Abstract
We previously showed that the authentic 5′ splice site (5′ss) of the first exon in the human β-globin gene is intrinsically stronger than a cryptic 5′ss located 16 nucleotides upstream. Here we examined by mutational analysis the contribution of individual 5′ss nucleotides to discrimination between these two 5′ss. Based on the in vitro splicing efficiencies of a panel of 26 wild-type and mutant substrates in two separate 5′ss competition assays, we established a hierarchy of 5′ss and grouped them into three functional subclasses: strong, intermediate, and weak. Competition between two 5′ss from different subclasses always resulted in selection of the 5′ss that belongs to the stronger subclass. Moreover, each subclass has different characteristic features. Strong and intermediate 5′ss can be distinguished by their predicted free energy of base-pairing to the U1 snRNA 5′ terminus (ΔG). Whereas the extent of splicing via the strong 5′ss correlates well with the ΔG, this is not the case for competition between intermediate 5′ss. Weak 5′ss were used only when the competing authentic 5′ss was inactivated by mutation. These results indicate that extensive complementarity to U1 snRNA exerts a dominant effect for 5′ss selection, but in the case of competing 5′ss with similarly modest complementarity to U1, the role of other 5′ss features is more prominent. This study reveals the importance of additional submotifs present in certain 5′ss sequences, whose characterization will be critical for understanding 5′ss selection in human genes.
Keywords: pre-mRNA splicing, 5′ splice site, U1 snRNA, pseudouridine
INTRODUCTION
Accurate pre-mRNA splicing is crucial for the gene expression pathway in eukaryotes. Introns are excised from primary transcripts through two sequential transesterification reactions that result in the joining of exons. Both exon–intron boundaries, known as the 5′ and 3′ splice sites, are critical for the recognition of introns and for splicing catalysis (Brow 2002). The 5′ splice site (5′ss) motif consists of nine partially conserved nucleotides at the exon–intron boundary, spanning from positions −3 to +6 (i.e., the last 3 nucleotides [nt] of the upstream exon and the first 6 nt of the intron). The 5′ss consensus sequence in higher eukaryotes corresponds to perfect Watson–Crick base-pairing to the U1 snRNA 5′ terminus (Horowitz and Krainer 1994). This base-pairing plays a critical role in 5′ss selection (Zhuang and Weiner 1986; Séraphin et al. 1988; Siliciano and Guthrie 1988), although several interesting exceptions have been reported: (1) U6 snRNA and SR proteins can make up for the absence, or limiting amount, of U1 snRNA, restoring splicing in U1-depleted extracts (Crispino et al. 1994; Crispino and Sharp 1995; Tarn and Steitz 1994); (2) in vitro selection of functional 5′ss from random sequences yielded the same consensus sequence in nuclear extracts containing either wild-type or 5′-end-truncated human U1 snRNAs (Lund and Kjems 2002); and (3) in yeast, the U1 snRNP can bind to a 5′ss in the absence of the 5′ end of the U1 snRNA (Du and Rosbash 2001), possibly through the U1C polypeptide (Du and Rosbash 2002). Hence, some of the proteins that bind to, or in the vicinity of, the 5′ss (Zhang and Rosbash 1999) may have similar sequence specificity as the intact U1 snRNA and may facilitate splicing when U1 is compromised.
After U1 recognizes the 5′ss, a conformational rearrangement during spliceosome assembly results in the displacement of U1 by U6 snRNA, which base-pairs to positions +2 to +6 of the 5′ss, and by U5 snRNA, which contacts both exon borders through a conserved U-rich loop (Newman and Norman 1992). The replacement of U1 by U6 snRNP has been proposed to contribute to the high fidelity of the reaction (Staley and Guthrie 1999; Chen et al. 2001). The invariant ACAGA-box sequence in U6 snRNA base-pairs to the 5′ss positions +2 to +6 (Wassarman and Steitz 1992; Kandels-Lewis and Séraphin 1993; Lesser and Guthrie 1993). Importantly, there are both artificial and naturally occurring examples of binding of U1 and U6 to adjacent, nonoverlapping sequences within a pre-mRNA, and in this case the actual site of transesterification is defined by U6 (Hwang and Cohen 1996; Brackenridge et al. 2003). These findings indicate that, although the consensus 5′ss motif has very limited complementarity to the U6 ACAGA box, this complementarity can also contribute to general splice-site selection. In addition to U1 and U6, the Prp8 protein splicing factor has been shown to interact with nucleotides at both the 5′ss and the downstream 3′ss (Newman 1997) and it is thought to play a critical role in splice-site selection fidelity, as well as in catalysis (Maroney et al. 2000).
In higher eukaryotes, introns are usually much longer than exons. Because both splice-site consensus motifs are degenerate, many matches to each consensus are present along pre-mRNAs, but the vast majority of these sequences, known as pseudo splice sites, are never selected for splicing (Sun and Chasin 2000). Thus, a 5′ss is defined by other sequence elements in addition to the 9-nt motif. The members of the SR protein family bind to exonic splicing enhancers and stimulate recognition of both 5′ss and 3′ss that flank constitutive and alternative exons (Cartegni et al. 2002). Other widespread elements, known as exonic splicing silencers, are bound by different factors and promote skipping of the exons that harbor them (Ladd and Cooper 2002). Intronic sequences can also play a critical role in 5′ss recognition. For example, intronic G-triplets (McCullough and Berget 1997) are contacted by the U1 snRNA nucleotides 8–10, which normally base-pair to 5′ss positions −3 to −1, and facilitate recognition of certain 5′ss (McCullough and Berget 2000), whereas the protein TIA-1 recognizes U-rich sequences and facilitates binding of U1 to an upstream suboptimal 5′ss (Forch et al. 2002).
A different yet related problem is the selection between nearby competing 5′ss, which is relevant for both alternative 5′ss selection and cryptic 5′ss activation. Previous 5′ss competition assays revealed that the stability of the 5′ss:U1 RNA duplex dictates the choice between two nearby 5′ss (Eperon et al. 1986; Lear et al. 1990). In a recent compilation of cryptic 5′ss in human genes, we showed that as a general rule, cryptic 5′ss are intrinsically weaker than their neighboring authentic 5′ss (Roca et al. 2003). Both RNA and protein factors are involved in discrimination between competing 5′ss. Genetic analyses revealed that U5 (Newman and Norman 1992), U6 (Kandels-Lewis and Séraphin 1993), and U1 (Alvarez and Wise 2001) are involved in cryptic 5′ss activation in budding and fission yeast. In Caenorhabditis elegans, a dominant, allele-specific suppressor mutation in the sup-39 gene affects the choice among two cryptic 5′ss and a mutant 5′ss, enabling splicing via the latter 5′ss (Roller et al. 2000). In mammalian pre-mRNAs, mutations in the conserved loop of U5 activate cryptic 5′ss (Cortes et al. 1993). In the human β-globin gene, a defect in U1 recruitment can explain why a cryptic 5′ss is not used in wild-type pre-mRNA, instead of the neighboring authentic 5′ss (Chabot and Steitz 1987). Several reports support the notion that U1 binding to a 5′ss is not necessarily followed by splicing at that site: (1) When 5′ss sequences are inserted at ectopic positions of a pre-mRNA, some of these 5′ss are not active, even though they are bound by U1 (Nelson and Green 1988); and (2) several U1 snRNP particles can be detected at nearby 5′ss, even when only one of them is used for the reaction, and protein splicing factors, such as SF2/ASF and hnRNP A1, influence U1 occupancy at these neighboring 5′ss (Eperon et al. 1993, 2000). Finally, changes in the levels of SR or hnRNP A/B proteins affect the relative use of cryptic and alternative 5′ss substrates (Krainer et al. 1990; Mayeda and Krainer 1992; Cáceres et al. 1994). However, no protein has been shown to activate a cryptic 5′ss in the context of a wild-type pre-mRNA.
The energetic stability of the RNA duplex between the 5′ss and the 5′ terminus of the U1 snRNA has been previously used as a method to predict the strength of a 5′ss (Mayeda and Ohshima 1988; Lear et al. 1990; Sorek et al. 2004). Other approaches to estimate the strength of a 5′ss are based on computational analyses of large sets of known 5′ss sequences. These methods are based on nucleotide frequency matrices (Shapiro and Senapathy 1987; Senapathy et al. 1990), neural networks (Brunak et al. 1991), and interdependencies between nucleotides at different positions of the consensus (Yeo and Burge 2003) (see below for descriptions of these methods). The matrices derived from 5′ss compilations not only include the sequence that is complementary to U1 snRNA, but might also comprise other interspersed sequence patterns recognized by other factors. Gene prediction programs that rely on local protein-coding information, in addition to the splicing signals, perform much better than those that only consider the latter (Thanaraj 2000). In fact, current methods to predict the intrinsic strength of a 5′ss remain inaccurate, as illustrated by the observation that different rankings of 5′ss scores are achieved by using different 5′ss scoring methods (Roca et al. 2003).
To obtain insights about the determinants of 5′ss selection, we experimentally addressed the contribution of different positions of a 5′ss sequence. We constructed a systematic panel of mutant 5′ss sequences based on the nucleotide differences between the human β-globin first exon authentic 5′ss and one of the nearby cryptic 5′ss. We analyzed the activation of splicing at several mutant 5′ss in two competition assays. We found that the mutant 5′ss could be sorted into three distinct subclasses: strong, intermediate, and weak. Strong and intermediate 5′ss were distinguishable by the predicted free energy of the 5′ss:U1 snRNA duplex. These results strongly suggest that the selection of 5′ss with modest complementarity to U1 snRNA relies on other sequence features embedded within the 5′ss sequence motif.
RESULTS
Design of a 5′ss competition assay
Several thalassemia-associated mutations in the first intron 5′ss of the human β-globin gene result in the activation of three cryptic 5′ss (Fig. 1A ▶), two upstream and one downstream of the natural site (Treisman et al. 1983). By definition, these cryptic 5′ss are completely silent in the context of a wild-type 5′ss. In a previous analysis of the authentic 5′ss and the cryptic 5′ss located 16 nt upstream (−16 5′ss), we showed that the absolute competitive advantage of the authentic over the cryptic 5′ss is determined by the relative efficiencies of their 9-nt 5′ss sequences (Roca et al. 2003). The sequences surrounding these 5′ss elements only had a minor influence on the discrimination between these two 5′ss. In the present study, we carried out a molecular dissection of the individual nucleotides that are critical for the dramatic splicing differences between these two 5′ss. We analyzed 26 mutants of the −16 cryptic 5′ss using in vitro splicing in HeLa cell nuclear extract, and determined how effectively they can compete with different versions of the authentic 5′ss. For consistency with previous nomenclature for this type of competition assays (Eperon et al. 1986; Lear et al. 1990), we refer to the authentic (+1) 5′ss as the “reference 5′ss”, and to the various mutant sequences at −16 as the “test 5′ss”.
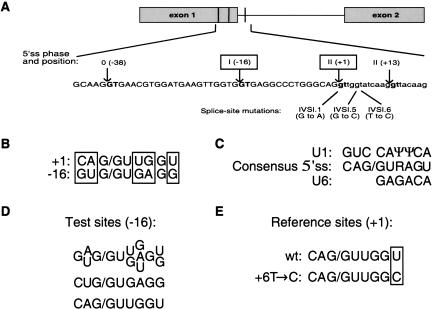
FIGURE 1.
(A) Diagram of the relevant human β-globin fragment including exons 1 and 2 (gray boxes) and intron 1. Vertical lines represent the cryptic 5′ss, whose GT dinucleotides are shown in bold along the sequence. The arrows indicate the cleavage/ligation sites, and the positions relative to the natural site (G at +1) are shown. The two 5′ss analyzed in this study are boxed. The phase, or position of the intron within a codon, is given in Roman numerals, and the number in parenthesis is the relative position of the splice site from the authentic splice site (+1). Exonic DNA sequence is shown in upper-case and intronic sequence in lowercase. The thalassemia mutations are shown below the sequence, with the position and nucleotide substitution indicated in each case. (B–E) Summary of the experimental design. (B) Alignment of the β-globin authentic 5′ss downstream of exon 1 (+1) and the cryptic 5′ss at position -16. The five distinctive nucleotides between these two sequences are indicated by the open boxes. (C) The human consensus 5′ss is shown (R = purine) aligned with the regions of U1 and U6 snRNAs that base-pair to the different positions of the 5′ss. (D) Summary of the combinations of test 5′ss, with the possible nucleotides at each position. Twenty-four mutants were generated, representing all possible permutations of the indicated nucleotides at the four variable positions. Two more test sites were generated and are shown separately: a single nucleotide change at position -3, and a copy of the authentic 5′ss at position -16. (E) The two reference 5′ss are shown, with the distinctive nucleotides boxed. The wild-type +1 5′ss was used as a reference site for competition scheme I. A weakened +1 5′ss, with a T→C transition at position +6, was used for competition scheme II.
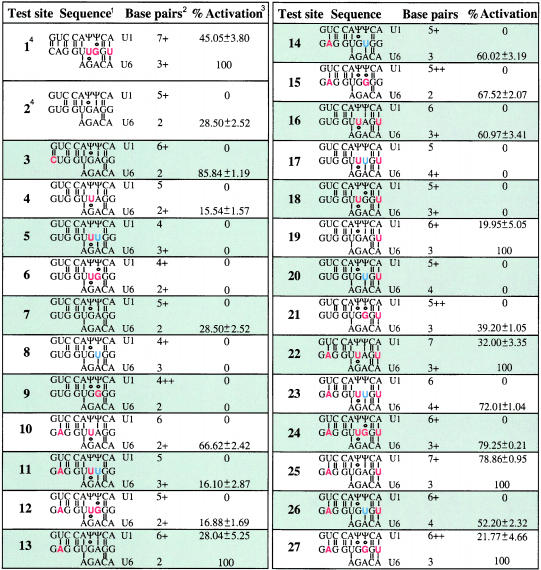
There are five nucleotide differences between the 5′ss at −16 and at +1, out of 9 nt that correspond to the 5′ss consensus sequence (Fig. 1B ▶). These five positions also correspond to the less conserved nucleotides in the 5′ss consensus sequence (Fig. 1C ▶; Shapiro and Senapathy 1987; Senapathy et al. 1990), so the above competition assay is also useful to study the relative contribution of these positions to 5′ss selection. We tried to activate the 5′ss at −16 by partially converting it to the sequence of the authentic 5′ss at +1. First, the authentic and −16 cryptic 5′ss placed at position −16 were designated as test sites 1 and 2, respectively. Second, position −3, which also differs between the two 5′ss, was analyzed separately (mutant 3), because the contribution of this position is often ignored in current nucleotide frequency matrices, even though it shows sequence conservation in the 5′ss consensus sequence (Shapiro and Senapathy 1987; Senapathy et al. 1990). Third, we individually mutated four of the distinctive nucleotides of the −16 5′ss, at positions −2, +3, +4, and +6, to the sequence found at the corresponding positions at the +1 5′ss, and we also constructed all the possible combinations of these changes (Fig. 1D ▶). The collection of mutants was expanded by introducing a further nucleotide substitution at position +4 (A→U), so as to extend the complementarity to U6 snRNA. For the other positions (+3 and +6), either the −16 or the +1 5′ss nucleotide is already complementary to U6. These combinations gave rise to 24 mutant constructs, numbered 4–27 (Table 1 ▶). Test site 7 represents a combination identical to the cryptic 5′ss (test site 2).
TABLE 1.
Complementarity of the mutant test 5′ splice sites (5′ss) to U1 and U6 snRNAs, and percentage of activation for competition schemes I and II
1The potential base-pairing for each mutant 5′ss to U1 and U6 snRNAs is shown above and below the 5′ss sequence, according to the conventional nomenclature for RNA base pairs (Leontis and Westhof 2001). Red letters indicate nucleotide changes to match the authentic 5′ss, and blue letters are nucleotide changes introduced to enhance complementarity to U6.
2Numbers indicate potential Watson–Crick base pairs to U1 (upper) and U6 (lower), and + indicates a G·ψ wobble base pair.
3% activation refers to the extent of splicing via the test site relative to splicing via both test and reference sites. Standard deviations are only shown when > 0. Values are shown for both competition scheme I (upper number) and II (lower number).
4Test sites 1 and 2 are the authentic and cryptic 5′ss, respectively, and were described previously (Roca et al. 2003); test sites 7 and 2 are equivalent.
Competition scheme I: β-Globin mutant 5′ss at position −16 vs. wild-type 5′ss at +1
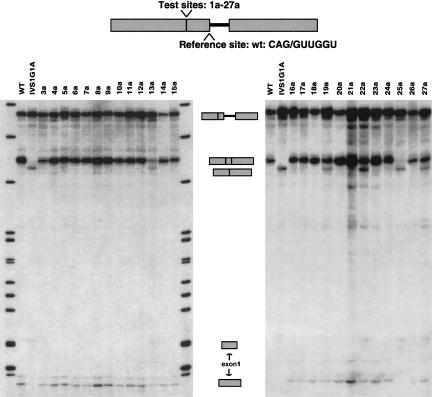
We sought to determine which nucleotide changes at the test 5′ss (mutants 1a–27a) can activate splicing at this site in its normal context, that is, in competition with the authentic 5′ss as a reference site (Fig. 1E ▶). In the context of wild-type β-globin pre-mRNA, the cryptic 5′ss at position −16 remains totally silent, and it cannot be activated by increasing the levels of splicing factors such as SF2/ASF or hnRNP A1 (Krainer et al. 1990; Mayeda and Krainer 1992; Cáceres et al. 1994). Fig. 2 ▶ shows representative in vitro splicing reactions with this series of mutant 5′ss at position −16. InTable 1 ▶ we show the percentage of splicing via the test 5′ss, in relation to total splicing (splicing via the test 5′ss + splicing via the reference 5′ss). Means and standard deviations from three independent experiments are provided for each mutant sequence (Table 1 ▶).
FIGURE 2.
In vitro splicing of the panel of mutants at the −16 test 5′ss, in competition with the wild-type authentic 5′ss at +1 (competition scheme I). Precursors, intermediates, and products are indicated between the gels. Boxes represent exons, and the line represents intron 1. The number above each lane identifies each mutant, whose sequence is shown in Table 1 ▶. For instance, mutant 3a corresponds to mutant test site number 3 (Table 1 ▶) in competition with a wild-type +1 5′ss (indicated in the top diagram). Wild-type (WT) and thalassemia mutant (IVS1G1A) β-globin substrates serve as controls for the position of the mRNAs that are generated by splicing via the +1 and the −16 5′ss.
A G→C substitution at position −3 (mutant 3a in Fig. 2 ▶ andTable 1 ▶), despite introducing a more conserved nucleotide in the consensus motif, as well as introducing an extra predicted G·C base pair to U1, was not sufficient to activate this test 5′ss. Of the remaining five single-nucleotide mutations, the only ones that activated the test site to some extent were U→A at position −2 (mutant 13a) and G→U at +6 (19a). This effect correlates with enhanced base-pairing of these mutant sites with U1 snRNA. On the other hand, mutants 4a (+3 G→U) and 9a (+4 A→G), which failed to activate the −16 5′ss, have a reduced base-pairing potential with U1 at the test site. Thus, of the six single point mutations at the test 5′ss, three result in increased base-pairing to U1, and two of these activated splicing via this site.
The −16 5′ss mutants with combinations of mutations at positions −2, +3, +4, and +6 also confirmed the above trend. The strongest activation was seen with mutant 25a (Fig. 2 ▶;Table 1 ▶), in which the only changes are U→A at −2 and G→U at +6, and very little splicing (20%) occurred via the wild-type reference site at +1. In this case, the nearly perfect match to the consensus 5′ss results in very effective competition against the authentic 5′ss. Some mutants in the positions that improve complementarity to U1 (−2 and +6), such as triple mutants 22a (−2 U→A /+3 G→U /+6 G→U) and 27a (−2 U→A /+4 A→G / +6 G→U), resulted in some use of the −16 5′ss, whereas others, such as quadruple mutant 24a (−2 U→A / +3 G→U / +4 A→G / +6 G→U) did not activate this splice site. Mutations at positions +3 and +4 decrease the complementarity to U1 snRNA. Mutants at these positions did not show any activation of the −16 5′ss, except for two cases in which mutations at −2 or +6 compensate for the loss of complementarity to U1 snRNA (mutants 22a and 27a). Activation of the panel of mutant test sites correlated with expanded complementarity to U1 snRNA, with the exception of the position −3 mutant (mutant 3a).
There was no correlation between the extent of complementarity of the various −16 5′ss with U6 snRNA and activation of splicing via the test site. For instance, mutant 13a has limited complementarity to U6 snRNA (2 bp) but nevertheless spliced via the −16 site (Fig. 2 ▶; Table 1 ▶; 28% of splicing via the test site), whereas mutant 26a, with four predicted base pairs to U6, spliced exclusively via the +1 site. Expanded complementarity to U6 snRNA was not necessarily detrimental for splicing at the −16 site, as seen with mutants 19a, 22a, 25a, and 27a. U5 snRNA base-pairing to the exonic nucleotides of the 5′ss is not sequence-specific, and the presence of the U5 U-rich loop that is involved in this base-pairing is dispensable in vitro (O’Keefe et al. 1996). We cannot draw any conclusions about the contribution of the 5′ss:U5 base-pairing to 5′ss selection on the basis of the present analysis, because we only mutated two exonic 5′ss nucleotides, at positions −2 and −3.
The relative levels of splicing via the test and reference sites for a given mutant were highly reproducible, regardless of variations in the overall splicing efficiency between experiments. Remarkably, the overall splicing efficiency for all the mutants that showed activation of the test site was about half of that observed with the rest of the mutants. It was shown previously that pre-mRNAs with two 5′ss situated <40 nt apart show a severely decreased splicing efficiency (Cunningham et al. 1991; Eperon et al. 1993). This trend, which was also seen with the second competition scheme (see below), presumably reflects the simultaneous binding of U1 at both sites, leading to steric hindrance.
The results of this first competition scheme allowed us to subdivide the pool of mutant test 5′ss at position −16 into two subclasses (Table 2 ▶): “strong 5′ss”, which are the six mutant 5′ss that showed some activation of splicing via these sites in competition with a wild-type reference site; and the remaining mutant 5′ss, which showed no activation at all. This second group will be further subdivided on the basis of the next competition scheme (see below).
TABLE 2.
Activation of the test 5′ splice sites (5′ss) and their 5′ss scores
| Mutant | Sequence | % Test site C.S. Ia | % Test site C.S. IIa | MAX ENTb | MDD | MM | S&S | NN | ΔGc |
| Strong | |||||||||
| 25 | GAG/GUGAGU | 78.86 | 100.00 | 10.03 | 14.48 | 10.90 | 93.71 | 0.99 | −14.2 |
| 1 | CAG/GUUGGU | 45.05 | 100.00 | 8.08 | 11.68 | 7.20 | 80.10 | 0.83 | −14.0 |
| 22 | GAG/GUUAGU | 32.00 | 100.00 | 7.15 | 12.78 | 6.80 | 87.26 | 0.99 | −11.9 |
| 13 | GAG/GUGAGG | 28.04 | 100.00 | 8.41 | 11.78 | 9.20 | 89.35 | 0.91 | −12.0 |
| 27 | GAG/GUGGGU | 21.77 | 100.00 | 7.07 | 12.98 | 7.57 | 83.42 | 0.85 | −13.4 |
| 19 | GUG/GUGAGU | 19.95 | 100.00 | 8.95 | 12.88 | 8.31 | 86.21 | 0.99 | −12.1 |
| Intermediate | |||||||||
| 3 | CUG/GUGAGG | 0.00 | 85.84 | 8.30 | 12.08 | 7.67 | 84.99 | 0.85 | −9.9 |
| 24 | GAG/GUUGGU | 0.00 | 79.25 | 6.36 | 11.28 | 6.23 | 76.96 | 0.59 | −11.9d |
| 23 | GAG/GUUUGU | 0.00 | 72.01 | 6.36 | 10.78 | 5.70 | 76.43 | 0.66 | −9.8 |
| 15 | GAG/GUGGGG | 0.00 | 67.52 | 4.41 | 9.88 | 5.87 | 79.05 | 0.13 | −11.2 |
| 10 | GAG/GUUAGG | 0.00 | 66.62 | 4.36 | 8.98 | 5.11 | 82.89 | 0.37 | −9.7 |
| 16 | GUG/GUUAGU | 0.00 | 60.97 | 4.94 | 7.68 | 4.22 | 79.75 | 0.87 | −9.8 |
| 14 | GAG/GUGUGG | 0.00 | 60.02 | 4.48 | 8.78 | 4.62 | 78.53 | 0.15 | −7.6 |
| 26 | GAG/GUGUGU | 0.00 | 52.20 | 6.14 | 12.48 | 6.31 | 82.89 | 0.95 | −10.8 |
| 21 | GUG/GUGGGU | 0.00 | 39.20 | 4.29 | 9.18 | 4.98 | 75.91 | 0.09 | −11.3 |
| 7 | GUG/GUGAGG | 0.00 | 28.50 | 6.13 | 10.48 | 6.62 | 81.84 | 0.54 | −9.9 |
| 12 | GAG/GUUGGG | 0.00 | 16.88 | 2.53 | 7.08 | 4.54 | 72.60 | 0.11 | −9.7 |
| 11 | GAG/GUUUGG | 0.00 | 16.10 | 3.53 | 5.98 | 4.01 | 72.07 | 0.10 | −7.6 |
| 4 | GUG/GUUAGG | 0.00 | 15.54 | 0.95 | 5.28 | 2.52 | 75.39 | 0.12 | −7.6 |
| Weak | |||||||||
| 20 | GUG/GUGUGU | 0.00 | 0.00 | 3.60 | 9.38 | 3.72 | 75.39 | 0.20 | −8.7 |
| 18 | GUG/GUUGGU | 0.00 | 0.00 | 2.44 | 3.98 | 3.64 | 69.45 | 0.01 | −9.8 |
| 17 | GUG/GUUUGU | 0.00 | 0.00 | 2.70 | 4.18 | 3.12 | 68.93 | 0.01 | −7.7 |
| 5 | GUG/GUUUGG | 0.00 | 0.00 | −1.33 | 1.78 | 1.42 | 64.57 | 0.02 | −5.5 |
| 9 | GUG/GUGGGG | 0.00 | 0.00 | 0.43 | 6.78 | 3.29 | 71.55 | 0.04 | −9.1 |
| 8 | GUG/GUGUGG | 0.00 | 0.00 | 0.74 | 6.98 | 2.03 | 71.02 | 0.04 | −6.5 |
| 6 | GUG/GUUGGG | 0.00 | 0.00 | −2.58 | 1.58 | 1.95 | 65.09 | 0.03 | −7.6 |
Strong, intermediate, and weak 5′ss are indicated. Bold numbers indicate the score used as a threshold to distinguish strong and intermediate
5′ss, and underlined numbers indicate scores for intermediate 5′ss that are higher than the threshold.
aC.S. I and C.S. II refer to competition schemes I and II, respectively.
bSee text for definitions of the 5′ss scoring methods.
cΔG values in Kcal/mol.
dIn this case, the value is equal to the threshold.
Competition scheme II: β-Globin mutant 5′ss at position −16 vs. weakened 5′ss at +1
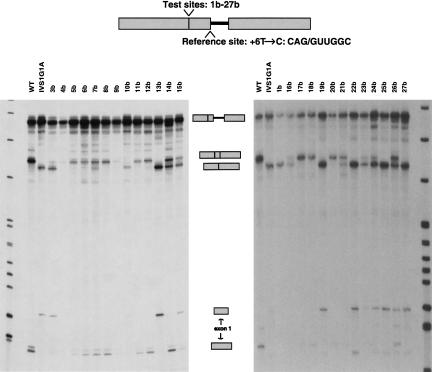
Next we analyzed the same panel of β-globin mutant 5′ss at −16 (mutants 1b–27b) in competition with a weakened 5′ss at +1 as a reference site (Fig. 1E ▶). We used a thalassemia-associated mutation that consists of a T-to-C transition at position +6 (+6T→C) of the authentic 5′ss, which has been shown to reduce, but not completely abrogate, splicing via this site and to activate the same three cryptic 5′ss as the +1G→A allele (Treisman et al. 1983; Krainer et al. 1984). The mutant RNAs were analyzed under identical splicing conditions as described above (Fig. 3 ▶; Table 1 ▶).
FIGURE 3.
In vitro splicing of the panel of mutants at the −16 test 5′ss, in competition with a weakened reference site (+6T→C) at +1 (competition scheme II). The number above each lane identifies each mutant, whose sequence is shown in Table 1 ▶. For instance, mutant 4b corresponds to mutant test site number 4 (Table 1 ▶) in competition with a weakened +1 5′ss. Other labeling is as in Fig. 2 ▶.
It is readily apparent that this second competition assay gave a generally higher degree of activation of mutant sequences at the test site than the previous assay. Nevertheless, the pattern of test 5′ss activation was remarkably consistent between the two assays: All six mutant test sites ranked in the group of strong 5′ss (Table 2 ▶) were used as the exclusive 5′ss (100% of total splicing) when placed in competition with the weakened +1 reference 5′ss. Moreover, none of the remaining test-site mutants spliced exclusively via the mutant test site. These findings indicate that, among all the permutations made to generate the pool of mutant test sites, these six sequences are the most efficient 5′ss.
The single G→C change at position −3 of the test 5′ss resulted in a strong increase in splicing via this site relative to total splicing, although to a lesser extent than that of the strong 5′ss mutants (mutant 3b in Fig. 3 ▶; Table 1 ▶). We conclude that matching the consensus 5′ss at position −3 does contribute to 5′ss selection, although not as much as matching it at each of the other four weakly conserved positions analyzed in this study.
The remaining mutant 5′ss were subdivided into two groups: (1) “Intermediate 5′ss” refers to those mutants that activated splicing via the test site at levels ranging from 15% to 86% (Table 2 ▶), with some use of the reference site; and (2) “weak 5′ss” are the seven mutant 5′ss that did not activate splicing via the test site (see below). The intermediate mutant 5′ss can be ranked on the basis of the percentage of splicing via the test site, although in some cases the differences are not statistically significant (for mutants 10b and 15b; 14b and 16b; and 4b, 11b, and 12b). The remaining mutants displayed consecutively in the gradation in Table 2 ▶ showed significantly different splicing percentages.
Weak 5′ss sequences could still be selected as functional 5′ss when the authentic 5′ss was completely inactivated by mutation, that is, a thalassemia-associated G→A transition at position +1 (see Supplementary Material, Fig. S1, at http://katahdin.cshl.org:9331/supplementary/roca2/abstract.html). The relative percentages of splicing via the test site varied among the different mutant sequences, but in most cases the level of activation was very low. Activation of the cryptic 5′ss at position −38 was also seen to varying extents, complicating the interpretation of these results. We conclude that the set of weak 5′ss represents a subclass of functional—although very inefficient—5′ss.
Contribution of G·ψ base pairs between the 5′ss and the U1 snRNA 5′ terminus
The two consecutive Us in U1 snRNA that base-pair with positions +3 and +4 of the 5′ss are post-transcriptionally modified to pseudouridine (ψ) (Reddy et al. 1981). Thus, the predicted base pairs between 5′ss positions +3 and +4 and U1 are actually A·ψ and G·ψ (wobble) base pairs. The contribution of these predicted base pairs to 5′ss selection was questioned by early studies that showed little or no effect of “suppressor” U1 snRNAs: U1 genes carrying compensatory mutations that restore base-pairing at these positions (Zhuang and Weiner 1986; Siliciano and Guthrie 1988). However, two recent reports showed a positive effect of suppressor U1 snRNAs that restored base-pairing at 5′ss positions +3 or +4 (Freund et al. 2003; Sorek et al. 2004). Here we address the contribution of G·ψ base pairs to 5′ss selection, based on the assumption that complementarity to U1 always contributes to some extent to 5′ss selection.
The preference for purines (G≈A) at position +3 of the human 5′ss consensus sequence (Shapiro and Senapathy 1987; Senapathy et al. 1990) is consistent with a G·ψ base pair with U1 snRNA being able to form at this position. The splicing patterns we observed for several mutant pairs differing only at +3—having either G or U—support the existence of a G·ψ base pair at +3 (Table 3 ▶). For instance, consider the pair of mutants 16 (U at +3) and 19 (G at +3). In competition scheme I, whereas mutant 19a spliced via this site (20% of total splicing), mutant 16a did not. In scheme II, whereas for 19b the test site was the only 5′ss used, for 16b the mutant 5′ss was only partially used (61% of total splicing). For each competition scheme, the informative pairs of mutant 5′ss are those with different percentages of splicing via the test site. The noninformative pairs are those in which both test sites were completely inactive (0% of total splicing) or in which both test sites were used exclusively instead of the reference site (100% for both). Out of a total of 12 pairs, three were informative in both competition schemes, and six were informative in only one scheme. The results of these nine informative mutant pairs suggest that a G·ψ base pair at +3 of the mutant 5′ss conferred a competitive advantage in 5′ss selection, compared with the equivalent test sites with a U at +3.
TABLE 3.
Analysis of the contribution of G·ψ base pairs at 5′ss position +3 to 5′ss selection
| Mutant | Sequence | U1 bpa | U6 bpa | ΔG | C.S. I | C.S. II |
| 7 | GUG/GUGAGG | 5+ | 2 | −9.9 | 0.00 | 28.50 |
| 4 | GUG/GUUAGG | 5 | 2+ | −7.6 | 0.00 | 15.54 |
| 8 | GUG/GUGUGG | 4+ | 3 | −6.5 | 0.00 | 0.00 |
| 5 | GUG/GUUUGG | 4 | 3+ | −5.5 | 0.00 | 0.00 |
| 9 | GUG/GUGGGG | 4++ | 2 | −9.1 | 0.00 | 0.00 |
| 6 | GUG/GUUGGG | 4+ | 2+ | −7.6 | 0.00 | 0.00 |
| 13 | GAG/GUGAGG | 6+ | 2 | −12.0 | 28.04 | 100.00 |
| 10 | GAG/GUUAGG | 6 | 2+ | −9.7 | 0.00 | 66.62 |
| 14 | GAG/GUGUGG | 5+ | 3 | −7.6 | 0.00 | 60.02 |
| 11 | GAG/GUUUGG | 5 | 3+ | −7.6 | 0.00 | 16.10 |
| 15 | GAG/GUGGGG | 5++ | 2 | −11.2 | 0.00 | 67.52 |
| 12 | GAG/GUUGGG | 5+ | 2+ | −9.7 | 0.00 | 16.88 |
| 20 | GUG/GUGUGU | 5+ | 4 | −8.7 | 0.00 | 0.00 |
| 17 | GUG/GUUUGU | 5 | 4+ | −7.7 | 0.00 | 0.00 |
| 19 | GUG/GUGAGU | 6+ | 3 | −12.1 | 19.95 | 100.00 |
| 16 | GUG/GUUAGU | 6 | 3+ | −9.8 | 0.00 | 60.97 |
| 21 | GUG/GUGGGU | 5++ | 3 | −11.3 | 0.00 | 39.20 |
| 18 | GUG/GUUGGU | 5+ | 3+ | −9.8 | 0.00 | 0.00 |
| 25 | GAG/GUGAGU | 7+ | 3 | −14.2 | 78.86 | 100.00 |
| 22 | GAG/GUUAGU | 7 | 3+ | −11.9 | 32.00 | 100.00 |
| 26 | GAG/GUGUGU | 6+ | 4 | −10.8 | 0.00 | 52.20 |
| 23 | GAG/GUUUGU | 6 | 4+ | −9.8 | 0.00 | 72.01 |
| 27 | GAG/GUGGGU | 6++ | 3 | −13.4 | 21.77 | 100.00 |
| 24 | GAG/GUUGGU | 6+ | 3+ | −11.9 | 0.00 | 79.25 |
Bold nucleotides indicate the mutations introduced at the test 5′ss. Bold numbers indicate the percentage of splicing via the test site for the informative pairs of mutant 5′ss. See Table 2 ▶ for definitions.
aPredicted base pairs to U1 and U6 snRNAs, respectively. Numbers indicate Watson–Crick base pairs, and + signs indicate G·ψ base pairs.
However, there was one exception to this overall trend: In the last pair of mutants in Table 3 ▶, mutant 26b, which has a G at +3, spliced significantly more poorly via the test site than mutant 23b (difference of 20%), which differs by having a U at +3. A noncanonical U·ψ base pair was proposed to form at the +4 position of yeast 5′ss (Chen et al. 2001) and at +3 in human 5′ss (Sorek et al. 2004). However, none of the remaining mutants in our set with a U at +3 or +4 positions enhanced splicing via the −16 5′ss, compared with having A or G at these positions (Tables 3 ▶, 4 ▶). Taken together, the results from our panel of mutants did not provide consistent evidence in favor of noncanonical U·ψ base pairs contributing to the definition of a 5′ss.
TABLE 4.
Analysis of the contribution of G·ψ base pairs at 5′ss position +4 to 5′ss selection
| Mutant | Sequence | U1 bpa | U6 bpa | ΔG | C.S. I | C.S. II |
| 4 | GUG/GUUAGG | 5 | 2+ | −7.6 | 0.00 | 15.54 |
| 6 | GUG/GUUGGG | 4+ | 2+ | −7.6 | 0.00 | 0.00 |
| 5 | GUG/GUUUGG | 4 | 3+ | −5.5 | 0.00 | 0.00 |
| 7 | GUG/GUGAGG | 5+ | 2 | −9.9 | 0.00 | 28.50 |
| 9 | GUG/GUGGGG | 4++ | 2 | −9.1 | 0.00 | 0.00 |
| 8 | GUG/GUGUGG | 4+ | 3 | −6.5 | 0.00 | 0.00 |
| 10 | GAG/GUUAGG | 6 | 2+ | −9.7 | 0.00 | 66.62 |
| 12 | GAG/GUUGGG | 5+ | 2+ | −9.7 | 0.00 | 16.88 |
| 11 | GAG/GUUUGG | 5 | 3+ | −7.6 | 0.00 | 16.10 |
| 13 | GAG/GUGAGG | 6+ | 2 | −12.0 | 28.04 | 100.00 |
| 15 | GAG/GUGGGG | 5++ | 2 | −11.2 | 0.00 | 67.52 |
| 14 | GAG/GUGUGG | 5+ | 3 | −7.6 | 0.00 | 60.02 |
| 16 | GUG/GUUAGU | 6 | 3+ | −9.8 | 0.00 | 60.97 |
| 18 | GUG/GUUGGU | 5+ | 3+ | −9.8 | 0.00 | 0.00 |
| 17 | GUG/GUUUGU | 5 | 4+ | −7.7 | 0.00 | 0.00 |
| 19 | GUG/GUGAGU | 6+ | 3 | −12.1 | 19.95 | 100.00 |
| 21 | GUG/GUGGGU | 5++ | 3 | −11.3 | 0.00 | 39.20 |
| 20 | GUG/GUGUGU | 5+ | 4 | −8.7 | 0.00 | 0.00 |
| 22 | GAG/GUUAGU | 7 | 3+ | −11.9 | 32.00 | 100.00 |
| 24 | GAG/GUUGGU | 6+ | 3+ | −11.9 | 0.00 | 79.25 |
| 23 | GAG/GUUUGU | 6 | 4+ | −9.8 | 0.00 | 72.01 |
| 25 | GAG/GUGAGU | 7+ | 3 | −14.2 | 78.86 | 100.00 |
| 27 | GAG/GUGGGU | 6++ | 3 | −13.4 | 21.77 | 100.00 |
| 26 | GAG/GUGUGU | 6+ | 4 | −10.8 | 0.00 | 52.20 |
Bold nucleotides indicate the mutations introducted at the test 5′ss. Bold numbers indicate the percentage of splicing via the test site for the informative pairwise comparisons of mutant 5′ss. See Table 2 ▶ for definitions.
aPredicted base pairs to U1 and U6 snRNAs, respectively. Numbers indicate Watson–Crick base pairs, and + signs indicate G·ψ base pairs.
In contrast to position +3, position +4 of the consensus 5′ss motif shows preference for A but not G nucleotides. Nevertheless, we obtained evidence for the occurrence of G·ψ base pairs between the 5′ss at +4 and U1, and their contribution to 5′ss activation. We sorted the mutants into groups of three, in which the only difference is the presence of A, G, or U at position +4 (Table 4 ▶), so as to compare the effect of having a putative G·ψ base pair at +4 versus a noncanonical U·ψ base pair or an A·ψ base pair. For all groups, the mutants with an A at +4 activated splicing via the test site at least as efficiently as those with a G, and conversely, mutants with a G at +4 activated the test site at least as well as those with a U. In 11 out of 16 groups—considering the two competition schemes separately—having an A at +4 conferred stronger splice-site activation than having a G. In five groups, having a G was significantly better than having a U. The informative pairwise comparisons are those in which the two mutants gave different percentages of splicing via the test site. In only four groups out of 16, all the pairwise comparisons were informative, such as for the group of mutants 13, 14, and 15 for competition scheme II. We conclude that at position +4 of the 5′ss motif, a G·ψ contributes to 5′ss selection, but significantly less than an A·ψ base pair.
Calculation of the predicted strength of the different test 5′ss by available computational methods
We calculated the scores for all our 5′ss sequences by different methods, to see which of the currently available algorithms best explains the results of the above in vitro splicing analyses (Table 2 ▶). We used the Shapiro and Senapathy (S&S) consensus matrix, the Neural Network (NN), the First Order Markov Model (MM), the Maximum Dependence Decomposition Model (MDD), and the Maximum Entropy Model (MAXENT) (see Materials and Methods for references and a description of these algorithms). The predicted free energy of the 5′ss:U1 snRNA duplex (ΔG) was also calculated using the RNA duplex free energy parameters known as the Turner rules (Serra and Turner 1995). The general trends obtained with the various methods were similar; that is, the scores for mutant sites at the top of the overall ranking were higher than the scores for 5′ss at the bottom.
The six “strong” 5′ss are those that were activated when in competition with the wild-type reference site, and that were exclusively used when in competition with a weakened reference site. Therefore, we expected the scores for all six mutant 5′ss to be higher than those of the remaining 5′ss. If this were the case, a threshold to distinguish between these two groups of splice sites could be established, which would correspond to the lowest score among the six strong 5′ss. However, for five out of the six 5′ss scoring methods, there were mutant sequences other than these six strong 5′ss that had a score above the threshold (Table 2 ▶). The most stringent threshold to discriminate between strong and intermediate 5′ss could be established by using the ΔG model, with a value of −11.9 Kcal/mol, even though mutant 24 is an intermediate 5′ss with the same predicted free energy.
However, ΔG could not be used to stringently distinguish between intermediate and weak 5′ss. Some of the weak 5′ (mutants 9, 17, 18, and 20 in Table 2 ▶) had a higher stability for the predicted base-pairing to U1 than some of the intermediate 5′ss (mutants 4, 11, and 12). A discriminating threshold could not be obtained by any of the other scoring methods used here.
The scores for each mutant test site were compared with the percentage of activation of splicing via this site in competition with the two reference sites (competition schemes I and II). We calculated the Pearson’s correlation coefficient (r-value) between the score and the percentage of activation of splicing via the mutant test site in competition with either reference site (Table 5 ▶). The r-values for the full set of test 5′ss in this study were similar for all six methods, ranging from 0.558 to 0.713 for scheme I, and from 0.811 to 0.881 for scheme II.
TABLE 5.
Pearson’s correlation coefficients of the percentage of activation of test 5′ss and their scores
| MAXENT | MDD | MM | S&S | NN | ΔGa | ||
| C.S. I | All | 0.605 | 0.558 | 0.713 | 0.646 | 0.599 | 0.702 |
| Strong | 0.670 | 0.594 | 0.656 | 0.523 | 0.219 | 0.704 | |
| C.S. II | All | 0.881 | 0.852 | 0.873 | 0.860 | 0.860 | 0.811 |
| Intermediate | 0.743 | 0.715 | 0.630 | 0.606 | 0.559 | 0.481 |
Bold numbers indicate the highest r-value for the corresponding set of 5′ss (each row). See Table 2 ▶ for definitions.
aCorrelation coefficients are given as absolute numbers to facilitate comparison.
Interestingly, when we considered the correlation coefficients for strong and intermediate 5′ss separately, we obtained an indication of the determinants of competition between 5′ss (Table 5 ▶). The ΔG correlation coefficient for the strong 5′ss (scheme I) was the highest (r = 0.704), although the corresponding r-values with MAXENT and MM were close to this value. Bearing in mind that all the methods gave high scores for the 5′ss with high complementarity to U1, we conclude that for the group of strong 5′ss, ΔG satisfactorily explains the results of the splicing assays. This finding also suggests that complementarity to U1 is the dominant parameter in determining the extent of splicing via a strong 5′ss.
In contrast, for the group of intermediate 5′ss (scheme II), the correlation coefficient for ΔG was by far the lowest (r = 0.481). Instead, the 5′ss scoring methods that take into account interdependencies between positions, such as MAXENT (0.743) and MDD (0.715), gave the highest r-values for the intermediate 5′ss set. This finding strongly suggests that for those 5′ss with limited base-pairing to the U1 snRNA 5′ end, other sequence patterns make a dominant contribution to 5′ss selection.
DISCUSSION
Subclasses of 5′ss
The results presented in this study expand the findings obtained in previous 5′ss competition assays. Eperon and colleagues analyzed a number of heterogeneous sequences—authentic, alternative, and cryptic 5′ss—from different genes (Eperon et al. 1986; Lear et al. 1990). They found that the level of activation of the different test 5′ss correlated with the predicted free energy of the 5′ss:U1 snRNA duplex. In contrast, in another competition assay, Mayeda and Ohshima (1988) found that a perfect 5′ss consensus sequence gave less efficient splicing in vitro, relative to the reference site, than another sequence with deviations from the consensus, probably due to an effect of the flanking sequences. Our results support the notion that different subclasses of U2-dependent 5′ss exist, which have distinct features. Strong 5′ss have a high degree of complementarity to the U1 snRNA 5′ terminus, and selection of these sites is dominant when in competition with 5′ss of other subclasses. Intermediate 5′ss have a more limited predicted base-pairing to U1, and they likely contain other sequence patterns that contribute to their recognition. Finally, weak 5′ss correspond to very suboptimal sequences that are only used when not in competition with a functional, authentic 5′ss.
We found that strong and intermediate 5′ss have distinguishing features, in that the extent of splicing activation correlated with different sequence patterns for each class. For the strong 5′ss, the stability between the 5′ss:U1 RNA duplex was a major determinant for the levels of 5′ss selection. For the intermediate 5′ss, subtle differences in the free energy of the duplex had a more modest effect on 5′ss selection, and instead other sequence features, which probably partially overlap with the 5′ss consensus motif, likely play an important role.
Determinants of 5′ss selection in mammalian 5′ss
Mutations that activated splicing via the test site when in competition with a strong 5′ss (competition scheme I) correlated with a significant enhancement of the stability of the RNA duplex between the 5′ss and the U1 snRNA 5′ terminus, regardless of their complementarity to U6 snRNA (Tables 1 ▶, 2 ▶). This observation is consistent with previous studies showing that complementarity to U1 plays a pivotal role in 5′ss selection (Zhuang and Weiner 1986; Séraphin et al. 1988; Siliciano and Guthrie 1988). We found no evidence for the contribution of enhanced base-pairing to U6 snRNA to 5′ss activation. There are reported cases in which U6, rather than U1, ultimately dictates the position of the 5′ss (Hwang and Cohen 1996; Brackenridge et al. 2003). In these cases, selection of the transesterification site by U6 depends on a very close U1-binding site.
The modest correlation between the percentage of splicing via the intermediate 5′ss and the scores of these sequences using the ΔG model (Table 5 ▶) indicates that the stability of the 5′ss:U1 RNA duplex explains poorly the experimental data for this 5′ss subclass. Instead, the r-values for the matrices that consider interdependencies between positions of the 5′ss—MAXENT and MDD—gave the strongest correlations among the six scoring tools used, suggesting that these methods capture some sequence patterns other than the complementarity to the U1 snRNA 5′ terminus. Further evidence for this notion was obtained by comparing the results obtained with MM versus ΔG. The first-order Markov model (MM) considers dependencies between adjacent pairs of nucleotides. Strikingly, the prediction of false positive 5′ss using MM is comparable to that obtained using a more sophisticated measurement of dependencies, such as a decision-tree approach equivalent to MDD (Cai et al. 2000). If the free energy of the 5′ss:U1 duplex were the only factor that determines 5′ss specificity, MM should recapitulate the adjacent base-stacking energies implicit in the Turner rules, and therefore, MM should perform just as well as the ΔG method. Interestingly, however, MM gave higher correlation coefficients than ΔG, both for the whole set and for the intermediate 5′ss subset, suggesting that the nearest-neighbor dependencies are not only due to base-stacking energies, but also to other sequence patterns.
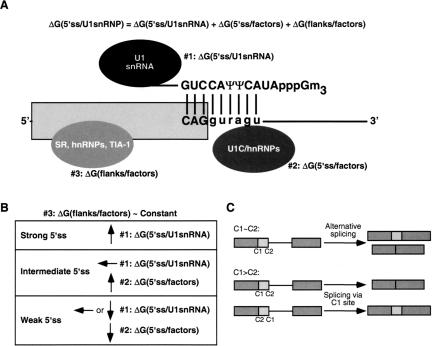
U1 snRNP binding to the 5′ss can be broken up into three thermodynamic parameters (Fig. 4A ▶): (1) the free energy of base-pairing between the 5′ss and the 5′ end of the U1 snRNA [ΔG(5′ss:U1snRNA)]; (2) the effects of other components—U1 snRNP polypeptides and other factors—that bind at the 5′ss and stabilize or destabilize the 5′ss:U1 RNA duplex [ΔG(5′ss/factors)]; and (3) the effects of the 5′ss flanking sequences, which might be bound by splicing factors or adopt secondary structures [ΔG(flanks/factors)] (McCullough and Berget 1997; Cartegni et al. 2002; Forch et al. 2002). The design of our competition schemes predetermines that ΔG(flanks/factors) not only has a minor influence (Roca et al. 2003), but also that this effect can be considered constant between all the test mutant 5′ss analyzed (Fig. 4B ▶). Among the three parameters, so far only the base-pairing between the U1 snRNA and the 5′ss can be estimated, using the ΔG values derived from the Turner rules (Serra and Turner 1995). Our data are consistent with the possibility that ΔG(5′ss:U1snRNA) represents the largest contribution to this interaction, and in comparison, the other two parameters become significant only when ΔG(5′ss:U1snRNA) is small.
FIGURE 4.
(A) Determinants of 5′ss selection in mammalian 5′ss. The thermodynamic parameters that contribute to the free energy of U1 snRNP binding to a 5′ss are shown. The diagram shows the putative factors that determine these three parameters at their respective locations (some of the factors in #3, such as TIA-1, actually bind in the downstream intron, but are shown bound to the exon for simplicity). The exon is represented as a box, the downstream intron as a line, and the base-pairing between a consensus 5′ss sequence and the U1 snRNA 5′ end is also shown. (B) Features of the distinct subclasses of 5′ss. The parameters in the previous equation that significantly contribute to 5′ss selection for each subclass are shown. The design of our β-globin competition analysis yields a roughly constant parameter #3 among the tested 5′ss sequences. Upward arrows indicate a high value or contribution for the corresponding parameter, downward arrows indicate a low value, and the horizontal arrow denotes an intermediate value. (C) Splicing pathways resulting from competition between 5′ss. C1 and C2 represent the competing 5′ss, which can belong to the same (C1 ~ C2) or to a different subclass (C1 > C2). Splicing precursors and products are represented as in Fig. 2 ▶.
Determinants of competition between 5′ss
Eperon and colleagues proposed that 5′ss choice depends on the intrinsic strength of the competing 5′ss, on the distance between them, and on their flanking sequences, which can encompass protein binding sites or secondary structures (Eperon et al. 1986). We propose an extension of this model for constitutive versus alternative 5′ss selection for nearby 5′ss, which relies on our experimental data and only takes into account the intrinsic strength of the 5′ss (Fig. 4C ▶). For a pre-mRNA in which the context does not play a prominent role in 5′ss recognition, such as β-globin, alternative 5′ss selection only happens when the two competing 5′ss belong to the same subclass (e.g., strong vs. strong). For substrates with two 5′ss from different subclasses, only the 5′ss from the higher subclass in the hierarchy is used. The observation that all tested strong 5′ss outcompete any intermediate 5′ss is consistent with previous data. In vitro selection experiments to isolate functional 5′ss from pools of random sequences resulted in closer matches to the consensus 5′ss as more iterative cycles were performed (Lund and Kjems 2002). Base-pairing to U1 was even extended to the nonconserved positions +7 and +8, indicating that those 5′ss with the best complementarity to U1 were selected most efficiently (Libri et al. 2002; Lund and Kjems 2002). Paradoxically, no differences in the selected sequences were found when the U1 snRNA 5′ terminus was cleaved by oligonucleotide-directed RNase H digestion.
When the two competing 5′ss belong to the same subclass, the relative usage of each site depends mostly on ΔG(5′ss:U1snRNA) for the strong 5′ss, or on ΔG(5′ss/factors) for the intermediate 5′ss. With other substrates, nearby cis-regulatory elements, and/or changes in the concentration of splicing factors (such as SR and hnRNP proteins), would modify the ΔG(flanks/factors) term, and thereby shift the relative usage of the competing 5′ss. Conceivably, these elements could completely silence one of the sites, even if this site occupies a higher position in this hierarchy (Mayeda and Ohshima 1988). It is also possible that in some cases, the final position of the 5′ss might be determined by splicing factors that do not influence U1 binding to the 5′ss. Known examples include pre-mRNAs in which several U1 snRNP particles bind simultaneously to two 5′ss, but only one site is selected for splicing (Eperon et al. 1993), or pre-mRNAs in which the transesterification site is dictated by U6 (Hwang and Cohen 1996; Brackenridge et al. 2003).
The contribution of G·ψ base pairs between the 5′ss and U1 to 5′ss selection
G·U wobble base pairs (Varani and McClain 2000) can presumably form in the context of helices involving a given 5′ss and either U1 or U6 snRNA, but their precise thermodynamic contribution compared with standard Watson–Crick base pairs in this particular context is not known. In other contexts, G·U base pairs can form either one or two hydrogen bonds, depending on the flanking nucleotides (Chen et al. 2000). Because the two consecutive U′s in U1 snRNA that are presumed to base-pair with positions +3 and +4 of the 5′ss are modified post-transcriptionally to pseudouridine (ψ) (Reddy et al. 1981), some of the predicted wobble base pairs are actually G·ψ base pairs. ψ can base-pair with either A or G in the context of an A-form RNA duplex, and examples of both are found in rRNA (Ofengand and Bakin 1997). ψ can contribute additional stability to an RNA duplex by promoting base stacking (Davis 1995) and by water-mediated hydrogen bonding to the phosphate backbone via its N-1 proton (Arnez and Steitz 1994). The effect of ψ substitution on uninterrupted RNA duplexes formed with consensus 5′ss and U1 undecamers has been studied by NMR and melting profiles (Hall and McLaughlin 1991). The presence of both ψ’s in the U1 undecamer—opposite two consecutive A’s in the 5′ss undecamer—resulted in only a 2°C increase in the TM. However, larger effects might be expected for natural 5′ss sequences, whose base-pairing to U1 snRNA usually involves multiple mismatches. In addition, the unpaired imino group of ψ, which projects into the major groove of a duplex, and the exocyclic amino group of G in a G·U or G·ψ wobble base pair, which projects into the minor groove, are distinctive structural features that could play important roles in 5′ss-selection specificity, for example, through recognition of base-paired regions by putative proofreading factors. For example, U1C might specifically recognize the modifications of the U1 snRNA 5′terminus. Wobble base pairs additionally cause structural perturbations in A-form helices, due to the distinctive glycosidic bond angles, and they project unique chemical features in the major groove, compared with the standard Watson–Crick base pairs (for review, see Varani and Mc-Clain 2000).
Our data strongly suggest that G·ψ base pairs between the 5′ss positions +3 and +4 and the U1 snRNA 5′ end contribute to 5′ss selection. Using an HIV system in which U1 binding to the SD4 5′ss stabilized the unspliced RNA, leading to synthesis of the env protein, it was likewise shown that a G·ψ base pair taking place at 5′ss position +3 contributed to the stability of the 5′ss:U1 duplex (Freund et al. 2003). However, that study did not find evidence for the contribution of G·ψ base pairs at +4, probably due to either the particular sequence of the mutant 5′ss that was used, or to the different assay used to monitor U1 binding to the 5′ss.
The comparison of mutants 23 and 26 was the only exception to the contribution of G·ψ base pairs to 5′ss selection in the present analysis. In the context of this mutant 5′ss pair, having a U at +3 correlated with a higher degree of splicing via the test site than having a G (Table 3 ▶). There are at least two possible explanations for this finding. (1) A noncanonical U·ψ base pair might occur in this context, as was suggested for the +4 position of 5′ss in Saccharomyces cerevisiae (Chen et al. 2001) and at +3 for human 5′ss (Sorek et al. 2004). Whether U·ψ base pairs can indeed form in the context of the 5′ss:U1 snRNA duplex likely depends on the overall architecture of the duplex. (2) The sequences found in either mutant 5′ss 23 or 26 could contain a protein-binding site that either enhances the rate of splicing via the test site in mutant 23, or reduces it in mutant 26. We note that mutant 26 not only showed robust splicing via the test site (52%), but also this 5′ss sequence is used as an authentic 5′ss in other genes (data not shown).
Proteins that bind at the 5′ss and affect 5′ss selection
The lack of correlation between the levels of test site activation and the degree of base-pairing to U6 snRNA makes it very unlikely that these sequence motifs act through differential U6 binding. Instead, various protein factors may differentially bind these sequences, or part of these sequences plus the flanking nucleotides, leading to the different levels of activation of the mutant 5′ss (Fig. 4A ▶). In yeast, up to eight polypeptides are known to bind around the 5′ss at an early stage of spliceosome assembly, and four of them bind to sites that overlap with the 9-nt 5′ss motif (Zhang and Rosbash 1999). One of them, the U1C protein, was shown to stabilize the 5′ss:U1 snRNA duplex (Chen et al. 2001). Iterative selection of binding sites using the yeast U1C protein resulted in a sequence with close similarity to the consensus 5′ss (Du and Rosbash 2002). Moreover, U1 snRNP can bind the 5′ss in the absence of the U1 snRNA 5′ end through the U1C polypeptide (Du and Rosbash 2001). In mice, the product of the scnm gene, encoding a putative U1C paralog, was genetically linked to 5′ss selection (Buchner et al. 2003). An attractive possibility is that U1C, SCNM, and maybe other unknown members of the U1C protein family differentially contribute to the recognition of specific subsets of intermediate 5′ss.
Alternatively, some of the intermediate and weak 5′ss sequences analyzed in our study may contain a high-affinity binding site for a protein that would compete to some extent with U1 base-pairing. This idea would explain the results in our competition assays with mutant 5′ss that were selected at a lower efficiency than expected from the calculated ΔG. It was previously shown that hnRNP A1 can reduce general 5′ss occupancy by the U1 snRNP (Eperon et al. 2000). Recently, the hnRNP H protein was shown to bind to a subset of 5′ss that contain a poly-G sequence, thereby competing with U1 base-pairing (Buratti et al. 2004). These investigators also showed that different RNA fragments encompassing 5′ss are bound by common and distinct polypeptides.
Among the proteins that play a role at a later stage of spliceosome assembly and/or in catalysis, the U5 snRNP-specific protein Prp8 is a strong candidate to play a role in 5′ss selection in our competition assay (Newman 1997). Prp8 can be cross-linked both to the conserved GU di-nucleotide at the 5′ss (Reyes et al. 1996; Maroney et al. 2000) and to the U1 snRNA (Wyatt et al. 1992) in an ATP-dependent manner. Moreover, some Prp8 alleles can suppress 5′ss mutations in yeast (Collins and Guthrie 1999; Siatecka et al. 1999). It has been proposed that the U4/U6·U5 tri-snRNP can act as a proofreading factor, ensuring the correct specification of the 5′ss (Crispino and Sharp 1995; Maroney et al. 2000). We hypothesize that a preference of the Prp8 protein for binding certain intermediate 5′ss could result in enhanced splicing via the test site.
Our study suggests that the contribution of submotifs within the 5′ss is important to 5′ss selection, especially for those 5′ss with limited complementarity to the U1 snRNA 5′ end. To identify 5′ss submotifs that are presumably bound by positive or negative splicing factors, it may be useful to separate the intermediate from the strong 5′ss, and the best tool to distinguish them appears to be the free energy of the 5′ss:U1 RNA duplex.
Here we presented the analysis of the splicing efficiencies of a panel of mutant 5′ss in two competition assays, using a sensitive and highly reproducible in vitro splicing technique. Careful measurements of the intrinsic differences of 5′ss sequences can also help explain small effects of 5′ss mutations on pre-mRNA splicing patterns. Usually, mutations that result in abnormal splicing are easily detectable by available tools, but subtler splice-site changes might remain unrecognized. This view is illustrated by a recent study describing a genetic modifier of a 5′ss mutation in the mouse sodium channel 8a gene, scn8a (Buchner et al. 2003). A mutation in the modifier gene, the above-mentioned scnm, led to only a 5% decrease in the synthesis of correctly spliced scn8a mRNA, but this subtle change was sufficient to transform a chronic movement disorder into a lethal neurological disease. Other minor variations in 5′ss sequences can have mild or severe consequences, depending on the genetic background.
MATERIALS AND METHODS
Cloning procedures
All β-globin substrates were inserted into a pcDNA3.1+ plasmid (Invitrogen), and all bear a mutation of the cryptic 5′ss at position +13—a T-to-C transition at +14—that inactivates this 5′ss. To generate the panel of mutants shown in Table 1 ▶, we used site-directed mutagenesis with oligonucleotides with the different mutations. For each pair of primers, 14–18 cycles of PCR with Pfu I Turbo (Stratagene) were performed. PCR products were digested with Dpn I (New England Biolabs), followed by transformation of competent Escherichia coli DH5α. The −3 position mutant of the β-globin −16 5′ss (mutant 3 in Table 1 ▶) was constructed separately by site-directed mutagenesis using primers carrying the single-nucleotide substitution (primer sequences available upon request). Mutants 4–27, which consist of permutations of different nucleotides at positions −2, +3, +4, and +6, were synthesized together using the following degenerate primers: β–16/4nt-F: 5′-ggtgaacgtggatgaagttggwggtkdgkccctgggcaggttggtatcaag-3′, and β–16/4nt-R: 5′-cttgataccaacctgcccagggmchmaccwccaacttcatccacgttcacc-3′ (where “W” is A or T, “K” is G or T, “D” is G, A, or T, “M” is A or C, and “H” is A, T, or C). Individual clones were sequenced with an ABI3700 automated sequencer.
A T-to-C mutation at position +6 of the β-globin authentic 5′ss (reference 5′ss) was introduced into the set of mutants at position −16 (mutants 1–27) to generate mutants 1b–27b, by overlap-extension PCR (primer sequences available upon request). Each PCR product was reintroduced into the pcDNA3.1+ plasmid by sub-cloning it into the HindIII and BamHI restriction sites. Similarly, a +1G→A mutation was introduced by overlap-extension PCR and cloned.
In vitro splicing experiments
Human β-globin splicing substrates were transcribed from PCR products using T7 RNA polymerase (Promega) as described (Roca et al. 2003). The downstream primer generates a PCR fragment that terminates at a position equivalent to a natural BamHI site 18 nt upstream from the 3′ end of exon 2. HeLa cell nuclear extracts were prepared and splicing reactions carried out as described (Mayeda and Krainer 1999a,b). For in vitro splicing reactions, 20 fmol of 32P-labeled, 7CH3-GpppG-capped T7 transcript was incubated in 12.5-μL splicing reactions with 30% (v/v) nuclear extract and 3.2 mM MgCl2, for 3 h at 30°C. All samples were analyzed by electrophoresis in 5.5 % polyacrylamide/7M urea gels. In some cases, the gel solutions were prepared in formamide, instead of water, to completely disrupt RNA secondary structures found in the splicing precursors, intermediates, and products from several mutants. Gels were exposed overnight onto X-OMAT film (Kodak), or exposed for 1 h using a FUJI PhosphorImager screen and quantified with FUJI-MacScan. Three independent in vitro splicing reactions were performed for each mutant.
5′ss scoring methods
The Shapiro and Senapathy (S&S) consensus matrix is a nucleotide frequency or position-weight matrix, which reflects the degree of conservation at each position of the consensus 5′ss motif in an alignment of 1446 5′ss (Shapiro and Senapathy 1987; Senapathy et al. 1990). The mammalian 5′ss consensus sequence is MAG|GURAGU (M = A or C; R = purine), and spans from position −3 (the third nucleotide from the 3′-end of the upstream exon) to +6 (the sixth nucleotide in the intron). Although position −3 is often ignored in these matrices, we took it into account because of the significant preference for C or A at this position. The S&S is one of the most commonly used 5′ss-scoring methods, and it assumes independence between individual positions of the 9-nt motif.
We also calculated the scores of the 5′ss by the neural network (NN) method (Brunak et al. 1991; http://www.fruitfly.org/seq_tools/splice.html). The NN algorithm is a machine-learning approach that recognizes sequence patterns once it is trained with a set of DNA sequences encompassing authentic 5′ss.
To take into account dependencies between positions of the 5′ss motif, we used three different algorithms developed by Burge and colleagues (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html): The first-order Markov model (MM) only considers dependencies between adjacent positions; the Maximum Dependence Decomposition model (MDD) is a decision-tree approach that emphasizes the strongest dependencies in the early branches of the tree (Burge 1998); and the Maximum Entropy Model (MAXENT) can monitor the importance of dependencies between different positions by using a maximum-entropy distribution consistent with low-order marginal constraints estimated from available data (Yeo and Burge 2003).
To calculate the free-energy parameters for the stability of the RNA duplexes between the various 5′ss sequences and the U1 snRNA 5′-terminus (ΔG), we used the Turner energy rules as described in http://www.bioinfo.rpi.edu/~zukerm/rna/energy/ (11/3/2000 update). These empirical rules are based on measurements with synthetic oligoribonucleotides and reflect the contribution of hydrogen bonding, base stacking, mismatches, and Watson–Crick or G·U base pairs (Serra and Turner 1995). The investigators reported that these nearest-neighbor rules work very well for Watson–Crick base pairs, satisfactorily well for G·U base pairs flanked by Watson–Crick base pairs, but less reliably for mismatches, noncanonical base pairs, and consecutive G·U base pairs. Other limitations of these measurements might be derived from the undetermined energy corrections that should be applied to the ends of a short RNA duplex. The U1 snRNA 5′ terminus, which base-pairs to the 5′ss, has two U′s that are post-transcriptionally modified to pseudouridines (ψ) (Reddy et al. 1981). ψ is a regioisomer of uridine in which the uracil is bound to the ribose through the C5 carbon, instead of the N1 nitrogen (Hall and McLaughlin 1991). The atoms involved in the Watson–Crick hydrogen bonds with A are conserved between U and ψ. The Turner rules have not thus far addressed ψ base-pairing to A or G nucleotides. However, a comparison of RNA duplex undecamers with either two consecutive Us or two ψs in one of the strands showed no significant differences in their thermodynamic properties (Hall and McLaughlin 1991). Thus, our ΔG calculations treated G·ψ and A·ψ base pairs as if they were G·U and A·U base pairs, respectively.
Acknowledgments
We are grateful to the members of our lab for helpful advice and discussions. We especially thank Akila Mayeda for helpful suggestions, Jim Duffy for help in preparing the figures, and Michelle Hastings for critically reading the manuscript. X.R. and A.R.K. acknowledge support from NCI grant CA13106.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2040605.
REFERENCES
- Alvarez, C.J. and Wise, J.A. 2001. Activation of a cryptic 5′ splice site by U1 snRNA. RNA 7: 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnez, J.G. and Steitz, T.A. 1994. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudouridines in stabilization of RNA structure. Biochemistry 33: 7560–7567. [DOI] [PubMed] [Google Scholar]
- Brackenridge, S., Wilkie, A.O., and Screaton, G.R. 2003. Efficient use of a ‘dead-end’ GA 5′ splice site in the human fibroblast growth factor receptor genes. EMBO J. 22: 1620–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Brunak, S., Engelbrecht, J., and Knudsen, S. 1991. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 220: 49–65. [DOI] [PubMed] [Google Scholar]
- Buchner, D.A., Trudeau, M., and Meisler, M.H. 2003. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science 301: 967–969. [DOI] [PubMed] [Google Scholar]
- Buratti, E., Baralle, M., De Conti, L., Baralle, D., Romano, M., Ayala, Y.M., and Baralle, F.E. 2004. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHβ genes. Nucleic Acids Res. 32: 4224–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. 1998. Modeling dependencies in pre-mRNA splicing signals. In Computational methods in molecular biology (eds. S.L. Salzberg et al.), chapter 8, pp. 129–164. Elsevier Science, Philadelphia, PA.
- Cáceres, J.F., Stamm, S., Helfman, D.M., and Krainer, A.R. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265: 1706–1709. [DOI] [PubMed] [Google Scholar]
- Cai, D., Delcher, A., Kao, B., and Kasif, S. 2000. Modeling splice sites with Bayes networks. Bioinformatics 16: 152–158. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285–298. [DOI] [PubMed] [Google Scholar]
- Chabot, B. and Steitz, J.A. 1987. Recognition of mutant and cryptic 5′ splice sites by the U1 small nuclear ribonucleoprotein in vitro. Mol. Cell. Biol. 7: 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., McDowell, J.A., Kierzek, R., Krugh, T.R., and Turner, D.H. 2000. Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G·U pairs: One hydrogen bond for each G·U pair in r(GGCGUGCC)(2) and two for each G·U pair in r(GAGUGCUC)(2). Biochemistry 39: 8970–8982. [PubMed] [Google Scholar]
- Chen, J.Y., Stands, L., Staley, J.P., Jackups, R.R.J., Latus, L.J., and Chang, T.H. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7: 227–232. [DOI] [PubMed] [Google Scholar]
- Collins, C.A. and Guthrie, C. 1999. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes & Dev. 13: 1970–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes, J.J., Sontheimer, E.J., Seiwert, S.D., and Steitz, J.A. 1993. Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5′ splice sites in vivo. EMBO J. 12: 5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino, J. and Sharp, P. 1995. A U6 snRNA:pre-mRNA interaction can be rate-limiting for U1-independent splicing. Genes & Dev. 9: 2314–2323. [DOI] [PubMed] [Google Scholar]
- Crispino, J.D., Blencowe, B.J., and Sharp, P.A. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265: 1866–1869. [DOI] [PubMed] [Google Scholar]
- Cunningham, S.A., Else, A.J., Potter, B.V., and Eperon, I.C. 1991. Influences of separation and adjacent sequences on the use of alternative 5′ splice sites. J. Mol. Biol. 217: 265–281. [DOI] [PubMed] [Google Scholar]
- Davis, D.R. 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 23: 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H. and Rosbash, M. 2001. Yeast U1 snRNP-pre-mRNA complex formation without U1snRNA-pre-mRNA base pairing. RNA 7: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419: 86–90. [DOI] [PubMed] [Google Scholar]
- Eperon, L.P., Estibeiro, J.P., and Eperon, I.C. 1986. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature 324: 280–282. [DOI] [PubMed] [Google Scholar]
- Eperon, I.C., Ireland, D.C., Smith, R.A., Mayeda, A., and Krainer, A.R. 1993. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 12: 3607–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon, I.C., Makarova, O.V., Mayeda, A., Munroe, S.H., Cáceres, J.F., Hayward, D.G., and Krainer, A.R. 2000. Selection of alternative 5′ splice sites: Role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell Biol. 20: 8303–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forch, P., Puig, O., Martinez, C., Séraphin, B., and Valcárcel, J. 2002. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 21: 6882–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund, M., Asang, C., Kammler, S., Konermann, C., Krummheuer, J., Hipp, M., Meyer, I., Gierling, W., Theiss, S., Preuss, T., et al. 2003. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res. 31: 6963–6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, K.B. and McLaughlin, L.W. 1991. Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry 30: 1795–1801. [DOI] [PubMed] [Google Scholar]
- Horowitz, D.S. and Krainer, A.R. 1994. Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet. 10: 100–106. [DOI] [PubMed] [Google Scholar]
- Hwang, D.Y. and Cohen, J.B. 1996. U1 snRNA promotes the selection of nearby 5′ splice sites by U6 snRNA in mammalian cells. Genes & Dev. 10: 338–350. [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis, S. and Séraphin, B. 1993. Involvement of U6 snRNA in 5′ splice site selection. Science 262: 2035–2039. [DOI] [PubMed] [Google Scholar]
- Krainer, A.R., Maniatis, T., Ruskin, B., and Green, M.R. 1984. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36: 993–1005. [DOI] [PubMed] [Google Scholar]
- Krainer, A.R., Conway, G.C., and Kozak, D. 1990. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell 62: 35–42. [DOI] [PubMed] [Google Scholar]
- Ladd, A.N. and Cooper, T.A. 2002. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 3: reviews0008.1–0008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear, A.L., Eperon, L.P., Wheatley, I.M., and Eperon, I.C. 1990. Hierarchy for 5′ splice site preference determined in vivo. J. Mol. Biol. 211: 103–115. [DOI] [PubMed] [Google Scholar]
- Leontis, N.B. and Westhof, E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser, C.F. and Guthrie, C. 1993. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science 262: 1982–1988. [DOI] [PubMed] [Google Scholar]
- Libri, D., Ducongé, F., Levy, L., and Vinauger, M. 2002. A role for the ψ-U mismatch in the recognition of the 5′ splice site of yeast introns by the U1 small nuclear ribonucleoprotein particle. J. Biol. Chem. 277: 18173–18181. [DOI] [PubMed] [Google Scholar]
- Lund, M. and Kjems, J. 2002. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA 8: 166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney, P.A., Romfo, C.M., and Nilsen, T.W. 2000. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell 6: 317–328. [DOI] [PubMed] [Google Scholar]
- Mayeda, A. and Krainer, A.R. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68: 365–375. [DOI] [PubMed] [Google Scholar]
- ———. 1999a. Mammalian in vitro splicing assays. Methods Mol. Biol. 118: 315–321. [DOI] [PubMed] [Google Scholar]
- ———. 1999b. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 118: 309–314. [DOI] [PubMed] [Google Scholar]
- Mayeda, A. and Ohshima, Y. 1988. Short donor site sequences inserted within the intron of β-globin pre-mRNA serve for splicing in vitro. Mol. Cell. Biol. 8: 4484–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough, A. and Berget, S. 1997. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 17: 4562–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. An intronic splicing enhancer binds U1 snRNPs to enhance splicing and select 5′ splice sites. Mol. Cell. Biol. 20: 9225–9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, K.K. and Green, M.R. 1988. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes & Dev. 2: 319–329. [DOI] [PubMed] [Google Scholar]
- Newman, A.J. 1997. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 16: 5797–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A. and Norman, C. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68: 743–754. [DOI] [PubMed] [Google Scholar]
- O’Keefe, R.T., Norman, C., and Newman, A.J. 1996. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell 86: 679–689. [DOI] [PubMed] [Google Scholar]
- Ofengand, J. and Bakin, A. 1997. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266: 246–268. [DOI] [PubMed] [Google Scholar]
- Reddy, R., Henning, D., and Busch, H. 1981. Pseudouridine residues in the 5′-terminus of uridine-rich nuclear RNA I (U1 RNA). Biochem. Biophys. Res. Commun. 98: 1076–1083. [DOI] [PubMed] [Google Scholar]
- Reyes, J.L., Kois, P., Konforti, B.B., and Konarska, M.M. 1996. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA 2: 213–225. [PMC free article] [PubMed] [Google Scholar]
- Roca, X., Sachidanandam, R., and Krainer, A.R. 2003. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 31: 6321–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller, A., Hoffman, D., and Zahler, A. 2000. The allele-specific suppressor sup-39 alters use of cryptic splice sites in Caenorhabditis elegans. Genetics 154: 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy, P., Shapiro, M.B., and Harris, N.L. 1990. Splice junctions, branch point sites, and exons: Sequence statistics, identification, and applications to genome project. Methods Enzymol. 183: 252–278. [DOI] [PubMed] [Google Scholar]
- Séraphin, B., Kretzner, L., and Rosbash, M. 1988. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 7: 2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra, M.J. and Turner, D.H. 1995. Predicting thermodynamic properties of RNA. Methods Enzymol. 259: 242–261. [DOI] [PubMed] [Google Scholar]
- Shapiro, M.B. and Senapathy, P. 1987. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 15: 7155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka, M., Reyes, J.L., and Konarska, M.M. 1999. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes & Dev. 13: 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano, P.G. and Guthrie, C. 1988. 5′ Splice site selection in yeast: Genetic alterations in base-pairing with U1 reveal additional requirements. Genes & Dev. 2: 1258–1267. [DOI] [PubMed] [Google Scholar]
- Sorek, R., Lev-Maor, G., Reznik, M., Dagan, T., Belinky, F., Graur, D., and Ast, G. 2004. Minimal conditions for exonization of intronic sequences: 5′ Splice site formation in Alu exons. Mol. Cell 14: 221–231. [DOI] [PubMed] [Google Scholar]
- Staley, J. and Guthrie, C. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3: 55–64. [DOI] [PubMed] [Google Scholar]
- Sun, H. and Chasin, L.A. 2000. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 20: 6414–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn, W. and Steitz, J. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes & Dev. 8: 2704–2717. [DOI] [PubMed] [Google Scholar]
- Thanaraj, T.A. 2000. Positional characterisation of false positives from computational prediction of human splice sites. Nucleic Acids Res. 28: 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman, R., Orkin, S.H., and Maniatis, T. 1983. Specific transcription and RNA splicing defects in five cloned β-thalassaemia genes. Nature 302: 591–596. [DOI] [PubMed] [Google Scholar]
- Varani, G. and McClain, W.H. 2000. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 1: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman, D.A. and Steitz, J.A. 1992. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science 257: 1918–1925. [DOI] [PubMed] [Google Scholar]
- Wyatt, J.R., Sontheimer, E.J., and Steitz, J.A. 1992. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 6: 2542–2553. [DOI] [PubMed] [Google Scholar]
- Yeo, G. and Burge, C.B. 2003. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. In Proceedings of the 7th Annual International Conference on Computational Molecular Biology (RECOMB03) (eds. W. Miller et al.), pp. 322–331. ACM Press, New York. [DOI] [PubMed]
- Zhang, D. and Rosbash, M. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes & Dev. 13: 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, Y. and Weiner, A.M. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46: 827–835. [DOI] [PubMed] [Google Scholar]