Abstract
The U5 snRNP plays an essential role in both U2- and U12-dependent splicing. Here, we have characterized a 52-kDa protein associated with the human U5 snRNP, designated U5-52K. Protein sequencing revealed that U5-52K is identical to the CD2BP2, which interacts with the cytoplasmic portion of the human T-cell surface protein CD2. Consistent with it associating with an snRNP, immunofluorescence studies demonstrated that the 52K protein is predominantly located in the nucleoplasm of HeLa cells, where it overlaps, at least in part, with splicing-factor compartments (or “speckles”). We further demonstrate that the 52K protein is a constituent of the 20S U5 snRNP, but is not found in U4/U6.U5 tri-snRNPs. Thus, it is the only 20S U5-specific protein that is not integrated into the tri-snRNP and resembles, in this respect, the U4/U6 di-snRNP assembly factor Prp24p/p110. Yeast two-hybrid screening and pulldown assays revealed that the 52K protein interacts with the U5-specific 102K and 15K proteins, suggesting that these interactions are responsible for its integration into the U5 particle. The N-terminal two-thirds of 52K interact with the 102K protein, whereas its C-terminal GYF-domain binds the 15K protein. As the latter lacks a proline-rich tract, our data indicate that a GYF-domain can also engage in specific protein–protein interactions in a polyproline-independent manner. Interestingly, the U5-102K protein has been shown previously to play an essential role in tri-snRNP formation, binding the U4/U6-61K protein. The interaction of 52K with a tri-snRNP bridging protein, coupled with its absence from the tri-snRNP, suggests it might function in tri-snRNP assembly.
Keywords: RNA splicing, U5 snRNP proteins, U4/U6.U5 tri-snRNP assembly thesis, GYF-domains, protein–protein interactions
INTRODUCTION
Pre-mRNA splicing is carried out by the spliceosome, a complex machinery that consists of five snRNAs and >150 proteins (for review, see Burge et al. 1999). This machinery has to be assembled de novo on each intron to be spliced. To facilitate this process, a major portion of the required proteins are preassembled into “small nuclear ribonucleoprotein” (snRNP) modules, each centred on one or more small nuclear RNA (snRNA) molecule (Will and Lührmann 2001). First, the snRNPs U1 and U2 interact, through RNA base-pairing, with the intron at the 5′ splice site and the branch point sequence, respectively. To this prespliceosomal complex A, the U4/U6 di-snRNP and the U5 snRNP are added as a single tri-snRNP; the resulting spliceosome is called the B complex. This matures in a series of rearrangements and eventually reaches its catalytically active conformation (the B* complex). During this step, U1 is displaced from the 5′ splice site and U6 base pairs with the same region. The duplex between the U4 and U6 snRNAs, which is present in the U4/U6 snRNP, is likewise disrupted to allow U2-U6 base-pairing. The RNA network that results from these rearrangements is thought to harbor the catalytic activity of the spliceosome (Nilsen 1998). After the first chemical step of the splicing reaction, further rearrangements take place to yield the C complex, which catalyzes the second step. The spliceosome is completely dismantled after the excision of each intron, and this process appears to leave many of the participating complexes altered, either in structure or composition. The U4/U6.U5 tri-snRNP, for example, is thought to dissociate into its individual subunits, the snRNPs U4, U5, and U6 (Staley and Guthrie 1998). Reentry into a new cycle therefore requires that these snRNPs regain their ability to unite as a tri-snRNP. It has been shown that the recycling of the U4/U6 di-snRNP requires a conserved auxiliary factor, which is termed Prp24p in yeast and p110 in humans, and which associates with U6 snRNPs to facilitate the annealing of U4 and U6 snRNAs (Raghunathan and Guthrie 1998; Bell et al. 2002). Interestingly, this protein dissociates from the U4/U6 di-snRNP before, or concomitantly with, its association with the U5 snRNP, and therefore is not part of the U4/U6-U5 tri-snRNP.
Much less is known about the recycling of the U5 snRNP during splicing. Various forms of U5 snRNPs have been described in nuclear extracts of HeLa cells. An abundant form is the 20S U5 snRNP, which is thought to join the U4/U6 di-snRNP to yield the 25S U4/U6.U5 tri-snRNP. The 20S U5 particle exhibits a rich diversity of proteins and contains, in addition to the seven Sm proteins, at least eight U5-specific polypeptides, most of which are evolutionarily highly conserved and have counterparts in yeast cells (Will and Lührmann 2001). The list of U5-specific proteins includes the DEXH/DEAD-box proteins 200K (Brr2p in yeast) and 100K (Prp28p), the ribosomal elongation factor EF-2 homolog and GTP-binding protein 116K (Snu114p), the 102K (Prp6p) protein containing a tetratricopeptide repeat (TPR) domain, the WD40 protein 40K, the 15K protein (Dib1p) containing a thioredoxin-like fold, and the 220K protein (Prp8p), whose sequence is evolutionarily extremely conserved but does not appear to contain known consensus motifs (Hodges et al. 1995). All of these proteins have also been found in the purified human U4/U6.U5 tri-snRNP (Will and Lührmann 2001). Biochemical and genetic approaches revealed that several of the U5-specific proteins play important roles in splicing. For example, the 200K (Brr2p), 116K (Snu114p), and the 100K (Prp28p) proteins are involved in mediating several of the RNA structural rearrangements that occur during spliceosome maturation and splicing catalysis. The 220K (Prp8p) protein contacts all of the important cis-acting intron consensus sequences during splicing (Siatecka et al. 1999) and is thought to govern the functional activities of other U5-specific proteins in the maturation of the spliceosome (Staley and Guthrie 1998; Brow 2002). The 102K protein (Prp6p) seems to play a major role in the formation of the tri-snRNP, as it interacts with the U4/U6-specific 61K protein (Prp31p) and thereby forms a bridge between the two particles (Makarova et al. 2002; Schaffert et al. 2004).
Recently, a novel 35S RNP particle containing U5 snRNA was identified in nuclear extracts from HeLa cells, and its protein composition was characterized by mass spectrometry. This particle contained—in addition to the established U5-specific proteins (220K, 200K, 116K, and 40K)—a group of splicing factors (such as the human Prp19 protein complex) that are stably integrated into the spliceosome during the transformation of complex B into the catalytically activated B* spliceosome, indicating that the 35S U5 snRNP complex represents a post-spliceosomal disassembly intermediate of the splicing machinery (Makarov et al. 2002; Makarova et al. 2004). Interestingly, several proteins that are stoichiometrically present in 20S U5 snRNPs—namely, the U5-specific 102K, 100K, and 15K proteins—are completely absent in the 35S U5 RNP particle (Makarov et al. 2002). Thus, the protein composition of the U5 snRNP changes within the framework of the spliceosome, and the resulting U5 particle has to be extensively reformed before it can build a U4/U6.U5 tri-snRNP and then reenter the spliceosome. Very little is known about these remodeling steps or the proteins possibly involved.
In earlier work we showed that a protein with an apparent molecular mass of 52 kDa is copurified in stoichiometric amounts with 20S U5 snRNPs of HeLa cells, and we therefore termed it U5-52K (Bach et al. 1989; Behrens and Lührmann 1991). The starting point of the present report is a biochemical characterization of the 52K protein. Interestingly, the sequence of the 52K protein is identical to that of a protein already identified as interacting through its C-terminal domain (the so-called GYF domain) with oligo-proline stretches in the cytoplasmic part of the human T-cell surface protein CD2, and accordingly termed CD2-binding protein 2 (CD2BP2) (Nishizawa et al. 1998; Freund et al. 1999). We further show that (1) the endogeneous 52K protein of HeLa cells is located in the nucleus and overlaps, at least in part, with splicing-factor compartments (the so-called “speckles”); and (2) that the identified 52K protein (CD2BP2) is indeed a constituent of the 20S U5 snRNP but (3) is not found in purified U4/U6.U5 tri-snRNPs, and is thus the only 20S U5-specific protein that is not integrated into the tri-snRNP (a property shared with the U4/U6 di-snRNP assembly factor Prp24p/p110). Finally, two-hybrid screening and biochemical analyses reveal that the 52K protein interacts with the U5-specific 102K and 15K proteins. These interactions are presumably responsible for its integration into the U5 particle. Interestingly, it is the GYF-domain that interacts with the 15K protein; since this protein does not exhibit a proline-rich tract, we infer that the GYF domain of the 52K protein must be capable of undergoing protein–protein interactions that do not involve polyproline.
RESULTS
Primary structure of the 52K protein
Oligopeptides from the 52K protein were prepared and microsequenced (see Materials and Methods). Eleven peptide sequences were identified and were used to search for entries in the NCBI database. The search identified one entry that contained all 11 peptide sequences. This protein, CD2BP2, has been reported to bind specifically to the cytoplasmic portion of the human T-cell surface protein CD2 (Nishizawa et al. 1998). Since such a role is not easily reconcilable with being part of nuclear RNPs, we wanted to make sure that this database entry really represented the U5-associated 52K protein. The oligopeptides sequenced were sufficiently numerous, long (in all, 108 out of 341 amino acid residues), and uniformly distributed over the length of the EST (indicated by black bars in Fig. 1 ▶) to exclude any likelihood that another protein might contain the same peptides. Still, the calculated mass of CD2BP2 is 37.6 kDa, far short of the 52 kDa estimated from SDS-polyacrylamide gels for the U5-associated protein (Bach et al. 1989; Behrens and Lührmann 1991). However, this apparent discrepancy is due to aberrant migration of the protein on SDS gels owing to its acidic nature (calculated pI = 4.49), since the product of translation of the CD2BP2 cDNA in vitro migrates in an identical manner (Nishizawa et al. 1998). Finally, we raised antibodies against the protein encoded by this cDNA (see Materials and Methods). These antibodies specifically recognize the 52K protein, and in Western blots analyzing proteins from purified snRNPs and from HeLa nuclear extracts they stain a single band that migrates at the same position in SDS-polyacrylamide gels (data not shown, but see also the blot shown in Fig. 3 ▶, below). The fact that a single band is stained also argues against the presence of different U5-52K isoforms in HeLa cells.
FIGURE 1.
Evolutionary conservation of the 52K protein. The figure shows the alignment of the human 52K protein (Homo sapiens, acc no. NP_006101) with the respective sequences from zebra fish (Danio rerio, NP_957255), frog (Xenopus laevis, AAH45270), fruit fly (Drosophila melanogster, Q9VKV5), nematode (Caenorhabdus elegans, NP_499455), mold (Neurospora crassa, XP_327141), yeast (Saccharomyces cerevisiae, P38852), and wall cress (Arabidopsis thaliana, AAN18131); species are indicated by their initials. In the first block, changed residues and serines (D, E, K, R, S) are highlighted in bold letters. In all other blocks, residues that are identical in at least six out of eight sequences are shown against a black background, and residues conserved in at least six sequences against gray. The consensus sequence of the first conserved domain is shown below in the second block, using the following abbreviations for conserved residues: s, small polar, G/S/D/N; h, hydrophobic, V/I/L/M; a, acidic, D/E; b, basic, K/R. The GYF domain is highlighted by a black bar, and the peptides identified by microsequencing of the human protein are indicated with thin lines above the sequence.
FIGURE 3.
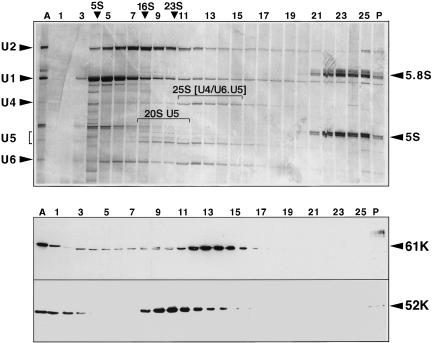
Cosedimentation of the 52K protein with U5 snRNP. One-hundred microliters of HeLa nuclear extract was centrifuged in a linear 10%–30% glycerol gradient. The gradient was harvested from top to bottom, and RNA as well as protein was isolated from each fraction. The RNA was separated by PAGE (10% polyacrylamide/7 M urea) and visualized by staining with silver (top). Proteins were separated on a 10% polyacrylamide–SDS gel, transferred onto a membrane, and immunostained with antibodies against the U4/U6 61K and 52K proteins (bottom).
As an snRNP polypeptide, the 52K protein is expected to be conserved throughout the eukaryotic kingdom. Therefore, we searched the nonredundant database, and were able to identify homologs in vertebrates, insects, nematodes, fungi, and plants. Alignment of the 52K sequences (Fig. 1 ▶) shows that amino acid similarity is found throughout the entire length of the protein, with local concentration of identity in several regions. First, in the N-terminal region, all 52K homologs are particularly rich in charged residues and serines. A second conserved region of about 40 amino acid residues is found directly after the charged domain. This putative domain does not exhibit detectable homology with other proteins, and thus its sequence offers no clues as to its functional significance. Further toward the C terminus, two shorter stretches of clustered identity are observed, followed finally by the C-terminal GYF motif (indicated by a black bar). The GYF motif forms a well-described domain that has the ability to bind a proline-rich tandem repeat in the cytoplasmic tail of CD2 (Freund et al. 1999, 2002), and all of the residues that play a direct part in this binding (Tyr285, Trp287, Pro298, Phe299, Met304, Trp307, Tyr312, and Phe313) are well conserved. Only the putative plant ortholog fails to exhibit a discernible GYF domain. Nevertheless, the high degree of conservation of all other characteristic sequence elements provides strong evidence that we have identified the correct plant U5 snRNP protein. It should be noted that other proteins with a similar GYF domain have been identified in Saccharomyces cerevisiae (Nishizawa et al. 1998). Of these GYF domain proteins, Snu40p/Lin1p (shown in Fig. 1 ▶) can be clearly identified as the homolog of 52K, because it shares the other characteristic sequence motifs and also associates with the yeast U5 snRNP (Stevens et al. 2001).
Predominant location of the 52K protein in the cell nucleus
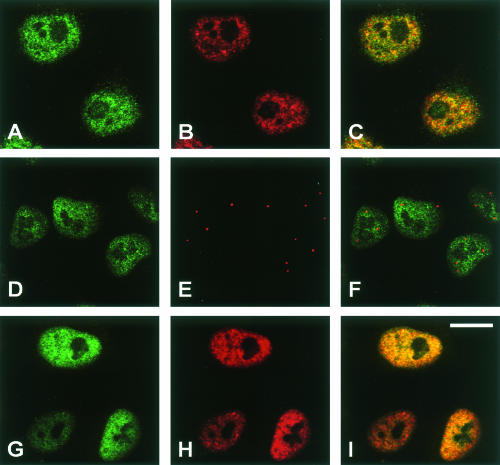
As an snRNP-associated protein, 52K should be found predominantly in the nucleus. Therefore, we investigated the intracellular distribution of 52K. Figure 2A ▶ shows the result of staining with anti-52K and Texas Red-conjugated anti-rabbit IgG secondary antibodies. The 52K-specific signal is seen almost exclusively in the nucleus, with dark nucleolar regions. Controls with preimmune anti-52K sera gave very weak and blurred images (not shown). As is typical of snRNP proteins (Lamond and Spector 2003), the protein is not distributed uniformly throughout the nucleoplasm, but rather it is localized in so-called “speckles.” However, double staining with the additional antibody Y12, which recognizes the common Sm proteins present in all snRNPs (Fig. 2B ▶), revealed only a partial overlap, with many speckles containing either more snRNPs (green in the overlaid picture; Fig. 2C ▶) or more of the 52K protein (red in the overlaid picture). Next we investigated whether the 52K protein accumulates in coiled bodies, where many snRNP components are found. Coiled or Cajal bodies (CBs) are subnuclear structures that contain components of many nuclear RNPs (Lamond and Spector 2003) and are assembly stations for the spliceosomal tri-snRNP (Schaffert et al. 2004, and refs. therein). Thus, we double stained cells with anti-52K antibodies and a second antibody against coilin, a marker of the CBs. As shown in Figure 2D–F ▶, there is little if any overlap between the two proteins. Where the 52K protein is seen in a CB region (yellow in the overlay, Fig. 2F ▶), the two proteins appear to be localized in structures of distinct shape, which are fortuitously in close proximity within one optical z-section.
FIGURE 2.
Predominant location of the 52K protein in the cell nucleus. HeLa cells were grown on coverslips, fixed, and stained with antibodies specific for the 52K protein (A,D). Alternatively, cells were transfected with a plasmid encoding an HA-tagged 52K fusion protein and stained with anti-HA antibodies (G). Cells were double stained with either anti-Sm antibody Y12 (B,H) or with anticoilin antibodies (E), and were examined by confocal fluorescence microscopy. (C,F,I) These panels show the overlaid pictures of the respective preceding two panels, with signals appearing yellow when they coincide. Scale bar = 15 μm.
To confirm the observed nuclear localization, we carried out a similar experiment by introducing an HA-tagged 52K fusion protein into HeLa cells (see Materials and Methods) and staining it with rat anti-HA antibodies and Texas Red-conjugated antirat IgG secondary antibodies. Again, the cells were also double stained with the Y12 anti-Sm antibody. As seen in Figure 2G–I ▶, the HA-tagged version of the 52K protein is also largely confined to the nucleus, confirming the results obtained with anti-52K antibodies. However, the speckled pattern observed with the anti-HA antibodies is less clear. We attribute this difference to the over-expression of the HA-52K fusion protein in these cells; excess 52K protein that is not incorporated into proper complexes is expected to diffuse freely in the nucleoplasm. We conclude that the 52K protein is predominantly located in the nuclei of HeLa cells, in agreement with a role as an snRNP protein. However, it does not colocalize perfectly with the majority of the spliceosomal snRNPs (see Discussion).
Detection of the 52K protein in the 20S U5 snRNP, but not in the U4/U6.U5 tri-snRNP
The 52K protein is readily visible in immunoaffinity-purified 20–25S snRNPs (Behrens and Lührmann 1991). To investigate whether the 52K protein is associated with both 20S U5 snRNPs and 25S tri-snRNPs, we loaded HeLa nuclear extract onto a glycerol gradient and analyzed the distribution of the 52K protein by SDS-PAGE followed by Western blotting. For determination of the sedimentation coefficients, RNA was isolated from aliquots of each fraction, separated by PAGE, and visualized by silver staining. As shown in Figure 3 ▶ (top panel), the 12S U1 snRNP peaks in fraction 5, the 20S U5 snRNP in fraction 10, the 25S U4/U6.U5 tri-snRNP in fraction 13, and the 40S small ribosomal subunit in fraction 23. The bottom panel shows the distribution of the 52K protein: The bulk of it peaks in the 20S and not in the 25S region of the gradient. Thus, it seems to be associated solely with the U5 snRNP and is not part of the U4/U6.U5 tri-snRNP. The 61K protein, in contrast, which is part of the U4/U6 snRNP and the U4/U6.U5 tri-snRNP, but not the 20S U5, cosediments with the U4/U6 and U5 snRNAs in a distinct peak, three fractions further down the gradient. A minor fraction of the 52K protein is found in the top three fractions of the gradient. This is probably free, monomeric 52K protein or 52K present in a very small complex (for comparison, the U1 snRNP in fraction 5 has a molecular weight of ca. 300 kDa). Finally, a negligible fraction of the 52K protein is found in the pellet of the gradient and probably arises through the formation of large, nonspecific aggregates and/or technical limitations of the gradient harvesting procedure (a similarly small proportion of the 61K protein was also found in the pellet fraction). We conclude that almost all of the 52K protein is present in a 20S complex, presumably the U5 snRNP, and the 52K protein appears to be excluded from the tri-snRNP.
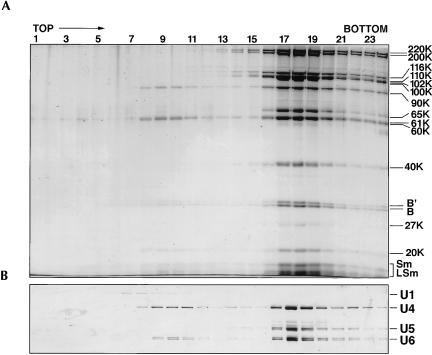
To test the latter conclusion, we immunoaffinity purified the 25S tri-snRNP from a mixture of spliceosomal snRNPs using an antipeptide antibody against the 61K protein. With this antibody it is possible to elute bound tri-snRNPs with the cognate 61K peptide under native conditions. The eluted tri-snRNPs were fractionated by glycerol gradient centrifugation and the protein and RNA content of each fraction analyzed by PAGE (Fig. 4 ▶). Most of the complexes migrated in the 25S region (fractions 17–19) and contained stoichiometric amounts of the U4, U5, and U6 snRNAs (Fig. 4B ▶), as well as U5-specific proteins (220K, 200K, 116K, 102K, 100K, 40K) and U4/U6-specific proteins (90K, 61K, 60K, 20K) (Fig. 4A ▶). In contrast, the band corresponding to the 52K protein was not visible in any of the fractions analyzed. Small amounts of the U4 and U6 snRNAs, and U4/U6-specific proteins (90K, 61K, 60K, and 20K) were detected in fractions 8–10; these fractions thus contained 13S U4/U6 snRNPs. The 52K protein was also absent in this snRNP particle. These results clearly demonstrate that the 52K protein is not a component of the 25S U4/U6.U5 tri-snRNP, and indicates that it dissociates from the U5 snRNP during the formation of the tri-snRNP particle. On the basis of the results of gradient fractionation followed by immunoblotting (Fig. 3 ▶) and immunoaffinity purification (Fig. 4 ▶), we conclude that the 52K protein is an integral protein of the U5 snRNP but is absent from the tri-snRNP. It is thus a protein that associates with a module of the tri-snRNP, but not with the entire particle (see also Discussion).
FIGURE 4.
Gradient centrifugation analysis of immunoaffinity-purified 25S U4/U6.U5 tri-snRNPs. Human 25S U4/U6.U5 tri-snRNPs were isolated from a mixture of anti-m3G purified HeLa spliceosomal snRNPs with antipeptide antibodies against the U4/U6-61K protein and subjected to 10%–30% glycerol gradient centrifugation. Distribution of (A) protein and (B) snRNA across the gradient. Proteins were separated by SDS-PAGE and visualized by staining with Coomassie. RNA was fractionated by denaturing PAGE and stained with silver. Gradient fraction numbers are indicated at the top and the identities of tri-snRNP proteins and snRNAs are shown on the right.
Specific binding of the 52K protein to the U5 snRNP 15K and 102K proteins
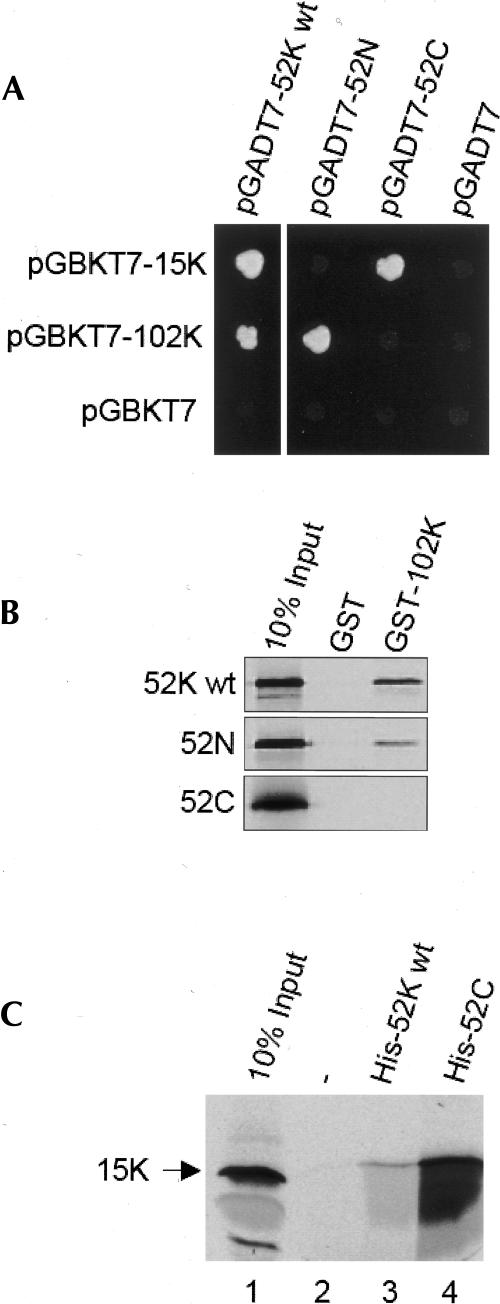
We next examined how the 52K protein may be integrated specifically into the U5 snRNP. To study the interactions of the 52K protein with other proteins, we performed two-hybrid analyses in yeast. In this technique, one potential binding partner is fused to the DNA-binding domain of the GAL4 transcription factor (“bait”), and the other potential binding partner is fused to the activation domain of Gal4 (“prey”). If the two partners do, in fact, bind to one another, then transformation of yeast cells with plasmids encoding both fusion proteins restores the activity of Gal4, and hence activates reporter genes that are driven by their respective promoter within the Gal4 cascade. Here, we chose the essential Ade2 and His3 proteins as reporters; Gal4 activity induces their synthesis, and hence permits growth on selective medium. As bait, we tested all of the U5 snRNP-specific proteins (15K, 40K, 100K, 102K, 116K, 200K, and 220K), U4/U6-snRNP-specific proteins (15.5K, 20K, 60K, 61K, and 90K) and the 27K, 65K, and 110K proteins which are present in the U4/U6.U5 tri-snRNP, but are not stably associated with single U5 or U4/U6 snRNP particles. Of all of these proteins, only the U5 snRNP 15K and 102K proteins were found to bind to the 52K protein (Fig. 5A ▶, left column, and data not shown). In contrast, none of the tri-snRNP or U4/U6 snRNP-specific proteins interacted with the 52K protein (data not shown).
FIGURE 5.
Association of the 52K protein with the U5-specific 15K and 102K proteins. (A) Two-hybrid analyses. Plasmid pairs were co-transformed into yeast host AH109 as indicated in the figure, and the interactions of the full-length (left panel) or truncated 52K protein (right panel) with 15K and 102K proteins were examined by growth on selective medium SD/-Leu-Trp-His-Ade at 30°C for 3 days. The empty vector pGADT7 or pGBKT7 served as a negative control. (B) GST pull-down assay. In vitro [35S]-labeled, c-Myc-tagged 52K constructs were incubated with 102K protein fused to a GST tag and precipitated by glutathione-Sepharose. Coprecipitated protein was analyzed by SDS-PAGE and visualized by fluorography. Ten percent of the translation products were loaded as input. (C) His-Tag pull-down assay. In vitro [35S]-labeled HA-tagged 15K protein was incubated with His-tagged wild-type 52K protein or the C-terminal truncation mutant and precipitated with Talon beads. Proteins were analyzed as in (C). Ten percent of the translation products was loaded as input.
The 52K protein contains a known protein–protein interaction motif, namely the GYF domain in the protein’s C-terminal 86 residues (Freund et al. 1999, 2002). Therefore, we next asked whether this C-terminal fragment is necessary, or possibly even sufficient, for the interaction with the 15K or 102K protein. For this purpose, full-length 52K was split into two fragments: 52N (amino acids 1–255) lacks the GYF motif, while 52C (amino acids 256–341) contains only this domain. As shown in Figure 5A ▶ (right column), the 15K protein binds to 52C, but not 52N. In contrast, the 102K protein binds to 52N, but not 52C. The negative controls with empty prey (pGADT7) or bait vector (pGBKT7) showed no cell growth on the selective medium, confirming that none of the constructs used here induced the reporter gene on its own.
To confirm the interactions observed in the two-hybrid system, we performed pull-down assays. As shown in Figure 5B ▶, the GST–102K fusion protein, but not GST alone, efficiently precipitated the radiolabeled 52K protein from reaction mixtures in which it had been generated by translation in vitro. The GST–102K protein also interacted with the N-terminal two-thirds of the 52K protein, but not with the GYF domain (Fig. 5B ▶). In a second set of experiments we over-expressed His-tagged 52K protein and determined whether it binds 15K protein prepared by translation in vitro. Indeed, a significant amount of the 15K protein was coprecipitated with the His-tagged 52K protein, but not with beads alone (Fig. 5C ▶, lanes 2,3). Consistent with the two-hybrid results, the C-terminal GYF domain also precipitates the 15K protein, and the efficiency of this precipitation was even higher than that observed with the full-length protein (cf lanes 3 and 4). In conclusion, the data from both the yeast two-hybrid system and the in vitro binding assays indicate that the 52K protein associates with the U5 snRNP particle by interacting with the 102K and 15K proteins. Interestingly, two different domains of the 52K protein are involved in these interactions, suggesting that they can occur simultaneously.
DISCUSSION
We report here the sequence of the U5 snRNP 52K protein, and describe its subnuclear localization, as well as protein–protein interactions that likely integrate this protein into the U5 snRNP particle. We conclude that the 52K protein is identical to the GYF domain containing protein CD2BP2 (Nishizawa et al. 1998) that has the ability to bind to polyproline domains in the C terminus of the T-cell marker CD2. In agreement with it being part of a spliceosomal snRNP, the 52K protein is found predominantly in the nucleoplasm of HeLa cells. It is a component of the 20S U5 snRNP, but not of other U snRNPs. In particular, it is not found in the U4/U6.U5 tri-snRNP. Thus, it is the only 20S U5-specific protein that is not part of the tri-snRNP, suggesting a role of the 52K protein in the assembly of the tri-snRNP. We identified two specific interactions of the 52K protein with U5-associated proteins; namely, the 15K and 102K proteins: interestingly, two different domains of the 52K protein are involved in these interactions. While the 102K protein binds the N-terminal two-thirds of the 52K protein, the 15K protein binds to the C-terminal GYF domain.
The following evidence supports the conclusion that the 52K protein described here is indeed a U5 snRNP-specific protein. First of all, antibodies against recombinant protein expressed in Escherichia coli recognize only one protein of the appropriate size, this protein cofractionates with U5 snRNP (Behrens and Lührmann 1991), and is also part of highly purified 20S U5 particles (as detected by SDS-PAGE; data not shown). Second, immunofluorescence studies with the same antibodies demonstrate that the 52K protein is found predominantly in the nucleus, and this finding was confirmed with an exogenous HA-tagged 52K protein (Fig. 2 ▶). While this manuscript was in preparation, other workers reported finding GFP-tagged 52K protein in the nuclei of COS cells (Kofler et al. 2004). Third, the 52K protein is stably associated with the purified U5 snRNP particle, and we have identified two specific 52K binding interactions with other U5 snRNP proteins. Therefore, unspecific aggregation of the 52K protein with snRNPs, e.g., during extract preparation, seems highly unlikely. Finally, the 52K protein has a homolog in the yeast S. cerevisiae that is stably associated with the yeast U5 snRNP (Stevens et al. 2001), indicating that the 52K protein is functionally conserved during evolution.
Database searches revealed that the sequence of the 52K protein is identical to that of a previously described protein, CD2BP2, which interacts with CD2, a surface marker of T cells, which facilitates activation of T lymphocytes on binding to its ligand CD58 (Nishizawa et al. 1998). Since CD2 resides in the plasma membrane of lymphocytes, our characterization of the same protein as a component of nuclear U5 snRNPs raises interesting questions as to the mechanism(s) by which the 52K/CD2BP2 protein may exert different functions. In this respect it is interesting to note that the 52K protein is phosphorylated (Behrens and Lührmann 1991) and posttranslational modification might well create isoforms with different association behavior. In any case, the T-cell-specific function must be a second role that the 52K protein acquired late in evolution: the 52K protein appears universally conserved in eukaryotes (Fig. 1 ▶), whereas the T-cell mediated immunity is found only in vertebrates. Indeed, CD2 association may not be the only alternative function of the protein. The yeast ortholog of the 52K protein, Lin1p, interacts with factors involved in chromosome segregation, and Lin1p has therefore been proposed to play a role in this process (Bialkowska and Kurlandzka 2002).
The distribution of the 52K protein in glycerol gradients (Fig. 3 ▶) and its behavior in immunoaffinity purification experiments (Fig. 4 ▶) both demonstrate that it associates only with the U5 snRNP. This raises the question of which contacts of the 52K protein with other components of the U5 snRNP might mediate its specific interaction with this particle. Recently, it was shown that the GYF domain of CD2BP2/52K interacts strongly with a particular polyproline motif that is also found in the common snRNP protein SmB. The GYF domain overexpressed in mammalian cells was shown to pull down SmB from cell extracts, suggesting that the 52K protein might interact with all splicing snRNPs (Kofler et al. 2004). These findings are not easily reconcilable with our observation that the 52K protein does not associate with the other spliceosomal snRNPs, which each contain a set of the Sm proteins. In particular, the Sm proteins of the 12S U1 snRNP are largely exposed to the solvent (Stark et al. 2001). Nonetheless, we do not observe that the 52K protein cofractionates with U1 in glycerol gradients. The yeast ortholog of the 52K protein, Lin1p, interacts with the yeast U5 snRNP protein Prp8p, presumably through the extensive polyproline stretches at the N-terminus of Prp8p (Bialkowska and Kurlandzka 2002). We do not detect an interaction of the 52K protein with the human Prp8/220K protein, nor with any of six overlapping fragments of the protein, at least by two-hybrid analysis, and indeed, the proline-rich N-terminus of Prp8 is absent in the mammalian Prp8 protein. Instead, employing yeast two-hybrid analysis and in vitro binding assays, we find specific contacts of the 52K protein with the 20S U5-specific 102K and 15K proteins. These two interactions may well occur simultaneously, as different domains of the 52K protein are involved; the N-terminal two-thirds bind to 102K while the C-terminus comprising the GYF domain binds to the 15K protein (Fig. 5 ▶). We therefore believe that these contacts are responsible for the specific integration of the 52K protein into the 20S U5 particle.
Considering that the GYF domain has been characterized as a polyproline-targeting molecule (Nishizawa et al. 1998; Freund et al. 1999, 2003), our observation that it forms a specific complex with the 15K protein came as a surprise, as 15K is devoid of potential proline-rich binding motifs (Reuter et al. 1999). Thus, our data indicate for the first time that a GYF domain can also engage in specific protein–protein interactions in a proline-independent manner.
The 52K protein is the only U5 snRNP protein that is not present in the [U4/U6.U5] tri-snRNP particle. Because the same is true of its yeast ortholog (Stevens et al. 2001), this characteristic property appears to be universally conserved. As U5 snRNP enters the spliceosome as part of the [U4/U6.U5] tri-snRNP, the 52K protein does not appear to have a function in the splicing reaction itself. To elucidate its function, we tried depletion in vivo by RNAi, but our preliminary data did not reveal any striking phenotype. In particular, the siRNAs that we assayed did not inhibit growth of HeLa cells, as they would have been expected to be if the cells had been depleted of an essential splicing factor. Consistent with this result, deletion of the yeast homolog of the 52K protein (Lin1p) is not lethal (Bialkowska and Kurlandzka 2002). This suggests that the 52K protein functions in a pathway that is auxiliary or redundant for the splicing process, and it is tempting to speculate that this function may have something to do with snRNP assembly/recycling; the 52K protein is the only U5 snRNP protein that leaves the 20S U5 snRNP when the latter is integrated into the U4/U6.U5 tri-snRNP. In this respect, it resembles the yeast protein Prp24p (p110 in humans), which is a component of the U6 and U4/U6 snRNPs but not of the tri-snRNP (Jandrositz and Guthrie 1995; Bell et al. 2002). Indeed, Prp24p acts as an assembly factor, being required for the formation of the U4/U6 di-snRNP and tri-snRNP particles (Raghunathan and Guthrie 1998; Bell et al. 2002). As mentioned above, recent experimental evidence indicates that the tri-snRNP is assembled in the CBs (Schaffert et al. 2004), and consistent with a function in tri-snRNP assembly, CBs are enriched in hPrp24 (Stanek et al. 2003). Therefore, the apparent absence of the 52K protein in the Cajal bodies (Fig. 2 ▶) seems to argue against a role for this protein in the assembly of the tri-snRNP. However, if the 20S U5 snRNPs were stored outside of the CBs and their integration into the tri-snRNP were rate-limiting in vivo, then the steady-state concentration of the 52K protein in the CBs would be very low. Since the 20S U5 snRNP is not a ready-to-use splicing factor and such factors are typically stored in the so-called splicing factor compartments, SFC (Lamond and Spector 2003), our finding that the location of the 52K protein overlaps only partly with that of the SFCs would be consistent with this hypothesis. Possibly, the 52K protein covers a protein–protein interaction site that is necessary for the association of the U5 and U4/U6 snRNPs. In support of this idea, the 52K protein interacts with the 102K protein in the U5 snRNP (Fig. 5 ▶). The 102K protein forms part of the bridge between the U5 and U4/U6 particles (Makarova et al. 2002; Schaffert et al. 2004) and may well need to be masked to prevent premature incorporation of U5 particles, which are not fully assembled, into the tri-snRNP (Makarov et al. 2000). Alternatively, the 52K protein could act as a chaperone for the 15K and 102K proteins. These two proteins form a dimer independently of the 52K protein (S. Liu, H.-P. Vornlocher, R. Rauhut, and R. Lührmann, in prep.), which 52K contacts via two independent interactions. Interestingly, both 15K and 102K are not present in the 35S U5 snRNP, which very likely represents a post-spliceosomal U5 intermediate in the spliceosomal disassembly process (Makarov et al. 2002). Therefore, these proteins appear to leave the U5 part of the tri-snRNP before or during the catalytic steps of the splicing reactions. Since two-thirds of the 102K protein consist of tetratricopeptide repeats, which are protein motifs capable of accommodating several simultaneous protein–protein interactions, it is conceivable that this protein needs a chaperone to avoid unfavorable interactions or aggregation before it is reincorporated into the 20S U5 snRNP and then into the 25S U4/U6.U5 tri-snRNP. It will be interesting to see, in future studies, what role the 52K protein may play in the dynamic restructuring of U5 snRNP-particles in the context of the nuclear pre-mRNA splicing machinery.
MATERIALS AND METHODS
Identification of the 52K protein, antisera production, and Western blotting
Proteins were isolated from 20 mg snRNPs by phenol extraction and acetone precipitation, and were separated by preparative SDS/PAGE. Coomassie-stained bands of the 52K protein were excised from the gel, and partial amino acid sequences of tryptic peptides of the protein were determined by Edman microsequencing and electrospray mass spectrometry. Database searches using the TBLASTN program (Altschul et al. 1990) were performed on the BLAST server at the National Center for Biotechnology Information, identifying the CD2BP2 cDNA in the nonredundant (nr) database and various entries in the EST database. One EST with a full-length ORF (accession number N27053) was obtained commercially and sequenced in full.
BamHI and XhoI restriction sites were added by PCR to the open reading frame, excluding the start methionine, of EST N27053. This fragment was subsequently subcloned into the pGEX 4T-1 vector. Overexpression of the GST-fusion protein in E. coli BL21/pLysS was induced by incubation with isopropyl-β-d-thio-galactoside (0.4 mM). Overexpressed fusion protein was purified by chromatography on Glutathione Sepharose 4B (Amersham) and used for immunization of rabbits (0.5 mg in 0.5 mL with 0.5 mL Freund’s complete adjuvant) (Harlow and Lane 1998). For Western blot analysis, proteins were separated on an SDS/polyacrylamide gel, electroblotted onto a PVDF membrane, and immunostained by using ECL chemiluminescence as described by the manufacturer (Amersham Pharmacia).
Immunolocalization studies
HeLa SS6 cells were grown under standard conditions (D-MEM medium + 10% FCS + 1× pencillin–streptamycin). Since the anti-52K antibodies appear to recognize an epitope that is lost upon fixation in the 4% PFA normally used, the cells were fixed with only 1% PFA in PBS for 20 min. The cells were then permeabilized and denatured with 0.1% SDS in PBS for 20 min (SDS rather than Triton enhanced the signal, but was not essential), blocked with 10% FCS in PBS for 30 min, and incubated with specific antibodies (diluted 1:500 in PBS/FCS) for 1 h. The cells were washed four times for 5 min with PBS and the incubated for another hour with fluorophore-conjugated secondary antibodies directed against rabbit IgG and mouse IgG (Molecular Probes). The cells were washed again as described above, dried, and mounted on Moviol (Calbiochem). Optical sections (ca. 700-nm thick: pinhole at 1 Airy unit) were recorded on a Zeiss 510 confocal microscope.
In transfection experiments, the ORF encoding the 52K protein extended by an amino-terminal HA-Tag (MEYPYDVPDYAH) was excised from the pGADT7 plasmid (see below) as an NcoI/XhoI fragment and subcloned into a modified pcDNA3 vector (Invit-rogen), which contains, in the 5′ half of the multiple cloning site, the second intron of the human β-actin gene followed by a Kozak sequence for translation initiation. The plasmid was transiently introduced into HeLa SS6 cells with the Fugene transfection reagent as described by the manufacturer (Roche). Cells were fixed 18 h after transfection and examined by immunofluorescence as described above.
Ultracentrifugation and analysis of the distribution of the 52K protein
One-hundred microliters of HeLa nuclear extract was diluted with an equal volume of IP150 buffer (20 mM HEPES pH 7.9, 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT), loaded onto a 4 mL linear 10%–30% glycerol gradient prepared with IP150 buffer, and centrifuged in a Sorvall TH660 rotor for 2 h at 60,000 rpm. The gradient was fractionated from the top by hand into 24 175-μL aliquots. For RNA analysis, 90 μL of each fraction was extracted with phenol/chloroform, ethanol-precipitated, and subjected to electrophoresis on a 10% polyacrylamide/7 M urea gel followed by staining with silver. For Western blotting, 10 μL of 6× SDS loading buffer was added to 50 μL of each fraction, and the proteins were separated on a 10% polyacrylamide/SDS gel, electrotransferred onto a Hybond P membrane, and probed with the antibodies against the U4/U6.61K and 52K protein at a dilution of 1:1000.
Immunoaffinity selection of U4/U6.U5 tri-snRNPs
To purify U4/U6.U5 tri-snRNPs from a preparation of total spliceosomal snRNPs, antibodies were raised against a C-terminal peptide (amino acids 484–497; CAEFLKVKGEKSGLM) of the human U4/U6-specific 61K protein (Makarova et al. 2002). Antibodies were affinity-purified using a SulfoLink column (Pierce) containing the immobilized cognate peptide. Two-hundred-fifty microliters (bed volume) of protein A Sepharose (PAS) beads were incubated with 250 μg of affinity-purified anti-61K antibodies in 1.0 mL of PBS (pH 8.0) containing 0.5 mg acetylated BSA and 50 μg total yeast tRNA at 4°C for 1.5 h with head-over-tail rotation. The PAS beads were washed twice with 1 mL of PBS (pH 8.0) and subsequently equilibrated with IP250 buffer (20 mM HEPES [pH 7.9], 250 mM NaCl, 1.5 mM MgCl2) containing 0.05% NP-40; 1.5 mg of total snRNPs, immunoaffinity selected from HeLa nuclear extract using the anti-m3G monoclonal antibody H20 and eluted with 7-methylguanosine as previously described (Will et al. 1994), were diluted with an equal volume of IP250 buffer containing 0.05% NP-40 and incubated with the anti-61K-charged PAS beads at 4°C for 2 h with head-over-tail rotation. The beads were washed six times with 1 mL of IP250 buffer containing 0.05% NP-40. During the last wash, the beads were transferred to a fresh tube. The bound material was eluted by incubation with 800 μL of IP250 buffer containing 5% glycerol and 0.5 mg/mL of the 61K peptide for 40 min at 4°C. Four hundred microliters of the eluate were loaded onto a 4-mL linear 10%–30% glycerol gradient prepared with IP150 buffer (20 mM HEPES pH 7.9, 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT) and centrifuged in a Sorvall TH660 rotor for 13 h at 30,000 rpm. The gradient was fractionated from the top by hand into 24t 175-μL aliquots. For protein analysis, 10 μL of 6× SDS loading buffer was added to 60 μL of each fraction and loaded onto a 10% polyacrylamide/SDS gel. The proteins were visualized by staining with Coomassie brilliant blue (Fig. 4A ▶). For RNA analysis, 60 μL of each fraction was extracted with phenol/chloroform, ethanol precipitated and subjected to electrophoresis on a 14% polyacrylamide/7 M urea gel followed by staining with silver (Fig. 4B ▶). The 25S U4/U6.U5 tri-snRNPs peaked in fractions 17–19, whereas 13S U4/U6 snRNPs were found in fractions 8–10.
Two-hybrid analysis
For the 52K protein, the ORF was PCR-amplified from an EST with forward and reverse oligonucleotides designed to introduce an NdeI site at the start codon and an XhoI site downstream from the stop codon. The coding regions of the N-terminal and C-terminal parts of the 52K protein were amplified in a similar way (residues 1–255 and 256–341). All PCR products were blunt-end cloned into pNoTA/T7 by using the Prime-PCR cloning kit (5′ → 3′ Inc.). After the correct sequences of the cloned fragments had been confirmed, the NdeI/XhoI fragments were isolated and ligated into pGBKT7 (DNA-binding domain) and pGADT7 (activation domain) to produce transfecting plasmids for two-hybrid analysis. For the 15K protein, corresponding plasmids containing the coding sequence were constructed by cloning an NdeI/SalI fragment that covered the complete ORF of the 15K protein, from pXC35-p15 (Reuter et al. 1999). For the 102K protein, the cloning procedure is described elsewhere (Makarova et al. 2002).
The two-hybrid assay was performed according to the instructions supplied by the manufacturer (Clontech Laboratories Inc., “Matchmaker” system). Plasmids were cotransformed into strain AH109 and selected on double-dropout medium (synthetic dropout medium, -Leu, -Trp) to establish the presence of cotransformants; the assay was performed by transfer to quadruple-dropout medium (synthetic dropout medium, -Leu, -Trp, -His, -Ade). In each assay, the vectors pGADT7 and pGBKT7 were used as negative controls, and pGADT7-T/pGBKT7-53 was included as a positive control. Yeast media were purchased from Bio 101.
In vitro pull-down assays
The 102K full-length protein was subcloned into pGEX-6P-1(Amersham Pharmacia Biotech), and recombinant GST-102K protein was expressed in E. coli BL21 and purified by affinity chromatography on Glutathione Sepharose 4B as described by the manufacturer (Amersham Pharmacia Biotech). The cDNA encoding full-length or truncated 52K protein in the vector pGBKT7 was used directly as a transcription template to produce in vitro [35S]-labeled, c-Myc-tagged fusion protein. The reaction was performed with Promega’s TnT system and [35S]-methionine (1000 Ci/mmol, Amersham) according to the manufacturer’s instructions. For GST pull-down assay, lysates containing the GST–102K fusion protein were bound to Glutathione Sepharose 4B beads for 30 min at room temperature and washed three times with PBS and then four times with binding buffer (20 mM HEPES pH 7.9, 100 mM KCl, 2.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 1 mM DTT). Twenty-five-microliter aliquots of the GST-102K- or GST-bound beads (containing ~5 μg protein) were incubated with 10 μL of [35S]-labeled protein in 300 μL of binding buffer for 2 h at 4°C. Beads were washed five times with the same buffer. Bound proteins were eluted in SDS sample buffer and analyzed by SDS-PAGE followed by autoradiography.
For His pull-down assays, the full-length 52K protein and the truncated 52C protein were subcloned into the pET28a vector and the recombinant His-tagged proteins were expressed in E. coli BL21 and purified by affinity chromatography on Talon beads as described by the manufacturer (Qiagen). Approximately 6 μg of purified His-tagged 52K or 52C protein was incubated with 10 μL of [35S]-labeled HA-tagged 15K protein, which was produced from pGADT7-15K by in vitro translation as described above, and 30 μL Talon beads in 250 μL IP buffer (20 mM Tris.HCl pH 8.0, 150 mM NaCl, 0.5 mM β-ME, 0.5% NP-40) at 4°C for 2 h. In the control reaction, recombinant protein was omitted. Subsequently, beads were washed four times with 0.8 mL of IP buffer. Bound proteins were eluted in SDS sample buffer, analyzed by SDS-PAGE, and visualized by autoradiography.
Acknowledgments
We thank Irene Öchsner for excellent technical assistence and Cindy L. Will for critical comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemein-schaft (LU294/12-19) and the Fonds der Chemischen Industrie to R.L.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2300805.
REFERENCES
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bach, M., Winkelmann, G., and Lührmann, R. 1989. 20S small nuclear ribonucleoprotein U5 shows a surprisingly complex protein composition. Proc. Natl. Acad. Sci. 86: 6038–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens, S.E. and Lührmann, R. 1991. Immunoaffinity purification of a [U4/U6.U5] tri-snRNP from human cells. Genes & Dev. 5: 1439–1452. [DOI] [PubMed] [Google Scholar]
- Bell, M., Schreiner, S., Damianov, A., Reddy, R., and Bindereif, A. 2002. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 21: 2724–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialkowska, A. and Kurlandzka, A. 2002. Proteins interacting with Lin 1p, a putative link between chromosome segregation, mRNA splicing and DNA replication in Saccharomyces cerevisiae. Yeast 19: 1323–1333. [DOI] [PubMed] [Google Scholar]
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA world (eds. R.F. Gesteland et al.), 2d ed., pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Freund, C., Dötsch, V., Nishizawa, K., Reinherz, E.L., and Wagner, G. 1999. The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences. Nat. Struct. Biol. 6: 656–660. [DOI] [PubMed] [Google Scholar]
- Freund, C., Kühne, R., Yang, H., Park, S., Reinherz, E.L., and Wagner, G. 2002. Dynamic interaction of CD2 with the GYF and the SH3 domain of compartmentalized effector molecules. EMBO J. 21: 5985–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund, C., Kühne, R., Park, S., Thiemke, K., Reinherz, E.L., and Wagner, G. 2003. Structural investigations of a GYF domain covalently linked to a proline-rich peptide. J. Biomol. NMR 27: 143–149. [DOI] [PubMed] [Google Scholar]
- Harlow, E. and Lane, D.P. 1998. Antibodies. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hodges, P.E., Jackson, S.P., Brown, J.D., and Beggs, J.D. 1995. Extraordinary sequence conservation of the PRP8 splicing factor. Yeast 11: 337–342. [DOI] [PubMed] [Google Scholar]
- Jandrositz, A. and Guthrie, C. 1995. Evidence for a Prp24 binding site in U6 snRNA and in a putative intermediate in the annealing of U6 and U4 snRNAs. EMBO J. 14: 820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler, M., Heuer, K., Zech, T., and Freund, C. 2004. Recognition sequences for the GYF domain reveal a possible spliceosomal function of CD2BP2. J. Biol. Chem. 279: 28292–28297. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I. and Spector, D.L. 2003. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4: 605–612. [DOI] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Achsel, T., and Lührmann, R. 2000. The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J. Mol. Biol. 298: 567–575. [DOI] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., and Lührmann, R. 2002. Small nuclear ribonucleopro-tein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Makarova, O.V., Makarov, E.M., Liu, S., Vornlocher, H.P., and Lührmann, R. 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21: 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, O.V., Makarov, E.M., Urlaub, H., Will, C.L., Gentzel, M., Wilm, M., and Lührmann, R. 2004. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23: 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen, T.W. 1998. RNA–RNA interactions in nuclear pre-mRNA splicing. In RNA structure and function (eds. R.W. Simons and M. Grunberg-Manago), pp. 279–307. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Nishizawa, K., Freund, C., Li, J., Wagner, G., and Reinherz, E.L. 1998. Identification of a proline-binding motif regulating CD2-triggered T lymphocyte activation. Proc. Natl. Acad. Sci. 95: 14897–14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan, P.L. and Guthrie, C. 1998. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science 279: 857–860. [DOI] [PubMed] [Google Scholar]
- Reuter, K., Nottrott, S., Fabrizio, P., Lührmann, R., and Ficner, R. 1999. Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein. J. Mol. Biol. 294: 515–525. [DOI] [PubMed] [Google Scholar]
- Schaffert, N., Hossbach, M., Heintzmann, R., Achsel, T., and Lühr-mann, R. 2004. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 23: 3000–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka, M., Reyes, J.L., and Konarska, M.M. 1999. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes & Dev. 13: 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Stanek, D., Rader, S.D., Klingauf, M., and Neugebauer, K.M. 2003. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol. 160: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, H., Dube, P., Lührmann, R., and Kastner, B. 2001. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409: 539–542. [DOI] [PubMed] [Google Scholar]
- Stevens, S.W., Barta, I., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D., and Abelson, J. 2001. Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7: 1543–1553. [PMC free article] [PubMed] [Google Scholar]
- Will, C.L. and Lührmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Kastner, B., and Lührmann, R. 1994. Analysis of ribonucleoprotein interactions. In RNA processing I (eds. S.D. Higgins and B.D.Hames), pp. 141–177. Oxford University Press, New York.