the5′SS (Staley and Guthrie 1999) and the prp28–1 mutation has the effect of enhancing the U4–cs1 phenotype. The cold-sensitivity of prp28–1 is also suppressed by the temperature-sensitive allele prp8–9, but not by prp8–1 (Strauss and Guthrie 1991). Normally yPrp28 is an essential protein, but this can be overcome by a mutation in the yeast U1C gene, yhc1–1 (Chen et al. 2001). However, another prp8 allele, prp8–501 when combined with prp28Δ and yhc1–1, causes inviability, hence Prp28p becomes essential again. This loss of function by prp8–501 indicates that Prp8p and Prp28p act in concert to destabilize the U1/5′SS (L. Betts and T-H. Chang, in prep.). As region a of Prp8p overlaps with the region that interacts with the U1 snRNP proteins, Prp39 and Prp40, it may function by moderating the release of U1 snRNP from the 5′SS.
Eight suppressors of U4–cs1 map to a very highly conserved area in region b (608–687) of Prp8p. One mutation, H695P, grows slowly at all temperatures but this defect can be suppressed by the prp8–cat mutation L1634F, from region e (see below) (Kuhn and Brow 2000).
Region c (785–864) is defined by six prp8–cat substitutions. The same section used as bait in a yeast two-hybrid screen pulled out the U1 snRNP protein Exo84. Exo84p is also a component of the evolutionarily conserved exocyst complex, and therefore may be involved with the secretory pathway.
Region d (1091–1197) contains the newly identified RRM, eleven suppressors of U4–cs1 and all six suppressors of brr2–1. As already discussed, these mutations may affect RNA binding and U4/U6 unwinding.
Region e (1624–1875) is defined by 16 suppressors of U4–cs1, six of which fall into the highly conserved region now designated region 3.2 (1547–1660). Combinations of these alleles helped identify possible intra-protein interactions. The slow growth phenotype of H659P (region b) is suppressed by L1634F(e) (highlighted in Fig. 3B ▶) and weakly by P1688L(e). This suppression could be explained by a close proximity of the two substituted amino acids, and a leucine zipper between these regions was postulated (Kuhn and Brow 2000). The mild temperature-sensitive growth defect of V1860D(e) or T1861P(e) is enhanced by E788G(c). Loss of U4–cs1 suppression was achieved with pairwise combinations of E788G(c), D1094A(d), or V1098D(d) with V1860D(e) or T1861P(e) which suggests that numerous amino acids in the center of the protein may be in close proximity to residues in region e, postulated to occur via a coiled-coil interaction (Kuhn and Brow 2000).
Mutations in region e overlap with the regions defined as responsible for suppression of 5′SS and 3′SS mutations and for polypyrimidine tract recognition (see Fig. 7 ▶). The U4– cs1 suppressor, prp8–201 (T1861P) is close to some weak 5′SS suppressors (prp8-D143 (K1864E) and others downstream), and itself confers suppression of 5′ and 3′ splice site mutations (Collins and Guthrie 1999; Kuhn and Brow 2000). With this information, Kuhn and Brow proposed a model for Prp8p as a facilitator of U4/U6 unwinding only when the U6 ACAGA-box can correctly recognize the 5′ splice site. This proofreader hypothesis stems from the observation that prp8–201 may allow unwinding of U4/U6 in the presence of a hyperstable stem 1 (U4–cs1 ) and conversely has a synthetic-lethal phenotype with U4G14C, a mutation that destabilizes U4/U6 (Kuhn et al. 1999). Hence mutations in the newly defined RRM region can cause a relaxation in proof-reading and initiate U4/U6 unwinding even in the presence of U4–cs1 or brr2–1 possibly through allosteric changes with Brr2p. Mutations that exacerbate the U4–cs1 phenotype seem likely to affect proteins that function contrary to Prp8p. Indeed, mutations in BRR2 (brr2–1), PRP24, PRP31, and PRP28 enhance the U4–cs1 mutation (Kuhn and Brow 2000; Kuhn et al. 2002).
The U6 snRNA intramolecular stem loop (ISL) mutation, U6–UA, rescues U4–cs1 lethality at 20°C, presumably by destabilizing U4/U6 RNA base-pairing during spliceosome activation and stabilizing the ISL structure in U6 RNA so that it may preferentially participate in catalysis (Huppler et al. 2002; Kuhn et al. 2002). Thus, one way that Prp8p might regulate spliceosome activation is by modulating the stability of the U6 ISL. A subset of three adjacent prp8–cat alleles (y1860–1862), at the C terminus of region e, are synthetic lethal with U6–UA, which might indicate that region e of Prp8p could influence the formation and/or stability of the U6 ISL (Kuhn et al. 2002).
Other splicing factors, in addition to Brr2p, that have been implicated in U4 RNA release during spliceosome activation, include the U4/U6 snRNP protein Prp4 (Ayadi et al. 1997), the non-snRNP protein Prp19 (Tarn et al. 1993), the tri-snRNP proteins Prp38 and Spp381 (Xie et al. 1998; Lybarger et al. 1999) and the U5 snRNP protein Prp8 (Kuhn et al. 1999, 2002). In addition, deletion of the N-terminal domain of Snu114p (snu114Δ) causes a temperature-sensitive phenotype at 37°C and results in accumulation of stalled spliceosomes that contain U4 still base-paired with the U6 RNA, indicating that Snu114p is involved in the dissociation of the U4/U6 duplex (Bartels et al. 2002). Mutations that affect the ability of Snu114p to bind and hydrolyze GTP also prevent U4/U6 unwinding (Bartels et al. 2003). Therefore, the effects of prp8 mutations on U4/U6 unwinding may be indirect, and could involve physical links between Prp8p, Snu114p, and Brr2p.
Overall, Prp8p may act as a control switch that prevents U4/U6 unwinding until all factors are in place to support correct splicing (for review, see Murray and Jarrell 1999). Brr2p, Snu114p, Prp24p, Prp38p, and Prp31p are required, directly or indirectly, for unwinding U4/U6, and act antagonistically to Prp8p. As Brr2p is the only one of these proteins that supports ATPase activity and helicase activity, it seems likely that the other factors act upstream to promote the function of Brr2p.
Copper reporter assay
In yeast, the CUP1 gene confers resistance to the toxic effects of copper. The ACT1–CUP1 reporter in which part of the ACT1 gene, including the intron, is fused in frame to the CUP1 gene, confers a level of copper resistance that is proportional to the level of pre-mRNA splicing and thereby permits a simple, indirect assessment of splicing activity (Lesser and Guthrie 1993). A panel of ACT1–CUP1 reporters containing point mutations at the 5′SS, the branchpoint, polypyrimidine tract, or the 3′SS has allowed many different prp8 mutations to be evaluated as suppressors or enhancers of these splicing defects. Thus, individual amino acids or regions of yPrp8p that may function at the catalytic core of the spliceosome have been identified.
Ten prp8 alleles that can suppress various mutations in position 5′SS+2 mostly correspond to multiple amino acid changes, but three were single point mutations: prp8-D135 (E1817G), -D136 (N1869D), and -D143 (K1864E) (Collins and Guthrie 1999). In another study, weak suppressors of the 5′SS U + 2A identified three mutations in PRP8, namely; prp8–153, prp8–154, and prp8–155, of which T1982A was the causative alteration. All the 5′SS suppressors are within the 5′SS binding domain IV and near the region corresponding to the hPrp8–5′SS cross-linking site (y1966–1970), suggesting that this region of Prp8p may be involved in a functional interaction with the 5′SS in both mammalian and yeast systems (Siatecka et al. 1999).
The same approach identified a region in Prp8p responsible for polypyrimidine tract recognition; alleles prp8–101 and prp8–D102 to prp8–D107 were identified, that caused substitution of either E1960K/G or F1834L/S (Umen and Guthrie 1996). Residue 1960 of Prp8p may also facilitate 3′SS recognition, as the E1960K mutation, known as prp8–101, causes a decrease in cross-linking of Prp8p to the 3′ splice site (G-1) in vitro.
ACT1–CUP1 reporters containing branchpoint mutations efficiently undergo the first but not the second step of splicing. A genetic screen for suppressors of BP mutations, identified three new alleles of PRP8, prp8–161 (P986T), prp8–162 (V1870N), and the cold-sensitive prp8–160 (L1823P) (Query and Konarska 2004). In addition to BP mutations, these alleles as well as other, previously characterized prp8 alleles, including prp8-C121 to prp8-C125 (Umen and Guthrie 1996; Collins and Guthrie 1999) and prp8–156 (N1869D, T1982A), suppress the 5′SS U + 2A mutation and 3′SS - 2 or 3′SS - 3 mutations (UuG/ and gAG/). Primer extension on the ACT1–CUP1 reporter showed that these prp8 suppressors act by improving the second step of splicing (Query and Konarska 2004).
The prp8-C122 (W1575R) allele is able to suppress both the temperature-sensitive growth caused by a prp17Δ mutation, and the slow growth phenotype of a U6-A51 mutation, although other prp8 alleles had no effect (Collins and Guthrie 1999; Ben-Yehuda et al. 2000b). Mutation of U6-U57 to either C or G also resulted in lethality at 37°C (McPheeters 1996) that was efficiently suppressed by prp8–156 (N1869D, T1982A), and the BP suppressors prp8–161, and prp8–162 alleles (Query and Konarska 2004). Physical proximity (of an unknown region) of Prp8p to this part of U6 RNA was also indicated by the cross-link of Prp8p to U6 position 54 (Vidal et al. 1999).
In total, ~25 prp8 alleles suppress both 3′SS mutations and 5′SS mutations; some of these suppress BP mutations as well. The prp8–201 (T1861P) allele is unique in that it confers suppression of 5′SS-U2A, 3′SS and U4–cs1 mutations (Collins and Guthrie 1999; Kuhn and Brow 2000). All of these alleles are mapped onto the Prp8 protein in Figure 7 ▶. The nature of this cross suppression by so many different alleles is unknown however a number of hypotheses are suggested. Suppression may be caused by a relaxation of specificity around the mutated RNA rather than altered substrate recognition (Umen and Guthrie 1996).
prp8–101 is the only allele that is synthetically lethal with a cold-sensitive step-two mutation of PRP16 (prp16–301) (Umen and Guthrie 1995b). Other alleles of PRP16, prp16–1, prp16–101, prp16–302, and of U6 snRNA, U6-U57C and U6-G71U, suppress BP, 5′SS and 3′SS mutations by improving the first step of catalysis (Burgess et al. 1990; McPheeters 1996; Query and Konarska 2004). The temperature sensitivity of prp16-R686I (Hotz and Schwer 1998) was suppressed by prp8–156. Functional interactions between prp8–156 and prp16–101, and between prp8–156 and U6-U57C were tested with a variety of 5′SS, 3′SS, and BP mutations. With mutant pre-mRNAs that are inefficient for both the first and second steps of splicing (U2A, BP-C), suppression of both steps can be achieved by combining first and second step suppressors; for example, prp8–156 with prp16–101, or U6–U57C with one of three prp8 alleles—prp8–156, prp8–161, or prp8–162. Overall, the data suggest that Prp8 acts in combination with U6 and Prp16 to rearrange the spliceosome during the first to second step transition (Query and Konarska 2004). Suppression of SS/BP mutations results from altering the equilibrium between the first and second step conformations, where Prp8 may modulate many transitions in the spliceosome by physical interaction with multiple ATPases (Brow 2002; Query and Konarska 2004).
The broad distribution of 3′SS and 5′SS suppressor mutations also suggests that splice site selection may be controlled by factors that act at a distance from the catalytic center. Selection of U2 and U12 splice site signals in eukaryotic systems thus occurs by altering distal conformational preferences without the need to change the conserved catalytic core of the spliceosome (Query and Konarska 2004).
Prp8p and the second step of splicing
Six proteins are known to be required for the second step of splicing: Prp22, Prp16, Prp17 (only required at elevated temperatures), Prp18, Slu7, and Prp8 (which is required for both steps). Certain mutations in PRP8, known as scf-alleles, suppress the temperature-sensitive growth and splicing defects conferred by the prp17Δ allele. A second set of PRP8 mutations, known as syf alleles, are synthetic lethal with prp17Δ at the normally permissive temperature (Ben-Yehuda et al. 2000a). Using the copper reporter assay, the prp8-scf alleles were shown to enable greater growth than did wild-type PRP8 in the presence of the 3′SS mutation UAG>UUG. Conversely, the prp8-syf alleles act antagonistically to the prp8-scf alleles, reducing splicing in the presence of a UUG 3′SS (Ben-Yehuda et al. 2000b). Neither of the two groups of mutations showed any effect with introns bearing 5′SS, branchpoint, or uridine tract mutations. It was, therefore, proposed that Prp8p and Prp17p might function cooperatively in recognition of the 3′SS, and that the prp8–scf and prp8–syf alleles affect this process. These alleles map to the highly conserved region 3.2.
Prp22p has two important roles in splicing, an ATP-independent role in the second trans-esterification step, and an ATP-hydrolyzing role that is essential for releasing mRNA from the spliceosome. From a bank of helicase-deficient prp22 mutations, one, (F697A), was identified that was cold-sensitive and unable to release mRNA from spliceosomes (Schneider et al. 2003). A screen to identify suppressors of this mutation produced the heat-sensitive prp8–R1753K allele. Substituting this arginine with other amino acids, or alanine scanning those in close proximity (yY1751–F1756), resulted in suppression of other prp22 helicase-deficient mutations as well. The area of Prp8p centered around R1753 was postulated to detain mRNA in the spliceosome, until ATP hydrolysis by Prp22 promotes its release. In this model, prp22 suppression relies upon mutant Prp8 proteins having a reduced grasp on the mRNA within the spliceosome (Schneider et al. 2003).
We propose a possible mechanism for prp8 suppression: The Prp8 region yY1751–F1756 lies between the 5′SS holding domain IV and the conserved region 3.2 which we have postulated locks the 5′exon onto the 3′SS loading region 3.2. In order to unlock mRNA from the spliceosome these two domains must undergo a conformational rearrangement, and mutations in a possible hinge region could facilitate this.
U2 snRNA
In order to identify factors that impact on the function of U2 snRNA, Xu et al. (1998) performed a genetic screen based on an 11-nucleotide substitution in U2 snRNA that affected both the U2 intramolecular stem I, and an alternative structure, in which this region pairs with U6 to form the U2/U6 helix II. One of the mutations, slt21 (Synthetic Lethal with UTwo), that was isolated by this approach, was an allele of PRP8 (prp8–21), which, in the presence of wild-type U2 snRNA, caused temperature-sensitive growth. The synthetic lethality of the prp8–21 allele was apparently specific to the effect of the U2 mutation on U2 snRNA stem I, as analogous mutations in U6 that affected U2/U6 helix II did not display lethality with prp8–21. Further analysis of prp8–21 showed that it caused a severe growth defect in the presence of prp16–1 or slu7–100, and that it was synthetic lethal with mutations in U5 snRNA loop I (Frank et al. 1992; Xu et al. 1998). On the other hand, slt11/ecm2 mutations were shown to be synthetic lethal with substitution mutations in either U2 or U6 that affect U2/U6 helix II, suggesting that Prp8p and Ecm2p have related functions, with Prp8p being involved with U2 stem I and Ecm2 with U2/U6 helix II (Xu et al. 1998; Xu and Friesen 2001).
Suppressors of the temperature-sensitive prp8–1 allele
DED1 (SPP81) was first identified in a search for extragenic suppressors of the prp8–1 mutation (Jamieson et al. 1991). Ded1p is an RNA helicase (Iost et al. 1999) involved in initiation of the translation of all yeast mRNAs (Linder 2003) and localizes to the cytoplasm (Chuang et al. 1997). Nevertheless, Ded1p is present in the yeast penta-snRNP (Stevens et al. 2002), and the hDed1 homolog, Abstrakt, has turned up in the activated spliceosome (Makarov et al. 2002).
Prp8p AND KINASES
The human SR protein kinase, SRPK1, was purified in a multisubunit complex that contained hPrp8 (Lee et al. 2004). This complex was termed the toposome as it contained topoisomerase IIα(TOP IIα), a protein known to be involved in decatenating mitotic chromosomes, possibly suggesting an alternative function for Prp8p during mitosis (Lee et al. 2004), or alternatively that TOP II′ may be involved in transcription/splicing (Ajuh et al. 2000). hSRPK1 is the only member of the SR protein kinase family that is conserved between humans and budding yeast (ySky1p). Deletion of SKY1 is not by itself lethal, but is synthetic lethal with three prp8 alleles in the 3.1 region (prp8–39, prp8–C133, prp8–C134) and with all prp17 mutations including prp17Δ. The combination of prp17Δ and prp8–39 is also synthetic lethal. These three prp8 mutations are able to suppress 3′SS mutations, as determined by the ACT1–CUP1 assay (Dagher and Fu 2001), and deletion of SKY1 specifically suppresses these same 3′SS mutations. These observations suggest that Prp8p, Prp17p, and Sky1p function in a common pathway for 3′SS recognition during the second step of the splicing reaction. Conceivably, 3′SS recognition by Prp8p may be subject to regulation through phosphorylation by Sky1p, most likely involving region 3.1. However, neither yPrp8 nor hPrp8 has a consensus site that would permit phosphorylation at SR residues by the SR protein kinase family (Wang et al. 1998); therefore, this interaction is probably indirect. Also, unlike Prp8p, Sky1p is cytoplasmic (Siebel et al. 1999).
In a search for potential substrates of the Sky1p kinase, Sad1p, a factor involved in the assembly of newly synthesized U4 into the U4/U6 di-snRNP (Lygerou et al. 1999), was identified as a high-copy suppressor of the temperature sensitivity and splicing defects of prp8–39 (Dagher and Fu 2001). However, prp8–39 remains synthetically lethal with sky1Δ regardless of SAD1 overexpression, indicating that the temperature sensitivity and splicing phenotypes are functionally separate from the Sky1p interaction. These results suggest a novel functional link between Sad1p and the U5-associated Prp8p, either in the assembly of functional U5•U4/U6 tri-snRNP or during RNA rearrangements in the spliceosome (Dagher and Fu 2001).
In S. pombe, a mutation in sprp8spp42–1 suppresses a temperature-sensitive mutation, prp4–73, of sPrp4p kinase, and therefore Schmidt et al. (1999) proposed a possible role for Prp4p kinase in controlling the assembly of catalytic spliceosomes. The sprp8spp42–1 mutation, G2248D (Fig. 3C ▶; A. Kuhn, pers. comm.) lies in the MPN domain (Tran et al. 2003). Furthermore, the human ortholog, PRP4K is known to associate with the U5 snRNP and with chromatin remodeling proteins (Dellaire et al. 2002). Although sPrp4 homologs are found in most eukaryotes, none has been identified in S. cerevisiae.
There are, therefore, a number of tantalizing links between Prp8p and kinases; however, the functional significance is still not clear.
CHROMATIN, TRANSCRIPTION, AND SPLICING
Human Prp8
Associations of Prp8p and other splicing factors with transcriptional coactivators and corepressors have been reported. For example, hPrp8 coimmunoprecipitates with BRG1 (A. Östlund-Farrants and P. Ast, pers. comm.), a component of the conserved SWI/SNF remodeling complex that is implicated in the activation of several nuclear-receptor regulated genes. The mammalian PRP4 kinase (PRP4K) is also known to associate directly with chromatin remodeling proteins, including BRG1, and with the U5 snRNP proteins hPrp8, hBrr2, and hPrp6 (Dellaire et al. 2002), and the orthologous sPrp4 has genetic links with sPrp8p (Schmidt et al. 1999). Dellaire and colleagues concluded that PRP4K may couple transcriptional regulation to pre-mRNA splicing via the modulation of chromatin structure.
The transcriptional coactivator protein NCoA-62/hSKIP(yPrp45) is a remodeling protein that is involved with vitamin D-regulated promoters (Baudino et al. 1998; Zhang et al. 2001, 2003; Barry et al. 2003). Adding GST-SKIP to HeLa cell nuclear extracts allowed the purification of a protein complex containing amongst others, the splicing factors hPrp8, PSF (polypyrimidine tract-binding protein-associated splicing factor), hPrp28, h200K (yBrr2p), and h116K (ySnu114p). Hence, hPrp8 associates, probably indirectly, with nuclear receptor promoter regions of the vitamin D response element (Zhang et al. 2003). The yeast homolog of hSKIP, yPrp45p is known to have a role in splicing (Albers et al. 2003) and hSKIP was copurified with the catalytically active spliceosome and the U5/Prp19 complex (Makarov et al. 2002). All the human spliceosomal proteins were identified, including Prp8p and the same proteins that were found at the vitamin D response element namely, U5–200K, U5–116K, and PSF.
Evidence is accumulating for the coupling of pre-mRNA splicing with pre-mRNA synthesis by RNA polymerase II (RNAP II; Bentley 2002; Maniatis and Reed 2002; Neugebauer 2002; Proudfoot et al. 2002; Kornblihtt et al. 2004; Zorio and Bentley 2004 and references therein). The crystal structure of yeast RNAP II showed that the C-terminal domain (CTD) is directly adjacent to the exit groove for the pre-mRNA and so is perfectly positioned to affect processing of the nascent transcript (Cramer et al. 2001). Immunoprecipitation experiments with CC3 antibodies showed that RNAP II with the hyperphosphorylated CTD (RNAPIIO) is stably associated with U5 snRNP, U5•U4/U6 tri-snRNP, and pre-mRNA. Furthermore, hPrp8 was identified in these immunoprecipitations (Chabot et al. 1995; Vincent et al. 1996).
Kameoka et al. (2004) also showed that purified transcription elongation complexes contain RNAPIIO, PSF, p54nrb, hPrp8, and all five UsnRNAs, indicating that the entire spliceosome is associated with the elongating polymerase. PSF and p54nrb are multifunctional proteins; they bind to the CTD of RNAP II (Emili et al. 2002), interact with each other and bind to the U5 snRNA stem 1b (Peng et al. 2002), the same region that is known to be closely associated with Prp8 (Dix et al. 1998). PSF and hPrp8 cross-linked to the same 5′SS position +6 in the early and late stages of spliceosome assembly, respectively (Ismaili et al. 2001). PSF has also been purified as a human tri-snRNP protein (Teigelkamp et al. 1997), suggesting that PSF and p54γact as docking partners between the RNAPIIO and the spliceosome (Kameoka et al. 2004). In addition, RNAPII and up to 20 transcription factors have been found associated with the purified human spliceosome (Rappsilber et al. 2002; Zhou et al. 2002). A summary of the hPrp8–chromatin–transcription complexes is shown in Figure 6B ▶.
In higher eukaryotes, pre-mRNA splicing factors are found in nuclear organelles known as ‘speckles’ or inter-chromatin granule clusters (IGC) and localize to regions that contain little or no DNA. Pre-mRNA is predominantly localized outside IGCs in fibrillar structures known as perichromatin fibrils (PF). The IGCs appear to function as storage/assembly compartments that supply splicing factors and possibly, upon regulated phosphorylation/dephosphorylation (e.g., involving PRP4K), the proteins localize to the PFs where transcription and splicing take place (Lamond and Spector 2003). Proteomic analysis of IGCs identified 146 proteins, of which 54% were pre-mRNA splicing proteins including, hPrp8 and other U5 proteins, hSKIP, PSF, and RNAPII (Saitoh et al. 2004).
There is a significant overlap between the protein compositions of all the Prp8-containing complexes discussed above. Figure 6B ▶ summarizes the relationship between RNA splicing and the gene expression pathway.
Yeast Prp8
IGCs are not found in yeast (Potashkin et al. 1990), and the links between chromatin, transcription, and RNA splicing are not as clear, coming mainly from protein–protein interaction studies.
ySwi1p is a general RNAP II transcription factor that is part of the conserved SWI/SNF complex (Muchardt and Yaniv 2001) and functions in chromatin remodeling (Sudarsanam et al. 1999), yet it was selected in two-hybrid screens with several splicing factors as bait, including Prp8p, Brr2p, and the U1-associated proteins Snp1p and Mud2p (Fromont-Racine et al. 1997; van Nues and Beggs 2001). Also, the protein yLin1/Snu40, which has “links” with chromatin, shows a specific two-hybrid interaction with Prp8p residues 15–152 (Bialkowska and Kurlandzka 2002; A. Bialkowska, pers. comm.) that is reciprocal (R.J. Grainger and J.D. Beggs, unpubl.). However, yLin1/Snu40 associates under native conditions with full-length Prp8p (Gavin et al. 2002) only in the U5 snRNP (Stevens et al. 2001). Also, the yeast DNA-binding transcriptional repressor Gal80p, a protein involved in regulation of genes for galactose metabolism, identified a clone encoding the Prp8 peptide 200–469 via phage display (Hertveldt et al. 2003). As ChIP analysis showed yPrp8p to be weakly detectable at promoter regions (J. Görnemann and K. Neugebauer, pers. comm.), this might explain the interactions of yPrp8p with the DNA-associated proteins ySwi1, yGal80, and yLin1/Snu40.
Thus, despite the rather fragmented nature of these observations, the many reported links between the transcription and splicing machineries offer a tantalizing role for Prp8p as a factor that contributes to the coupling of splicing with the elongating RNAPII.
Prp8p AND THE CELL CYCLE
The dbf3 mutation was obtained in a screen for temperature-sensitive DNA synthesis mutations in S. cerevisiae, and subsequently the DBF3 gene was identified as PRP8 (Shea et al. 1994). It was reported that the mutation dbf3–1 showed no apparent defect in RNA metabolism; however, a splicing defect has subsequently been demonstrated by Northern analysis (Brown 1995). At restrictive temperatures, dbf3–1 cells arrest with buds and have an undivided nucleus with an ~2C content of DNA. On the other hand, prp8–1 rapidly ceases cell division at the restrictive temperature but shows no evidence of a specific cell cycle stage arrest. Specific suppression of dbf3–1 only, and of no other prp8 or dna (see below) alleles, was achieved with overexpression of the heat shock protein SSB1 (Shea et al. 1994).
Three other prp8 mutations, called dna39 alleles, were isolated in a separate screen for temperature-sensitive mutants defective in DNA synthesis (Dumas et al. 1982). Transformation of the dbf3–1 and of the three dna39 strains with the cloned PRP8 gene rescued the temperature-sensitive defect. However, diploids made by crossing dbf3–1 with prp8–1 grew at the restrictive temperature of 37°C, although this was not observed with prp8–2, prp8–3, prp8–7, prp8–8 or with three dna39 strains (Shea et al. 1994), indicating a highly specific dbf3–1/prp8–1 intra-allelic complementation. The dbf3–1 mutation alters a conserved proline, P1489L (Brown 1995).
Using a combination of synchronized cultures, temperature shifts and hydroxy urea experiments it was possible to ascertain that dbf3–1 affects the end of S-phase and dna39–3 affects the beginning of S-phase. An explanation for these observations could be that the dbf3–1 and dna39–3 are defective in the splicing of different intron-containing transcripts that are expressed at different times in the cell cycle. Recently, it was proposed that in certain splicing mutants, low levels of nuclear matrix proteins whose genes contain introns (e.g., α-tubulin may be responsible for cell cycle arrest; Burns et al. 2002; Dahan and Kupiec 2002) and the dbf3–1ts phenotype can be ameliorated by TUB1 cDNA (Brown 1995).
SUMMARY AND CONCLUSIONS
In Figure 7 ▶, the physical properties (protein sequence comparisons, amino acid motifs, and protein–protein interaction data) and genetic map of yPrp8p are presented side-by-side, with blue bands linking the two to facilitate their comparison. It is clear that mutations occur at higher densities in certain regions. In general, regions with a high density of genetic information correspond to regions with high sequence conservation, supporting these as regions with great functional significance. Of note, are regions 3.1 and 3.2 that contain many 5′SS and 3′SS suppressor mutations as well as genetic interactions with prp17Δ. Domain IV also contains a high density of genetic features, including suppressors of 5′SS, 3′SS, BP, U4–cs1, and snr6/U6 mutations, plus synthetic lethal interactions with prp28 mutations. These genetic links are compatible with roles for regions 3.1, 3.2, and IV at or close to the active site of the spliceosome. The RRM motif is dense with mutations that show U4–cs1 and brr2 interactions, possibly indicating a role for the RRM in modulating the unwinding of U4/U6; however, clusters of U4–cs1 suppressor mutations occur throughout the PRP8 genetic map. Currently, the MPN domain does not contain any informative mutations and the function of this region remains unknown.
The N-terminal region of yPrp8p has been reported to have physical interactions with U1 snRNP proteins (Prp39p and Prp40p) as well as with U5 snRNP proteins (Brr2p and Snu114p). As an Aar2–U5 snRNP complex that lacked the majority of known Prp8-associated proteins, including Brr2p, was coisolated with U1 snRNP (Gottschalk et al. 2001), it will be interesting to determine whether the U1 and U5 snRNP protein interactions with yPrp8p can coexist or are mutually exclusive.
The secretory protein Exo84p interacts with the U1 snRNP protein, Snp1p, and both of these interact with more central regions of yPrp8p. Significantly, anti-8.1 antibodies, which were raised against the part of yPrp8p with which Exo84p interacts, precipitate only free U5 snRNPs and apparently disrupt U5•U4/U6 tri-snRNPs (Lossky et al. 1987), suggesting that this region of Prp8p may be important for interaction with the U4/U6 di-snRNP. As Exo84p has been reported to be important for pre-spliceosome formation (Awasthi et al. 2001), we propose that the Exo84p interaction with this region of Prp8p may either promote or depend upon a conformational rearrangement that occurs when the tri-snRNP associates with the pre-spliceosome, making these parts of Prp8p available for interaction with components of the U1 snRNP, and thereby allowing Prp8p to regulate the exchange of U1 for U6 at the 5′SS.
Also, recalling the proposal that Sad1p may affect the assembly of functional tri-snRNP (Dagher and Fu 2001), suppression of prp8–39 by overproduction of Sad1p might be explained by an effect of Sad1p on the proposed intra-molecular coiled-coil interaction in Prp8p (Kuhn and Brow 2000), altering the interaction of the 3′SS fidelity region (where prp8–39 lies) with the anti-8.1 region (involved in tri-snRNP formation).
As noted earlier, the very large size of Prp8p has been conserved in addition to its sequence. After millions of years of evolution, Prp8p is the largest conserved nuclear protein spanning the eukaryotic taxa. This implicates Prp8p as the primordial splicing protein. The large size may be a manifestation of the multiple protein and RNA interactions in which Prp8p is involved. It can be considered to be performing a scaffold-like role in the spliceosome, holding on to many different components. However, clearly it is not an inert structure. Most likely, Prp8p undergoes conformational changes and modulates the activities of at least some of the factors with which it interacts, such as Brr2p. One region of Prp8, referred to here as region 3.2, is so astonishingly conserved in its DNA and protein sequences that it must have a vital role, probably at the catalytic center of the spliceosome.
Prp8p is central to the expression of all nuclear intron-containing mRNAs. In higher eukaryotes, it is responsible for processing thousands of transcripts in alternative splicing pathways, and in both U2 and U12 spliceosomes. It is important for the pathology of human disease, as all eukaryotic pathogens and parasites require Prp8p to functionally express their genes, in some cases via the trans-spliceosome. Retinitis Pigmentosa, a human genetic disorder that causes progressive blindness, positions Prp8p as a target for therapeutic medicine. Thus, studies of Prp8p in pathogenic species, its functional regulation and its role in the molecular basis of Retinitis Pigmentosa have potential relevance for diagnosis and treatment in healthcare.
Hopefully, analyses of conserved domains with predicted functions such as the RRM, the putative protein kinase C site and the MPN domain will clarify the role of Prp8p in these processes. New insights into Prp8ps functional relationships are a focus of research in many laboratories and we can anticipate exciting new developments in Prp8p research in the near future.
Acknowledgments
We are very grateful to Tamara Brenner, David Brow, John Brown, Magda Konarska, Andreas Kuhn, Alan Kutach, Reinhard Lührmann, Karla Neugebauer, Andy Newman, Erik Sontheimer, and Tommaso Villa for helpful comments on the manuscript, and to Beate Schwer and Tien-Hsien Chang for providing manuscripts prior to publication. We thank Alastair Kerr for help with the bioinformatic analysis of the RRM sequence. R.J.G. is supported by a grant to J.D.B. from the Wellcome Trust.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2220705.
REFERENCES
- Abovich, N. and Rosbash, M. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412. [DOI] [PubMed] [Google Scholar]
- Achsel, T., Ahrens, K., Brahms, H., Teigelkamp, S., and Luhrmann, R. 1998. The human U5–220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homolog of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18: 6756–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajuh, P., Kuster, B., Panov, K., Zomerdijk, J.C., Mann, M., and Lamond, A.I. 2000. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 19: 6569–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers, M., Diment, A., Muraru, M., Russell, C.S., and Beggs, J.D. 2003. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA 9: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvi, R.K., Lund, M., and Okeefe, R.T. 2001. ATP-dependent interaction of yeast U5 snRNA loop 1 with the 5′ splice site. RNA 7: 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroggio, X.I., Rees, D.C., and Deshaies, R.J. 2004. JAMM: A metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2: 0113–0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, J.S., Lyon, C.E., Fox, A.H., Leung, A.K., Lam, Y.W., Steen, H., Mann, M., and Lamond, A.I. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12: 1–11. [DOI] [PubMed] [Google Scholar]
- Anderson, G.J., Bach, M., Luhrmann, R., and Beggs, J.D. 1989. Conservation between yeast and man of a protein associated with U5 small nuclear ribonucleoprotein. Nature 342: 819–821. [DOI] [PubMed] [Google Scholar]
- Anthony, J.G, Weidenhammer, E.M., and Woolford Jr., J.L. 1997. The yeast Prp3 protein is a U4/U6 snRNP protein necessary for integrity of the U4/U6 snRNP and the U4/U6.U5 tri-snRNP. RNA 3:1143–1152. [PMC free article] [PubMed] [Google Scholar]
- Ast, G. and Weiner, A.M. 1997. A novel U1/U5 interaction indicates proximity between U1 and U5 snRNAs during an early step of mRNA splicing. RNA 3: 371–381. [PMC free article] [PubMed] [Google Scholar]
- Awasthi, S., Palmer, R., Castro, M., Mobarak, C.D., and Ruby, S.W. 2001. New roles for the Snp1 and Exo84 proteins in yeast pre-mRNA splicing. J. Biol. Chem. 276: 31004–31015. [DOI] [PubMed] [Google Scholar]
- Ayadi, L., Miller, M., and Banroques, J. 1997. Mutations within the yeast U4/U6 snRNP protein Prp4 affect a late stage of spliceosome assembly. RNA 3: 197–209. [PMC free article] [PubMed] [Google Scholar]
- Bach, M., Winkelmann, G., and Luhrmann, R. 1989. 20S small nuclear ribonucleoprotein U5 shows a surprisingly complex protein composition. Proc. Natl. Acad. Sci. 86: 6038–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, J.B., Leong, G.M., Church, W.B., Issa, L.L., Eisman, J.A., and Gardiner, E.M. 2003. Interactions of SKIP/NCoA-62, TFIIB, and retinoid X receptor with vitamin D receptor helix H10 residues. J. Biol. Chem. 278: 8224–8228. [DOI] [PubMed] [Google Scholar]
- Bartels, C., Klatt, C., Luhrmann, R., and Fabrizio, P. 2002. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 3: 875–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels, C., Urlaub, H., Luhrmann, R., and Fabrizio, P. 2003. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 278: 28324–28334. [DOI] [PubMed] [Google Scholar]
- Baudino, T.A., Kraichely, D.M., Jefcoat Jr., S.C., Winchester, S.K., Partridge, N.C., and MacDonald, P.N. 1998. Isolation and characterization of a novel coactivator protein, NCoA-62, involved in vitamin D-mediated transcription. J. Biol. Chem. 273: 16434–16441. [DOI] [PubMed] [Google Scholar]
- Beggs, J.D., Teigelkamp, S., and Newman, A.J. 1995. The role of PRP8 protein in nuclear pre-mRNA splicing in yeast. J. Cell Sci. Suppl. 19: 101–105. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda, S., Dix, I., Russell, C.S., McGarvey, M., Beggs, J.D., and Kupiec, M. 2000a. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 156: 1503–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda, S., Russell, C.S., Dix, I., Beggs, J.D., and Kupiec, M. 2000b. Extensive genetic interactions between PRP8 and PRP17/ CDC40, two yeast genes involved in pre-mRNA splicing and cell cycle progression. Genetics 154: 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, D. 2002. The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14: 336–342. [DOI] [PubMed] [Google Scholar]
- Bialkowska, A. and Kurlandzka, A. 2002. Proteins interacting with Lin1p, a putative link between chromosome segregation, mRNA splicing and DNA replication in Saccharomyces cerevisiae. Yeast 19: 1323–1333. [DOI] [PubMed] [Google Scholar]
- Birney, E., Kumar, S., and Krainer, A.R. 1993. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic. Acids Res. 21: 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody, E. and Abelson, J. 1985. The “spliceosome”: Yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 228: 963–967. [DOI] [PubMed] [Google Scholar]
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Brown, A. 1995. “Molecular genetic studies of the Prp8 splicing factor.” Ph.D. thesis, University of Edinburgh, Edinburgh, UK.
- Brown, J.D. and Beggs, J.D. 1992. Roles of PRP8 protein in the assembly of splicing complexes. EMBO J. 11: 3721–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G. and Dreyfuss, G. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNA by the spliceosome. In The RNA world (eds. R.F. Gesteland et al.), pp 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Burgess, S., Couto, J.R., and Guthrie, C. 1990. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell 60: 705–717. [DOI] [PubMed] [Google Scholar]
- Burns, C.G., Ohi, R., Mehta, S., O’Toole, E.T., Winey, M., Clark, T.A., Sugnet, C.W., Ares Jr., M., and Gould, K.L. 2002. Removal of a single α-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, M.I., Goodwin, T.J., and Poulter, R.T. 2001. A nuclear-encoded intein in the fungal pathogen Cryptococcus neoformans. Yeast 18: 1365–1370. [DOI] [PubMed] [Google Scholar]
- Chabot. B., Bisotto, S., and Vincent, M. 1995. The nuclear matrix phosphoprotein p255 associates with splicing complexes as part of the [U4/U6.U5] tri-snRNP particle. Nucleic. Acids Res. 23: 3206–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarova, C.F., Hims, M.M., Bolz, H., Abu-Safieh, L., Patel, R.J., Papaioannou, M.G., Inglehearn, C.F., Keen, T.J., Willis, C., Moore, A.T., et al. 2002. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 11: 87–92. [DOI] [PubMed] [Google Scholar]
- Chen, J.Y., Stands, L., Staley, J.P., Jackups Jr., R.R., Latus, L.J., and Chang, T.H. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7: 227–232. [DOI] [PubMed] [Google Scholar]
- Chiara, M.D., Gozani, O., Bennett, M., Champion-Arnaud, P., Palandjian, L., and Reed, R. 1996. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol. Cell. Biol. 16: 3317–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara, M.D., Palandjian, L., Feld Kramer, R., and Reed, R. 1997. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J. 16: 4746–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, K. and Reed, R. 1999. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature 402: 207–210. [DOI] [PubMed] [Google Scholar]
- Chuang, R.Y., Weaver, P.L., Liu, Z., and Chang, T.H. 1997. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science 275: 1468–1471. [DOI] [PubMed] [Google Scholar]
- Collins, C.A. and Guthrie, C. 1999. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes & Dev. 13: 1970–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. The question remains: Is the spliceosome a ribozyme? Nat. Struct. Biol. 7: 850–854. [DOI] [PubMed] [Google Scholar]
- Cramer, P., Bushnell, D.A., and Kornberg, R.D. 2001. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science 292: 1863–1876. [DOI] [PubMed] [Google Scholar]
- Dagher, S.F. and Fu, X.D. 2001. Evidence for a role of Sky1p-mediated phosphorylation in 3′ splice site recognition involving both Prp8 and Prp17/Slu4. RNA 7: 1284–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan, O. and Kupiec, M. 2002. Mutations in genes of Saccharomyces cerevisiae encoding pre-mRNA splicing factors cause cell cycle arrest through activation of the spindle checkpoint. Nucleic. Acids Res. 30: 4361–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Erkenez, A.C., Berson, E.L., and Dryja, T.P. 2002. Novel mutations in the PRPC8 gene, encoding a pre-mRNA splicing factor in patients with autosomal dominant Retinitis Pigmentosa. ARVO 2002: (online abstract).
- de la Cruz, J., Kressler, D., and Linder, P. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24: 192–198. [DOI] [PubMed] [Google Scholar]
- Dellaire, G., Makarov, E.M., Cowger, J.J., Longman, D., Sutherland, H.G., Luhrmann, R., Torchia, J., and Bickmore, W.A. 2002. Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol. Cell. Biol. 22: 5141–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix, I., Russell, C.S., O’Keefe, R.T., Newman, A.J., and Beggs, J.D. 1998. Protein–RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA 4: 1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, S., Zauner, S., Fraunholz, M., Beaton, M., Penny, S., Deng, L.T., Wu, X., Reith, M., Cavalier-Smith, T., and Maier, U.G. 2001. The highly reduced genome of an enslaved algal nucleus. Nature 410: 1091–1096. [DOI] [PubMed] [Google Scholar]
- Dumas, L.B., Lussky, J.P., McFarland, E.J., and Shampay, J. 1982. New temperature-sensitive mutants of Saccharomyces cerevisiae affecting DNA replication. Mol. Gen. Genet. 187: 42–46. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. 1998. Profile hidden Markov models. Bioinformatics 14: 755–763. [DOI] [PubMed] [Google Scholar]
- Emili, A., Shales, M., McCracken, S., Xie, W., Tucker, P.W., Kobayashi, R., Blencowe, B.J., and Ingles, C.J. 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8: 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., Laggerbauer, B., Lauber, J., Lane, W.S., and Luhrmann, R. 1997. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 16: 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J., Zhang, Y.Q., Li, P., Tong, C., Tan, L., and Zhu, Y.S. 2004. Interaction between plasminogen activator inhibitor type-2 and pre-mRNA processing factor 8. Acta Biochim. Biophys. Sin. 36: 623–628. [DOI] [PubMed] [Google Scholar]
- Favre, A. 1990. Photochemistry and the nucleic acids. John Wiley and Sons, New York.
- Favre, A., Saintome, C., Fourrey, J.L., Clivio, P., and Laugaa, P. 1998. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid-protein interactions. J. Photochem. Photobiol. B 42: 109–124. [DOI] [PubMed] [Google Scholar]
- Frank, D., Patterson, B., and Guthrie, C. 1992. Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol. Cell. Biol. 12: 5197–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund, C., Dotsch, V., Nishizawa, K., Reinherz, E.L., and Wagner, G. 1999. The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences. Nat. Struct. Biol. 6: 656–660. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Rain, J.C., and Legrain, P. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16: 277–282. [DOI] [PubMed] [Google Scholar]
- Fu, H., Reis, N., Lee, Y., Glickman, M.H., and Vierstra, R.D. 2001. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20: 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco, M.A., Anderson, G.J., Beggs, J., and Sharp, P.A. 1990. A mammalian protein of 220 kDa binds pre-mRNAs in the spliceosome: A potential homologue of the yeast PRP8 protein. Proc. Natl. Acad. Sci. 87: 3082–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331–336. [DOI] [PubMed] [Google Scholar]
- Gottschalk, A., Neubauer, G., Banroques, J., Mann, M., Luhrmann, R., and Fabrizio, P. 1999. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 18: 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk, A., Kastner, B., Luhrmann, R., and Fabrizio, P. 2001. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA 7: 1554–1565. [PMC free article] [PubMed] [Google Scholar]
- Hartmuth, K., Urlaub, H., Vornlocher, H.P., Will, C.L., Gentzel, M., Wilm, M., and Luhrmann, R. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. 99: 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L.H. 1967. Macromolecule synthesis in temperature-sensitive mutants of yeast. J. Bacteriol. 93: 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L.H., McLaughlin, C.S., and Warner, J.R. 1970. Identification of ten genes that control ribosome formation in yeast. Mol. Gen. Genet. 109: 42–56. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N., and Nakayama, K.I. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276: 33111–33120. [DOI] [PubMed] [Google Scholar]
- Hertveldt, K., Dechassa, M.L., Robben, J., and Volckaert, G. 2003. Identification of Gal80p-interacting proteins by Saccharomyces cerevisiae whole genome phage display. Gene 307: 141–149. [DOI] [PubMed] [Google Scholar]
- Hinz, M., Moore, M.J., and Bindereif, A. 1996. Domain analysis of human U5 RNA. Cap trimethylation, protein binding, and spliceosome assembly. J. Biol. Chem. 271: 19001–19007. [DOI] [PubMed] [Google Scholar]
- Hodges, P.E., Jackson, S.P., Brown, J.D., and Beggs, J.D. 1995. Extraordinary sequence conservation of the PRP8 splicing factor. Yeast 11: 337–342. [DOI] [PubMed] [Google Scholar]
- Hotz, H.R. and Schwer, B. 1998. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics 149: 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala, E., Dickerman, A.W., Garcia-Hernandez, M., Weems, D., Reiser, L., LaFond, F., Hanley, D., Kiphart, D., Zhuang, M., Huang, W., et al. 2001. The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic. Acids Res. 29: 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppler, A., Nikstad, L.J., Allmann, A.M., Brow, D.A., and Butcher, S.E. 2002. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat. Struct. Biol. 9: 431–435. [DOI] [PubMed] [Google Scholar]
- Imamura, O., Saiki, K., Tani, T., Ohshima, Y., Sugawara, M., and Furuichi, Y. 1998. Cloning and characterization of a human DEAH-box RNA helicase, a functional homolog of fission yeast Cdc28/Prp8. Nucleic. Acids Res. 26: 2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost, I., Dreyfus, M., and Linder, P. 1999. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274: 17677–17683. [DOI] [PubMed] [Google Scholar]
- Ismaili, N., Sha, M., Gustafson, E.H., and Konarska, M.M. 2001. The 100-kDa U5 snRNP protein (hPrp28p) contacts the 5′ splice site through its ATPase site. RNA 7: 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, and Sakaki Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde, E., Adam, S., Bauch, A., Bastuck, S., Huhse, B., Leutwein, C., Heurtier, M.A., Copley, R.R., Edelmann, A., Querfurth, E., et al. 1998. Transport of macromolecules between the nucleus and the cytoplasm. RNA 4: 351–364. [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.P., Lossky, M., and Beggs, J.D. 1988. Cloning of the RNA8 gene of Saccharomyces cerevisiae, detection of the RNA8 protein, and demonstration that it is essential for nuclear pre-mRNA splicing. Mol. Cell. Biol. 8: 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, S.A., Turner, W., and Schwer, B. 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8: 1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson, D.J., Rahe, B., Pringle, J., and Beggs, J.D. 1991. A suppressor of a yeast splicing mutation (prp8–1) encodes a putative ATP-dependent RNA helicase. Nature 349: 715–717. [DOI] [PubMed] [Google Scholar]
- Jones, M.H., Frank, D.N., and Guthrie, C. 1995. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl. Acad. Sci. 92: 9687–9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S., Licklider, L.J., Gygi, S.R., Grigorieff, N., and Moore, M.J. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kameoka, S., Duque, P., and Konarska, M.M. 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23: 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka, M.D., Duprat, S., Cornillot, E., Metenier, G., Thomarat, F., Prensier, G., Barbe, V., Peyretaillade, E., Brottier, P., Wincker, P., et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450–453. [DOI] [PubMed] [Google Scholar]
- Kay, B.K., Williamson, M.P., and Sudol, M. 2000. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14: 231–241. [PubMed] [Google Scholar]
- Kielkopf, C.L., Lucke, S., and Green, M.R. 2004. U2AF homology motifs: Protein recognition in the RRM world. Genes & Dev. 18: 1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.H. and Abelson, J. 1996. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA 2: 995–1010. [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H. and Lin, R.J. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 16: 6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, D.S. and Beggs, J.D. 1990. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic. Acids Res. 18: 6559–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, H., Tahira, T., Mizota, A., Adachi-Usami, E., Oshima, K., and Hayashi, K. 2003. Diagnosis of autosomal dominant retinitis pig-mentosa by linkage-based exclusion screening with multiple locus-specific microsatellite markers. Invest. Ophthalmol. Vis. Sci. 44: 1275–1281. [DOI] [PubMed] [Google Scholar]
- Konforti, B.B. and Konarska, M.M. 1994. U4/U5/U6 snRNP recognizes the 5′ splice site in the absence of U2 snRNP. Genes & Dev. 8: 1962–1973. [DOI] [PubMed] [Google Scholar]
- ———. 1995. A short 5′ splice site RNA oligo can participate in both steps of splicing in mammalian extracts. RNA 1: 815–827. [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt, A.R., de la Mata, M., Fededa, J.P., Munoz, M.J., and Nogues, G. 2004. Multiple links between transcription and splicing. RNA 10: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65: 367–409. [DOI] [PubMed] [Google Scholar]
- Krogan, N.J., Peng, W.T., Cagney, G., Robinson, M.D., Haw, R., Zhong, G., Guo, X., Zhang, X., Canadien, V., Richards, D.P., et al. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13: 225–239. [DOI] [PubMed] [Google Scholar]
- Kuhn, A.N. and Brow, D.A. 2000. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics 155: 1667–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, A.N., Li, Z., and Brow, D.A. 1999. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell 3: 65–75. [DOI] [PubMed] [Google Scholar]
- Kuhn, A.N., Reichl, E.M., and Brow, D.A. 2002. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. 99: 9145–9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesza, H., Simpson, G.G., Waugh, R., Beggs, J.D., and Brown, J.W. 1993. Detection of a plant protein analogous to the yeast spliceosomal protein, PRP8. FEBS Lett. 318: 4–6. [DOI] [PubMed] [Google Scholar]
- Laggerbauer, B., Achsel, T., and Luhrmann, R. 1998. The human U5–200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. 95: 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A.I. and Spector, D.L. 2003. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4: 605–612. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C. and Woolford Jr., J.L. 1983. Molecular cloning and analysis of the CRY1 gene: A yeast ribosomal protein gene. Nucleic. Acids Res. 11: 403–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.G., Hague, L.K., Li, H., and Donnelly, R. 2004. Identification of toposome, a novel multisubunit complex containing topoisomerase IIα. Cell Cycle 3: 638–647. [PubMed] [Google Scholar]
- Lesser, C.F. and Guthrie, C. 1993. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics 133: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. and Brow, D.A. 1996. A spontaneous duplication in U6 spliceosomal RNA uncouples the early and late functions of the ACAGA element in vivo. RNA 2: 879–894. [PMC free article] [PubMed] [Google Scholar]
- Lin, R.J., Lustig, A.J., and Abelson, J. 1987. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes & Dev. 1: 7–18. [DOI] [PubMed] [Google Scholar]
- Linder P. 2003. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol. Cell 95: 157–167. [DOI] [PubMed] [Google Scholar]
- Liu, X.Q. and Yang, J. 2004. Prp8 intein in fungal pathogens: Target for potential antifungal drugs. FEBS Lett. 572: 46–50. [DOI] [PubMed] [Google Scholar]
- Liu, Z.R., Laggerbauer, B., Luhrmann, R., and Smith, C.W. 1997. Crosslinking of the U5 snRNP-specific 116-kDa protein to RNA hairpins that block step two of splicing. RNA 3: 1207–1219. [PMC free article] [PubMed] [Google Scholar]
- Lossky, M., Anderson, G.J., Jackson, S.P., and Beggs, J. 1987. Identification of a yeast snRNP protein and detection of snRNP-snRNP interactions. Cell 51: 1019–1026. [DOI] [PubMed] [Google Scholar]
- Lucke, S., Klockner, T., Palfi, Z., Boshart, M., and Bindereif, A. 1997. Trans mRNA splicing in trypanosomes: Cloning and analysis of a PRP8-homologous gene from Trypanosoma brucei provides evidence for a U5-analogous RNP. EMBO J. 16: 4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren, K., Allan, S., Urushiyama, S., Tani, T., Ohshima, Y., Frendewey, D., and Beach, D. 1996. A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell 7: 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H.R., Moreau, G.A., Levin, N., and Moore, M.J. 1999. The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA 5: 893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig, A.J., Lin, R.J., and Abelson, J. 1986. The yeast RNA gene products are essential for mRNA splicing in vitro. Cell 47: 953–963. [DOI] [PubMed] [Google Scholar]
- Lybarger, S., Beickman, K., Brown, V., Dembla-Rajpal, N., Morey, K., Seipelt, R., and Rymond, B.C. 1999. Elevated levels of a U4/U6.U5 snRNP-associated protein, Spp381p, rescue a mutant defective in spliceosome maturation. Mol. Cell. Biol. 19: 577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou, Z., Christophides, G., and Seraphin, B. 1999. A novel genetic screen for snRNP assembly factors in yeast identifies a conserved protein, Sad1p, also required for pre-mRNA splicing. Mol. Cell. Biol. 19: 2008–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias, M.J., Wiesner, S., and Sudol, M. 2002. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 513: 30–37. [DOI] [PubMed] [Google Scholar]
- MacMillan, A.M., Query, C.C., Allerson, C.R., Chen, S., Verdine, G.L., and Sharp, P.A. 1994. Dynamic association of proteins with the pre-mRNA branch region. Genes & Dev. 8: 3008–3020. [DOI] [PubMed] [Google Scholar]
- MacMorris, M., Brocker, C., and Blumenthal, T. 2003. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA 9: 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair, G., Shi, H., Li, H., Djikeng, A., Aviles, H.O., Bishop, J.R., Falcone, F.H., Gavrilescu, C., Montgomery, J.L., Santori, M.I., et al. 2000. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA 6: 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maita, H., Kitaura, H., Keen, T.J., Inglehearn, C.F., Ariga, H., and Iguchi-Ariga, S.M. 2004. PAP-1, the mutated gene underlying the RP9 form of dominant retinitis pigmentosa, is a splicing factor. Exp. Cell Res. 300: 283–296. [DOI] [PubMed] [Google Scholar]
- Maita, H., Kitaura, H., Ariga, H., and Iguchi-Ariga, S.M. 2005. Association of PAP-1 and Prp3p, the products of causative genes of dominant retinitis pigmentosa, in the tri-snRNP complex. Exp. Cell. Res. 302: 61–68. [DOI] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., Luhrmann, R. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Makarova, O.V., Makarov, E.M., Liu, S., Vornlocher, H.P., and Luhrmann, R. 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/ U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21: 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malca, H., Shomron, N., and Ast, G. 2003. The U1 snRNP base pairs with the 5′ splice site within a penta-snRNP complex. Mol. Cell. Biol. 23: 3442–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Maroney, P.A., Yu, Y.T., Jankowska, M., and Nilsen, T.W. 1996. Direct analysis of nematode cis- and trans-spliceosomes: A functional role for U5 snRNA in spliced leader addition trans-splicing and the identification of novel Sm snRNPs. RNA 2: 735–745. [PMC free article] [PubMed] [Google Scholar]
- Maroney, P.A., Romfo, C.M., and Nilsen, T.W. 2000. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell 6: 317–328. [DOI] [PubMed] [Google Scholar]
- Martin, A., Schneider, S., and Schwer, B. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277: 17743–17750. [DOI] [PubMed] [Google Scholar]
- Martinez-Gimeno, M., Gamundi, M.J., Hernan, I., Maseras, M., Milla, E., Ayuso, C., Garcia-Sandoval, B., Beneyto, M., Vilela, C., Baiget, M., et al. 2003. Mutations in the pre-mRNA splicing-factor genes PRPF3, PRPF8, and PRPF31 in Spanish families with autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 44: 2171–2177. [DOI] [PubMed] [Google Scholar]
- Masciadri, B., Areces, L.B., Carpinelli, P., Foiani, M., Draetta, G., and Fiore, F. 2004. Characterization of the BUD31 gene of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 320: 1342–1350. [DOI] [PubMed] [Google Scholar]
- Masuda, K., Watanabe, I., Unoki, K., Ohba, N., and Muramatsu, T. 1995. Functional Rescue of photoreceptors from the damaging effects of constant light by survival-promoting factors in the rat. Invest. Ophthalmol. Vis. Sci. 36: 2142–2146. [PubMed] [Google Scholar]
- Maytal-Kivity, V., Reis, N., Hofmann, K., and Glickman, M.H. 2002. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, T.S. and Steitz, J.A. 2001. Proximity of the invariant loop of U5 snRNA to the second intron residue during pre-mRNA splicing. EMBO J. 20: 3577–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, W.H., Ohi, R., Smelkova, N., Frendewey, D., and Gould, K.L. 1999. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 19: 5352–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie, A.B., McHale, J.C., Keen, T.J., Tarttelin, E.E., Goliath, R., van Lith-Verhoeven, J.J., Greenberg, J., Ramesar, R.S., Hoyng, C.B., Cremers, F.P., et al. 2001. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum. Mol. Genet. 10: 1555–1562. [DOI] [PubMed] [Google Scholar]
- McPheeters, D.S. 1996. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA 2: 1110–1123. [PMC free article] [PubMed] [Google Scholar]
- McPheeters, D.S. and Muhlenkamp, P. 2003. Spatial organization of protein–RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 23: 4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters, D.S., Schwer, B., and Muhlenkamp, P. 2000. Interaction of the yeast DExH-box RNA helicase prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic. Acids Res. 28: 1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt, C and Yaniv, M. 2001. When the SWI/SNF complex remodels…the cell cycle. Oncogene 20: 3067–3075. [DOI] [PubMed] [Google Scholar]
- Murray, H.L. and Jarrell, K.A. 1999. Flipping the switch to an active spliceosome. Cell 96: 599–602. [DOI] [PubMed] [Google Scholar]
- Nakai, K. and Horton, P. 1999. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24: 34–36. [DOI] [PubMed] [Google Scholar]
- Neugebauer, K.M. 2002. On the importance of being co-transcriptional. J Cell Sci 115: 3865–3871. [DOI] [PubMed] [Google Scholar]
- Newman, A.J. and Norman, C. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68: 743–754. [DOI] [PubMed] [Google Scholar]
- Newman, A.J., Teigelkamp, S., and Beggs, J.D. 1995. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA 1: 968–980. [PMC free article] [PubMed] [Google Scholar]
- Nilsen, T.W. 2003. The spliceosome: The most complex macromolecular machine in the cell? BioEssays 25: 1147–1149. [DOI] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., and Heringa, J. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Ohi, M.D., Link, A.J., Ren, L., Jennings, J.L., McDonald, W.H., and Gould, K.L. 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi, M.D., Vander Kooi, C.W., Rosenberg, J.A., Chazin, W.J., and Gould, K.L. 2003. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 10: 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe, R.T. 2002. Mutations in U5 snRNA loop 1 influence the splicing of different genes in vivo. Nucleic. Acids Res. 30: 5476–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe, R.T. and Newman, A.J. 1998. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 17: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe, R.T., Norman, C., and Newman, A.J. 1996. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell 86: 679–689. [DOI] [PubMed] [Google Scholar]
- Paterson, T., Beggs, J.D., Finnegan, D.J., and Luhrmann, R. 1991. Polypeptide components of Drosophila small nuclear ribonucleo-protein particles. Nucleic. Acids Res. 19: 5877–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, R., Dye, B.T., Perez, I., Barnard, D.C., Thompson, A.B., and Patton, J.G. 2002. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 8: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Canadillas, J.M. and Varani, G. 2001. Recent advances in RNA–protein recognition. Curr. Opin. Struct. Biol. 11: 53–58. [DOI] [PubMed] [Google Scholar]
- Pietrokovski, S. 2001. Intein spread and extinction in evolution. Trends Genet. 17: 465–472. [DOI] [PubMed] [Google Scholar]
- Pinto, A.L. and Steitz, J.A. 1989. The mammalian analogue of the yeast PRP8 splicing protein is present in the U4/5/6 small nuclear ribonucleoprotein particle and the spliceosome. Proc. Natl. Acad. Sci. 86: 8742–8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potashkin, J.A., Derby, R.J., and Spector, D.L. 1990. Differential distribution of factors involved in pre-mRNA processing in the yeast cell nucleus. Mol. Cell. Biol. 10: 3524–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Query, C.C. and Konarska, M.M. 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell 14: 343–354. [DOI] [PubMed] [Google Scholar]
- Raghunathan, P.L. and Guthrie, C. 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8: 847–855. [DOI] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., Mann, M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R. and Chiara, M.D. 1999. Identification of RNA–protein contacts within functional ribonucleoprotein complexes by RNA site-specific labeling and UV crosslinking. Methods 18: 3–12. [DOI] [PubMed] [Google Scholar]
- Reichert, V.L., Le Hir, H., Jurica, M.S., and Moore, M.J. 2002. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes & Dev. 16: 2778–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, J.L., Kois, P., Konforti, B.B., and Konarska, M.M. 1996. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA 2: 213–225. [PMC free article] [PubMed] [Google Scholar]
- Reyes, J.L., Gustafson, E.H., Luo, H.R., Moore, M.J., and Konarska, M.M. 1999. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA 5: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, N., Spahr, C.S., Patterson, S.D., Bubulya, P., Neuwald, A.F., Spector, D.L., and Lamond, AI. 2004. Proteomic analysis of inter-chromatin granule clusters. Nuclear speckles: A model for nuclear organelles. Mol. Biol. Cell 15: 3876–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H., Richert, K., Drakas, R.A., and Kaufer, N.F. 1999. spp42, identified as a classical suppressor of prp4–73, which encodes a kinase involved in pre-mRNA splicing in fission yeast, is a homologue of the splicing factor Prp8p. Genetics 153: 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., Hotz, H.R., and Schwer, B. 2002a. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 277: 15452–15458. [DOI] [PubMed] [Google Scholar]
- Schneider, C., Will, C.L., Makarova, O.V., Makarov, E.M., and Luhrmann, R. 2002b. Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol. Cell. Biol. 22: 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., Campodonico, E., and Schwer, B. 2003. Motifs IV and V in the DEAH-box splicing factor Prp22 are important for RNA unwinding and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 279: 8617–8626. [DOI] [PubMed] [Google Scholar]
- Segault, V., Will, C.L., Polycarpou-Schwarz, M., Mattaj, I.W., Branlant, C., and Luhrmann, R. 1999. Conserved loop I of U5 small nuclear RNA is dispensable for catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol. Cell. Biol. 19: 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin, B., Abovich, N., and Rosbash, M. 1991. Genetic depletion indicates a late role for U5 snRNP during in vitro spliceosome assembly. Nucleic. Acids Res. 19: 3857–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea, J.E., Toyn, J.H., and Johnston, L.H. 1994. The budding yeast U5 snRNP Prp8 is a highly conserved protein which links RNA splicing with cell cycle progression. Nucleic. Acids Res. 22: 5555–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka, M., Reyes, J.L., and Konarska, M.M. 1999. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes & Dev. 13: 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel, C.W., Feng, L., Guthrie, C., and Fu, X.D. 1999. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc. Natl. Acad. Sci. 96: 5440–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer, E.J. and Steitz, J.A. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996. [DOI] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- ———. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3: 55–64. [DOI] [PubMed] [Google Scholar]
- Stefl, R., Skrisovska, L., and Allain, F.H. 2005. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep. 6: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, S.W. and Abelson, J. 1999. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. 96: 7226–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, S.W., Barta, I., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D., and Abelson, J. 2001. Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7: 1543–1553. [PMC free article] [PubMed] [Google Scholar]
- Stevens, S.W., Ryan, D.E., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D., and Abelson, J. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9: 31–44. [DOI] [PubMed] [Google Scholar]
- Strauss, E.J. and Guthrie, C. 1991. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes & Dev. 5: 629–641. [DOI] [PubMed] [Google Scholar]
- Sudarsanam, P., Cao, Y., Wu, L., Laurent, B.C., and Winston, F. 1999. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 18: 3101–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Muramatsu, H., Takagi, S., Fujisawa, H., Miyake, Y., and Muramatsu, T. 2001. A splicing factor, Prp8: Preferential localization in the testis and ovary in adult mice. J. Biochem. 129: 599–606. [DOI] [PubMed] [Google Scholar]
- Tarn, W.Y., Lee, K.R., and Cheng, S.C. 1993. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl. Acad. Sci. 90: 10821–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp, S., Newman, A.J., and Beggs, J.D. 1995a. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 14: 2602–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp, S., Whittaker, E., and Beggs, J.D. 1995b. Interaction of the yeast splicing factor PRP8 with substrate RNA during both steps of splicing. Nucleic. Acids Res. 23: 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp, S., Mundt, C., Achsel, T., Will, C.L., and Luhrmann, R. 1997. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA 3: 1313–1326. [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, H.J., Allen, M.D., Lowe, J., and Bycroft, M. 2003. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 42: 11460–11465. [DOI] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Umen, J.G. and Guthrie, C. 1995a. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes & Dev. 9: 855–868. [DOI] [PubMed] [Google Scholar]
- ———. 1995b. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA 1: 584–597. [PMC free article] [PubMed] [Google Scholar]
- ———. 1996. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics 143: 723–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki, K., Ohba, N., Arimura, H., Muramatsu, H., and Muramatsu, T. 1994. Rescue of photoreceptors from the damaging effects of constant light by midkine, a retinoic acid-responsive gene product. Invest. Ophthalmol. Vis. Sci. 35: 4063–4068. [PubMed] [Google Scholar]
- Unoki, K., Okubo, A., Arimura, H., Uehara, F., Muramatsu, H., Kadomatsu, K., and Muramatsu, T. 1995. Beneficial effect of a retinoic acid responsive gene product, midkine, on constant light-induced retinal damage in albino mice. Nippon Ganka Gakkai Zasshi 99: 636–641. [PubMed] [Google Scholar]
- Urlaub, H., Hartmuth, K., Kostka, S., Grelle, G., and Luhrmann, R. 2000. A general approach for identification of RNA–protein crosslinking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA–protein contacts in native U1 and U4/U6.U5 snRNPs. J. Biol. Chem. 275: 41458–41468. [DOI] [PubMed] [Google Scholar]
- van Lith-Verhoeven, J.J., van der Velde-Visser, S.D., Sohocki, M.M., Deutman, A.F., Brink, H.M., Cremers, F.P., and Hoyng, C.B. 2002. Clinical characterization, linkage analysis, and PRPC8 mutation analysis of a family with autosomal dominant retinitis pigmentosa type 13 (RP13). Ophthalmic Genet. 23: 1–12. [DOI] [PubMed] [Google Scholar]
- van Nues, R.W. and Beggs, J.D. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157: 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani, G. and Nagai, K. 1998. RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct. 27: 407–445. [DOI] [PubMed] [Google Scholar]
- Verdone, L., Galardi, S., Page, D., and Beggs, J.D. 2004. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr. Biol. 14: 1487–1491. [DOI] [PubMed] [Google Scholar]
- Vidal, V.P., Verdone, L., Mayes, A.E., and Beggs, J.D. 1999. Characterization of U6 snRNA–protein interactions. RNA 5: 1470–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan, U., Parker, R., Tamm, J., Iimura, Y., Rossi, J., Abelson, J., and Guthrie, C. 1986. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 5: 1683–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan, U., Company, M., and Abelson, J. 1989. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes & Dev. 3: 1206–1216. [DOI] [PubMed] [Google Scholar]
- Vincent, M., Lauriault, P., Dubois, M.F., Lavoie, S., Bensaude, O., and Chabot, B. 1996. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic. Acids Res. 24: 4649–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithana, E.N., Abu-Safieh, L., Allen, M.J., Carey, A., Papaioannou, M., Chakarova, C., Al-Maghtheh, M., Ebenezer, N.D., Willis, C., Moore, A.T., et al. 2001. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol. Cell 8: 375–381. [DOI] [PubMed] [Google Scholar]
- Wang, Y. and Guthrie, C. 1998. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA 4: 1216–1229. [PMC free article] [PubMed] [Google Scholar]
- Wang, H.Y., Lin, W., Dyck, J.A., Yeakley, J.M., Songyang, Z., Cantley, L.C., and Fu, X.D. 1998. SRPK2: A differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell. Biol. 140: 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Hobbs, K., Lynn, B. and Rymond, B.C. 2002. The Clf1p splicing factor promotes spliceosome assembly through N-terminal TPR contacts. J. Biol. Chem. 278: 7875–7883. [DOI] [PubMed] [Google Scholar]
- Whittaker, E. and Beggs, J.D. 1991. The yeast PRP8 protein interacts directly with pre-mRNA. Nucleic. Acids Res. 19: 5483–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker, E., Lossky, M., and Beggs, J.D. 1990. Affinity purification of spliceosomes reveals that the precursor RNA processing protein PRP8, a protein in the U5 small nuclear ribonucleoprotein particle, is a component of yeast spliceosomes. Proc. Natl. Acad. Sci. 87: 2216–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner, S., Stier, G., Sattler, M., and Macias, M.J. 2002. Solution structure and ligand recognition of the WW domain pair of the yeast splicing factor Prp40. J. Mol. Biol. 324: 807–822. [DOI] [PubMed] [Google Scholar]
- Wyatt, J.R., Sontheimer, E.J., Steitz, J.A., Newman, A.J., and Norman, C. 1992. Site-specific crosslinking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 6: 2542–2553. [DOI] [PubMed] [Google Scholar]
- Xie, J., Beickman, K., Otte, E., and Rymond, B.C. 1998. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 17: 2938–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D. and Friesen, J.D. 2001. Splicing factor slt11p and its involvement in formation of U2/U6 helix II in activation of the yeast spliceosome. Mol. Cell. Biol. 21: 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D, Field, D.J., Tang, S.J., Moris, A., Bobechko, B.P., and Friesen, J.D. 1998. Synthetic lethality of yeast slt mutations with U2 small nuclear RNA mutations suggests functional interactions between U2 and U5 snRNPs that are important for both steps of pre-mRNA splicing. Mol. Cell. Biol. 18: 2055–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Baudino, T.A., Dowd, D.R., Tokumaru, H., Wang, W., and MacDonald, P.N. 2001. Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription. J. Biol. Chem. 276: 40614–40620. [DOI] [PubMed] [Google Scholar]
- Zhang, C., Dowd, D.R., Staal, A., Gu, C., Lian, J.B., Van Wijnen, A.J., Stein, G.S., and MacDonald, P.N. 2003. Nuclear coactivator-62 kDa/Ski-ineracting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 37: 35325–35336. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. [DOI] [PubMed] [Google Scholar]
- Zorio, D.A. and Bentley, D.L. 2004. The link between mRNA processing and transcription: Communication works both ways. Exp. Cell. Res. 296: 91–97. [DOI] [PubMed] [Google Scholar]