Abstract
Human tyrosyl-tRNA synthetase from mitochondria (mt-TyrRS) presents dual sequence features characteristic of eubacterial and archaeal TyrRSs, especially in the region containing amino acids recognizing the N1-N72 tyrosine identity pair. This would imply that human mt-TyrRS has lost the capacity to discriminate between the G1-C72 pair typical of eubacterial and mitochondrial tRNATyr and the reverse pair C1-G72 present in archaeal and eukaryal tRNATyr. This expectation was verified by a functional analysis of wild-type or mutated tRNATyr molecules, showing that mt-TyrRS aminoacylates with similar catalytic efficiency its cognate tRNATyr with G1-C72 and its mutated version with C1-G72. This provides the first example of a TyrRS lacking specificity toward N1-N72 and thus of a TyrRS disobeying the identity rules. Sequence comparisons of mt-TyrRSs across phylogeny suggest that the functional behavior of the human mt-TyrRS is conserved among all vertebrate mt-TyrRSs.
Keywords: tRNA identity, cross-species aminoacylation, phylogeny, tRNA acceptor stem, tyrosylation
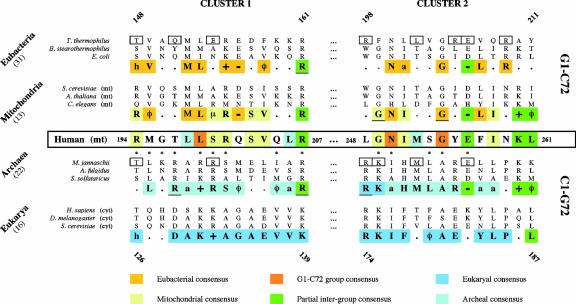
Tyrosyl-tRNA synthetase (TyrRS) is distinguished from other aminoacyl-tRNA synthetases by its ambiguous assignation to class I of synthetases (Bedouelle 2004). Indeed, this homodimeric enzyme with typical class I motifs binds its homologous tRNATyr across its inferface (Bedouelle and Winter 1986) via the major groove side of the amino acid acceptor stem, as occurs for class II synthetases (Lee and RajBhandary 1991; Bedouelle et al. 1993; Fechter et al. 2000; Yaremchuk et al. 2002). Furthermore, tyrosylation reactions are peculiar in that no cross-aminoacylation between eu-bacterial tRNATyr and eukaryal TyrRS is possible and vice-versa (Kleeman et al. 1997; Wakasugi et al. 1998). This strong and unusual phylogenetic barrier mainly relies on the properties of the N1-N72 base pair at the top of the acceptor stem that was assigned as the major tyrosine identity element in eukarya (C1-G72) (Fechter et al. 2000) and a minor element in eubacteria (G1-C72) (Quinn et al. 1995). Sequence alignments reveal strict conservation of the G1-C72 pair in all eubacterial tRNATyr species and its replacement by the reverse C1-G72 pair in eukaryal and archaeal tRNATyr molecules, an uncommon pair at this position in other tRNAs (Sprinzl and Vassilenko 2005). Recently, the crystallographic structures of Thermus thermophilus and Methanococcus jannaschii tRNATyr/TyrRS complexes brought to light the specific interactions established between the N1-N72 pair and TyrRS (Yaremchuk et al. 2002; Giegé 2003; Kobayashi et al. 2003). The differences seen in these interactions provide a structural support accounting for the absence of cross-tyrosylation. In this context, the correlation between two specific sequence clusters in the catalytic domain of TyrRSs and the N1-N72 identity pair can be recalled (Nair et al. 1997). Indeed, cluster 1, within the connective peptide domain, contains the main amino acids contacting the bases of the N1-N72 identity pair (Yaremchuk et al. 2002; Kobayashi et al. 2003). The functional role of these amino acids (Fig. 1 ▶) was already supported by peptide transplantation experiments enabling an identity switch in heterologous tyrosylation systems (Wakasugi et al. 1998). Likewise, cluster 2 is involved in N1 recognition (Yaremchuk et al. 2002; Kobayashi et al. 2003).
FIGURE 1.
Sequence comparison of the clusters in the catalytic domains of TyrRSs in the vicinity to identity elements in tRNATyr. The figure emphasizes the clusters of human mt-TyrRS and compares their sequences to those of TyrRSs from four phylogenetic groups ranked according to the nature of the N1-N72 base pair. For each group (eubacteria, mitochondria—but without vertebrate mitochondria—archaea, eukarya) three sequences are explicitly given. Among mitochondrial TyrRSs, those of Arabidopsis thaliana and Caenorhabditis elegans were annotated for the purpose of this work. Alignments were done with Tcoffee (Poirot et al. 2003) or 3DCoffee (Poirot et al. 2004) to take into account the known TyrRS crystallographic structures (T. thermophilus, Yaremchuk et al. 2002; M. jannaschii, Kobayashi et al. 2003; Bacillus stearothermophilus, Brick et al. 1989; Staphylococcus aureus, Qiu et al. 2001; and Homo sapiens mini-TyrRS, Yang et al. 2002). Numberings flanking the alignments correspond to those of TyrRS from T. thermophilus for eubacterial/mitochondrial TyrRSs and from M. jannaschii for archaeal/eukaryal TyrRSs; numbering of human mt-TyrRS is also given. The asterisks below the human mitochondrial sequence indicate strict conservation of amino acids in vertebrate mt-TyrRSs (see the text). Consensus sequences were established with up to 31 known or predicted sequences (exact numbers are indicated in parentheses); the displayed consensus residues are present in >70% of the analyzed sequences. Underlined amino acids are strictly conserved; amino acids with the same properties are depicted by (φ) for hydrophobic, (+) positively charged, (−) negatively charged, (a) aliphatic, (μ) small, and (h) with hydroxyl group. The consensus within the G1-C72 and C1-G72 groups and within the three domains of life or mitochondria are emphasized by colors (notice that the conserved amino acids crossing the G1-C72/C1-G72 barrier are in green). Squared residues are those found in direct contact with bases of either T. thermophilus tRNATyr (Yaremchuk et al. 2002) or M. jannaschi tRNATyr (Kobayashi et al. 2003).
On the other hand, beside a few studies on yeast mt-TyrRSs involved in splicing events (Akins and Lambowitz 1987; Kamper et al. 1992), nothing is known concerning other mt-TyrRSs, in particular in relation to tyrosine identity. In the frame of a global approach to study human mitochondrial synthetases, the human mt-TyrRS has been purified (Bonnefond et al. 2005). Sequence comparisons of this enzyme with other TyrRSs revealed a eubacterial character as could be anticipated from the close evolutionary relation between mitochondria and eubacteria (Gray et al. 2001). Surprisingly, human mt-TyrRS presents also archaeal sequence features in the two aforementioned clusters (Fig. 1 ▶). When comparing the residues in human mt-TyrRS homologuous to those interacting directly with the acceptor stem of tRNATyr, as seen in the crystal structures (Yaremchuk et al. 2002; Kobayashi et al. 2003), it appears that four among them cannot be linked to any phylogenetic group of TyrRSs (T197, L248, Y255, and N259), three are of mitochondrial type (R194, S200, and G249), M252 is of archaeal type, and E256 is conserved in almost all TyrRSs. It was previously suggested and experimentally verified that the sequence of these clusters relies on the specificity of the synthetase toward tRNA (Quinn et al. 1995). In the present case, the mosaic character of the human mt-TyrRS in the two clusters implies a different recognition of the tRNA than that found typically in eubacteria or eukarya. In other words, human mt-TyrRS would tyrosylate tRNATyr species disregarding the nature of their N1-N72 base pair. To verify this assumption, a series of tRNATyr molecules from different organisms, either wild type or mutated, were tested for their tyrosylation abilities (Fig. 2 ▶).
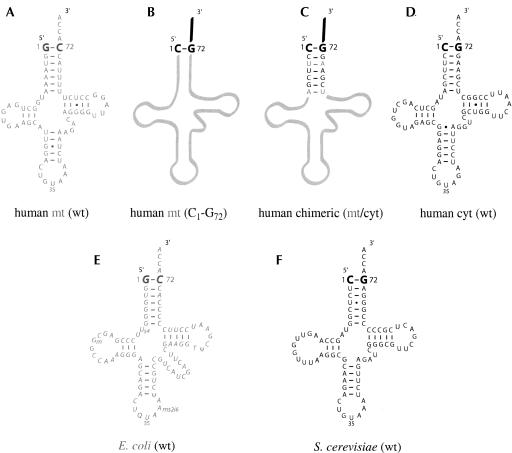
FIGURE 2.
Sequence of wild-type and mutated tRNATyr molecules. The nature of the N1-N72 base pair is emphasized in each molecule. Wild-type human mt-tRNATyr is depicted in A, its variant with the reversed C1-G72 pair in B, chimeric mt/cyt-tRNATyr with cyt-tRNATyr acceptor stem in C, and wild-type human cyt-tRNATyr in D. The sequence of native tRNATyr from E. coli (E) is displayed in italics. Mitochondrial and E. coli sequences are in gray whereas eukaryal (cytosolic) sequences are in black. Sequence data are taken from Sprinzl and Vassilenko (2005). All transcripts were produced in vitro using the “transzyme” method and purified as described in Fechter et al. (1998) whereas native tRNATyr from E. coli (E) was from Subriden.
It is known that human mt-TyrRS charges not only its cognate mitochondrial tRNATyr transcript but also, and even more efficiently, Escherichia coli tRNATyr, both molecules presenting a G1-C72 pair at the top of their acceptor stems (Bonnefond et al. 2005; Fig. 2A,E ▶; Table 1 ▶). The importance of this pair for eubacterial TyrRS recognition has been known since the 1970s (Celis et al. 1973) and was strengthened by studies on E. coli microhelices showing that a transversion of G1-C72 into C1-G72 reduces the efficiency of tyrosylation by the homologous enzyme (Quinn et al. 1995). Because of the mosaic character of human mt-TyrRS, a mutant of its cognate tRNATyr (Fig. 2B ▶) with the reverse eukaryal/archaeal type identity version (C1-G72) should also present tyrosylation capacity. This was verified by a kinetic analysis showing that permutation of the N1-N72 pair does not affect the overall tyrosylation efficiency (as defined by the kcat/KM ratio) of these tRNAs (Table 1 ▶). However, the individual kinetic parameters are modified, with reduced KM and kcat values, indicating an increased affinity of the mutant for mt-TyrRS compensated by a reduced turnover of the enzyme. This functional behavior demonstrates the noninvolvement of the N1-N72 pair in mitochondrial tyrosine identity in human, although the altered values of KM and kcat suggest that N1 and/or N72 play a role in the catalytic mechanism of tyrosylation. To our knowledge, this is the first case where identity rules are disobeyed with a determinant having lost its universal function. From the point of view of evolution, this conclusion may appear surprising since identity rules are ancient. To rationalize this contradiction, one can suggest that the tyrosine system in human mitochondria was under a subtle evolutionary process where mutations in the synthetase relaxed its specificity by removing or altering specific contacts with the primordial G1-C72 identity pair. This process should find appropriate conditions within mitochondria that are niches with a reduced set of tRNAs and a simplified and more permissive macromolecular environment. This view finds support in the overall different tyrosylation mechanism of tRNAs with G1-C72 or G1-C72 pairs.
TABLE 1.
Tyrosylation by human mt-TyrRS of wild-type tRNATyr molecules or their variants with mutations in the acceptor stem
| tRNATyr | KM (μM) | kcat (10−3 · s−1) | kcat/KM (10−3 · s−1 · μM−1) | La (x-fold) | |
| [G1-C72] group | |||||
| human mtb | (wt) | 4.8 | 46 | 9.6 | 1 |
| E. colib | (wt) | 0.52 | 210 | 400 | 0.024 |
| human mt (A73) | (var) | 24 | 0.054 | 0.002 | 4800 |
| [C1-G72] group | |||||
| human mt (C1-G72) | (var) | 0.48 | 3.4 | 7.1 | 1.3 |
| human chimeric (mt/cyt) | (var) | 2.2 | 30 | 13.6 | 0.7 |
| human cyt | (wt) | 48 | 100 | 2.1 | 4.6 |
| S. cerevisiae | (wt) | 1.3 | 1.7 | 1.3 | 7.4 |
tRNAs are ranked in two groups according to the nature of the N1-N72 pair. The C-terminal His-tagged human mt-TyrRS was cloned, expressed, and purified as described elsewhere (Bonnefond et al. 2005). Tyrosylation assays were performed at 37°C in 50 mM HEPES-NaOH (pH 7.6) buffer containing 25 mM KCl, 12 mM MgCl2, 2.5 mM ATP, 0.2 mg/mL bovine serum albumin, 1 mM spermine, and 10 μM L-[3H]-tyrosine at 49 Ci/mmol (Sohm et al. 2003). Apparent kinetic parameters were determined from Lineweaver–Burk plots in the presence of 75 or 1.5 μM TyrRS with transcript concentrations varying from 0.2 to 1.6 μM. tRNAs were renatured by heating at 60°C for 90 sec in water and slow cooling to room temperature before use. Experimental errors on kcat and KM varied at most by 20%. Experiments represent an average of at least two independent experiments.
aL values correspond to losses of efficiency relative to wild-type human mt-tRNATyr (the value <1 corresponds to a gain in efficiency).
bData taken from Bonnefond et al. (2005).
In all aminoacylation systems studied so far, part of the acceptor branch was always found involved in identity (Giegé et al. 1998; Beuning and Musier-Forsyth 1999). Therefore we first verified whether other elements of the acceptor stem participate in tyrosine identity. For that we constructed a chimeric molecule derived from wild-type human mt-tRNATyr with the whole stem replaced by that of human cytosolic (cyt) tRNATyr. This variant has a C1-G72 pair and only two (out of seven) base pairs in common with human mt-tRNATyr (U3-A70 and A7-U65) (Fig. 2C ▶). Based on the crystal structure of the T. thermophilus TyrRS/ tRNATyr complex (Yaremchuk et al. 2002), these two pairs would a priori not interact with mt-TyrRS, and therefore not participate in tyrosine identity. This is also the case for the other four remaining pairs, since the chimerical molecule is fully active in tyrosylation and, remarkably, displays kinetic parameters similar to those of wild-type mt-tRNATyr (Table 1 ▶). Thus, it can be concluded for an absence of tyrosine identity determinants in the double-stranded moiety of the acceptor branch in human mt-tRNATyr. This conclusion is supported by the efficient tyrosylation by human mt-TyrRS of tRNAs with important sequence variations in their acceptor stems, namely, human cyt-tRNATyr and yeast tRNATyr (Fig. 2D,F ▶; Table 1 ▶). In a next step we searched whether discriminator residue 73 is a tyrosine identity element. As found in other tyrosylation systems (Fechter et al. 2001), mutating A73 into G73 abolishes tyrosylation of human mt-tRNATyr (KM/kcat reduced 4800-fold; see Table 1 ▶). Summarizing, it appears that only residue A73 in the tRNA acceptor branch plays an identity role for tyrosylation of human mt-tRNATyr. This implies that other elements in the tRNA molecule, determinants in the acceptor branch or antideterminants, have acquired a more preponderant role in specifying tyrosylation in human mitochondria.
In an evolutionary perspective it would be surprising that the unprecedented properties of a TyrRS to charge indifferently eubacterial and eukaryal tRNATyr species are restricted to the human mitochondrial enzyme. Examination of mitochondrial TyrRS sequences revealed strong homologies between human and other vertebrates mt-TyrRSs. Nine putative mt-TyrRS sequences could be retrieved from the genomes of Canis familiaris, Danio rerio, Gallus gallus, Mus musculus, Pan troglodytes, Ratus norvegicus, Sus scrofa, Takifugu rubripes, and Tetraodon nigroviridis. The high sequence conservation in the clusters (Fig. 1 ▶) thus suggests the same dual functionality of the concerned TyrRSs toward the N1-N72 base pair. Whether the corresponding TyrRSs actually exhibit this property remains to be investigated.
Acknowledgments
We are indebted to A. Théobald-Dietrich and A. Fender for fruitful advice, and we thank M. Sissler and C. Florentz for stimulating discussions. This work was supported by the Centre National de la Recherche Scientifique and Université Louis Pasteur (Strasbourg).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7246805.
REFERENCES
- Akins, R.A. and Lambowitz, A.M. 1987. A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell 50: 331–345. [DOI] [PubMed] [Google Scholar]
- Bedouelle, H. 2004. Tyrosyl-tRNA synthetases. In Aminoacyl-tRNA synthetases (eds. M. Ibba et al.), Chapter 12, pp. 111–124. Landes Biosciences, Georgetown, TX.
- Bedouelle, H. and Winter, G. 1986. A model of synthetase/transfer RNA interaction as deduced by protein engineering. Nature 320: 371–373. [DOI] [PubMed] [Google Scholar]
- Bedouelle, H., Guez-Ivanier, V., and Nageotte, R. 1993. Discrimination between transfer-RNAs by tyrosyl-tRNA synthetase. Biochimie 75: 1099–1108. [DOI] [PubMed] [Google Scholar]
- Beuning, P.J. and Musier-Forsyth, K. 1999. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers 52: 1–28. [DOI] [PubMed] [Google Scholar]
- Bonnefond, L., Fender, A., Rudinger-Thirion, J., Giegé, R., Florentz, C., and Sissler, M. 2005. Towards the full set of human mitochondrial aminoacyl-tRNA synthetases: Characterization of AspRS and TyrRS. Biochemistry (in press). [DOI] [PubMed]
- Brick, P., Bhat, T.N., and Blow, D.M. 1989. Structure of tyrosyl-tRNA synthetase refined at 2.3 Å resolution. Interaction of the enzyme with tyrosyl adenylate intermediate. J. Mol. Biol. 208: 83–98. [DOI] [PubMed] [Google Scholar]
- Celis, J.E., Hooper, M.L., and Smith, J.D. 1973. Amino acid acceptor stem of E. coli suppressor tRNATyr is a site of synthetase recognition. Nat. New Biol. 244: 261–264. [DOI] [PubMed] [Google Scholar]
- Fechter, P., Rudinger, J., Giegé, R., and Théobald-Dietrich, A. 1998. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: Production and activity. FEBS Lett. 436: 99–103. [DOI] [PubMed] [Google Scholar]
- Fechter, P., Rudinger-Thirion, J., Théobald-Dietrich, A., and Giegé, R. 2000. Identity of tRNA for yeast tyrosyl-tRNA synthetase: Tyrosylation is more sensitive to identity nucleotides than to structural features. Biochemistry 39: 1725–1733. [DOI] [PubMed] [Google Scholar]
- Fechter, P., Rudinger-Thirion, J., Tukalo, M., and Giegé, R. 2001. Major tyrosine identity determinants in Methanococcus jannaschii and Saccharomyces cerevisiae tRNATyr are conserved but expressed differently. Eur. J. Biochem. 268: 761–767. [DOI] [PubMed] [Google Scholar]
- Giegé, R. 2003. Genetic code expansion. Nat. Struct. Biol. 10: 414–416. [DOI] [PubMed] [Google Scholar]
- Giegé, R., Sissler, M., and Florentz, C. 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26: 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, M.W., Burger, G., and Lang, B.F. 2001. The origin and early evolution of mitochondria. Genome Biol. 2: 1018.1–1018.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamper, U., Kück, U., Cherniak, A.D., and Lambowitz, A.M. 1992. The mitochondrial tyrosyl-tRNA synthetase of Podospora anserina is a bifunctional enzyme active in protein synthesis and RNA splicing. Mol. Cell. Biol. 12: 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeman, T.A., Wei, D., Simpson, K.L., and First, E.A. 1997. Human tyrosyl-tRNA synthetase shares amino acid sequence homology with a putative cytokine. J. Biol. Chem. 272: 14420–14425. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Nureki, O., Ishitani, R., Yaremchuk, A., Tukalo, M., Cusack, S., Sakamoto, K. and Yokoyama, S. 2003. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Biol. 10: 425–432. [DOI] [PubMed] [Google Scholar]
- Lee, C.P. and RajBhandary, U.L. 1991. Mutants of Escherichia coli initiator tRNA that suppress amber codons in Saccharomyces cerevisiae and are aminoacylated with tyrosine by yeast extracts. Proc. Natl. Acad. Sci. 88: 11378–11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, S., Ribas de Pouplana, L., Houman, F., Avruch, A., Shen, X., and Schimmel, P. 1997. Species-specific tRNA recognition in relation to tRNA synthetase contact residues. J. Mol. Biol. 269: 1–9. [DOI] [PubMed] [Google Scholar]
- Poirot, O., O’Toole, E., and Notredame, C. 2003. Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 31: 3503–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirot, O., Suhre, K., Abergel, C., O’Toole, E. and Notredame, C. 2004. 3DCoffee@igs: A web server for combining sequences and structures into multiple sequence alignment. Nucleic Acids Res. 32: W37–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X., Janson, C.A., Smith, W.W., Green, S.M., McDevitt, P., Johanson, K., Carter, P., Hibbs, M., Lewis, C., Chalker, A., et al. 2001. Crystal structure of Staphylacoccus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors. Protein Sci. 10: 2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, C.L., Tao, N., and Schimmel, P. 1995. Species-specific micro-helix aminoacylation by a eukaryotic pathogen tRNA synthetase dependent on a single base pair. Biochemistry 34: 12489–12495. [DOI] [PubMed] [Google Scholar]
- Sohm, B., Frugier, M., Brulé, H., Olszak, K., Przykorska, A., and Florentz, C. 2003. Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 328: 995–1010. [DOI] [PubMed] [Google Scholar]
- Sprinzl, M. and Vassilenko, K.S. 2005. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 33: D139–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi, K., Quinn, C.L., Tao, N., and Schimmel, P. 1998. Genetic code in evolution: Switching species-specific aminoacylation with a peptide transplant. EMBO J. 17: 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.L., Skene, R.J., McRee, D.E., and Schimmel, P. 2002. Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proc. Natl. Acad. Sci. 99: 15369–15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaremchuk, A., Kriklivyi, I., Tukalo, M., and Cusack, S. 2002. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 21: 3829–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]