Abstract
The Escherichia coli autoregulatory ribosomal protein L20 consists of two structurally distinct domains. The C-terminal domain is globular and sits on the surface of the large ribosomal subunit whereas the N-terminal domain has an extended shape and penetrates deep into the RNA-rich core of the subunit. Many other ribosomal proteins have analogous internal or terminal extensions. However, the biological functions of these extended domains remain obscure. Here we show that the N-terminal tail of L20 is important for ribosome assembly in vivo. Indeed, a truncated version of L20 without its N-terminal tail is unable to complement the deletion of rplT, the gene encoding L20. In addition, this L20 truncation confers a lethal-dominant phenotype, suggesting that the N-terminal domain is essential for cell growth because it could be required for ribosome assembly. Supporting this hypothesis, partial deletions of the N-terminal tail of the protein are shown to cause a slow-growth phenotype due to altered ribosome assembly in vivo as large amounts of intermediate 40S ribosomal particles accumulate. In addition to being a ribosomal protein, L20 also acts as an autogenous repressor. Using L20 truncations, we also show that the N-terminal tail of L20 is dispensable for autogenous control.
Keywords: autogenous control, protein structure, ribosomal protein, ribosome assembly, rRNA
INTRODUCTION
The recent unveiling of the atomic structure of the prokaryotic ribosome has made possible the construction of detailed models to help understand the functioning of this ribonucleoprotein complex (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000; Harms et al. 2001). The three-dimensional (3D) structure revealed that the shape of each ribosomal subunit corresponds to the shape of the ribosomal RNA (rRNA), whereas the ribosomal proteins (r-proteins) appear as relatively small molecules, sitting mainly on the surface of each subunit. This structural organization highlights the pivotal role played by rRNA in some of the fundamental processes of translation, such as the accurate decoding of genetic information (Ogle et al. 2001, 2002) or the peptidyl-transferase activity (Nissen et al. 2000). Nevertheless, r-proteins are required for both ribosome assembly, because they consolidate specific rRNA structures (Agalarov et al. 2000), and ribosome function, through their capacity to facilitate the dynamics of translation (Gao et al. 2003). More than one-third of the 54 r-proteins possess nonglobular extensions, protruding long distances from globular domains into the RNA-rich core of the ribosome and making extensive contacts with different regions of rRNA (Ban et al. 2000; Wimberly et al. 2000; Brodersen et al. 2002). However, the roles of these extensions remain largely unknown and the few that have been studied in some detail, such as the extended loops of Escherichia coli r-proteins S9, S13, L4, and L22, are apparently not required for ribosome assembly and/or function (Zengel et al. 2003; Hoang et al. 2004).
The eubacterial r-protein L20 is another example of a protein with a long extension. This protein is organized into two structurally distinct domains within the ribosome, each composed of about 60 amino acids (Harms et al. 2001). The C-terminal domain adopts a globular structure, formed from four α-helices, which contacts the helix 40–41 junction in domain II of 23S rRNA. In contrast, the N-terminal domain is composed of two α-helices forming a rather long tail that protrudes from the globular body of the protein into the large ribosomal subunit by contacting several regions of domains I and II in 23S rRNA. Interestingly, the structure of the C-terminal domain is similar in both free and ribosome-bound forms of L20, whereas the N-terminal domain is not folded in solution, presumably because of its strong basic character (Raibaud et al. 2002).
L20 is one of five ribosomal proteins essential for the first reconstitution step of ribosome assembly in vitro (Spillmann et al. 1977). However, it can be withdrawn from the mature 50S ribosomal subunit without effect on its activity (Nowotny and Nierhaus 1980). In addition, it can also replace the assembly initiator protein L24 at low temperature in vitro (Franceschi and Nierhaus 1988). L20 has been shown to be essential in vivo, as a deletion within its gene is lethal unless the wild-type copy of the gene is provided by a complementing plasmid (this article). Taken together, these data suggest that L20 might facilitate the proper 23S rRNA folding during the assembly of the large ribosomal subunit.
In addition to its scaffolding role in ribosome assembly, L20 also acts as an autogenous repressor. L20 is encoded by rplT, the distal gene of the IF3 operon in E. coli, which also contains the infC and rpmI genes, encoding translation initiation factor IF3 and r-protein L35, respectively. L20 directly inhibits the expression of rpmI at a translational level (Lesage et al. 1990), and indirectly that of its own gene, through translational coupling (Lesage et al. 1992). Repression requires the binding of L20 to two distinct sites in the leader of rpmI mRNA (Guillier et al. 2002). Interestingly, both sites display structural similarities with the L20-binding site in 23S rRNA allowing the protein to recognize the three RNA sites in similar way (Guillier et al. 2005).
To learn more about the functions of r-protein extensions, we focused our attention on the N-terminal tail of L20. We constructed truncated derivatives of the protein and examined their ability to function in both ribosome assembly and autogenous repression. In contrast to the extended loops of S9, S13, L4, and L22, which are apparently neither required for ribosome assembly and/or function nor L4-mediated autogenous control (Zengel et al. 2003; Hoang et al. 2004), we show here that the N-terminal extension of L20 is important for ribosome assembly, although dispensable for control.
RESULTS
Truncations in the N-terminal domain of L20 dramatically reduce growth rate
To investigate the role of the N-terminal domain of L20, we constructed three in-frame deletions removing increasing portions of this domain and compared the growth rate afforded by the resulting L20 truncations to that provided by the wild-type L20. The L20Δ1 truncation (Fig. 1 ▶), expressed from plasmid pBL20ecΔ6–20, was constructed by deleting the N-terminal α-helix, which spans amino acid residues 6–20. The L20Δ2 truncation, expressed from plasmid pBL20ecΔ6–29, was constructed by removing amino acid residues 6–29, which include the N-terminal α-helix together with its short proximal loop. Finally, the L20ΔN truncation, expressed from plasmid pBL20ecΔN, was constructed by removing amino acid residues 6–58, comprising most of the N-terminal domain. Wild-type L20 was expressed from the rplT gene cloned in plasmid pBL20ec. The wild-type and the three deleted alleles of rplT were cloned without the regulatory cis-acting sequences bound by L20 during autogenous repression. Therefore, the expression of the different rplT alleles borne by these plasmids is neither repressed by the wild-type chromosomal copy of L20 nor by their own products. In addition, all of these plasmids express a shortened lac repressor that is still able to repress transcription from a lac promoter, thus allowing synthesis of the different versions of L20 upon induction with IPTG. Using these plasmids, we first analyzed the ability of the different mutants to complement an E. coli strain (IBPC6801) that lacks the chromosomal copy of rplT. Since rplT is an essential gene, this strain is only viable if L20 is produced from a resident plasmid. Therefore, we first replaced the counterselectable resident plasmid pSGL6, which allows the production of L20, by each of the four tester plasmids described above. We then studied the phenotype of the resulting strains on LB plates at 37°C, under inducing and noninducing conditions. Under inducing conditions, L20Δ1 and L20Δ2 complemented the strain lacking rplT (Fig. 2A ▶, L20Δ1 and L20Δ2 sectors), although growth was somewhat reduced compared to that afforded by wild-type L20 (Fig. 2A ▶, L20 sector). However, without induction, i.e., when expressed at a limiting level, both deletions displayed severe complementation defects (Fig. 2B ▶, L20Δ1 and L20Δ2 sectors), compared to the wild type. These defects were suppressed by the production of wild-type L20 in trans from the compatible plasmid pSGL6 in which rplT was cloned without its cis-acting regulatory sequence (Fig. 2B ▶, L20Δ1 + L20 and L20Δ2 + L20 sectors). This was not the case if the N-terminal domain of L20 was produced alone in trans from the compatible plasmid pSUrplTW61Stop (Fig. 2B ▶, L20Δ1 + L20N and L20Δ2 + L20N sectors). Finally, substitution of the resident plasmid pSGL6 by plasmid pBL20ecΔN, which expresses the L20ΔN truncation, was not possible (data not shown), even under inducing conditions. Thus, the N-terminal domain of L20 is essential for cell growth, but fulfills this function only when covalently linked to the C-terminal domain of the protein. Expression of L20ΔN under inducing conditions conferred a dominant-lethal phenotype on the E. coli strain IBPC5311, which contains the wild-type chromosomal copy of rplT (Fig. 2C ▶, L20ΔN sector). This result is fully consistent with the essential role of the N-terminal domain of L20 in sustaining cell growth. The dominant-lethal phenotype conferred by L20ΔN is most likely due to its ability to inhibit the expression of the wild-type chromosomal rplT gene (see below), combined with its incapacity to sustain cell growth. Indeed, viability was restored when wild-type L20 is provided in trans from the compatible plasmid pSGL6, constitutively expressing L20 from the lac promoter (Fig. 2C ▶, L20ΔN + L20 sector). This was not the case when the L20 N-terminal domain alone was overproduced in trans from the compatible plasmid pSUrplTW61Stop (Fig. 2C ▶, L20ΔN + L20N sector), thus indicating that elimination of the dominant-lethal phenotype requires the full-length L20 protein.
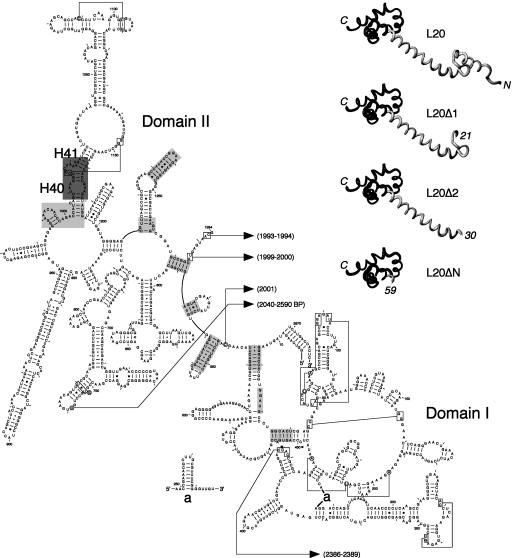
FIGURE 1.
Structure of the ribosome-bound form of L20 and its interactions with domains I and II of 23S rRNA. (Left) Secondary structure diagram of domains I and II of Deinococcus radiodurans 23S rRNA (available online at http://www.rna.icmb.utexas.edu). Domains I (nucleotides 15–535) and II (588–1274) were defined according to the secondary structure model for E. coli 23S rRNA (Noller et al. 1981). Helices are numbered according to the helix numbering of E. coli 23S rRNA (Yusupov et al. 2001). The regions interacting with the N-terminal and C-terminal domains of L20 are shown in light and dark gray backgrounds, respectively. The L20-contacting regions have been determined using the RasMol program (Sayle and Milner-White 1995) for rendering of the coordinates for the molecular structure of the D. radiodurans large ribosomal subunit (Harms et al. 2001) deposited at the Protein Data Bank under PDB accession number 1NKW. L20-contacting regions have been determined using a probing distance of 10 Å around the Cα backbone of the protein. Helices H40 and H41 containing the L20 C-terminal domain-binding site in 23S rRNA are indicated. (Upper right) Structure of the ribosome-bound D. radiodurans L20 (Harms et al. 2001) and structural models of the truncated forms of L20 (L20Δ1, L20Δ2, and L20ΔN) used in this study. The L20Δ1, L20Δ2 and L20ΔN truncations were constructed by deleting amino acid residues 6–20, 6–29, and 6–58, respectively. Amino acid residues 1–5 of L20 are present in the three deleted forms, so that the translation initiation efficiency is assumed to be the same as that of the wild-type L20. The structural models were drawn using the MolMol program (Koradi et al. 1996). The N- and C-terminal domains of L20 are in gray and black, respectively.
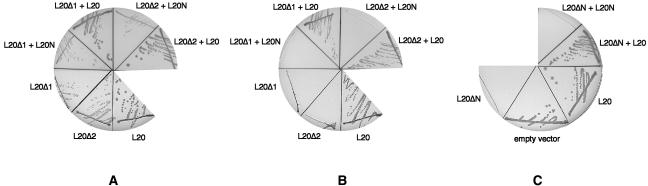
FIGURE 2.
Analysis of the growth phenotypes conferred by the L20Δ1, L20Δ2, and L20ΔN truncations. (A,B) L20Δ1 and L20Δ2 display complementation defects. The E. coli ΔrplT strain IBPC6801 was transformed with either plasmid pBL20ec, expressing wild-type L20 (L20 sector) or its derivatives pBL20ecΔ6–20 (L20Δ1 sector) or pBL20ecΔ6–29 (L20Δ2 sector) expressing the L20Δ1 and L20Δ2 truncations, respectively. The pBL20ecΔ6–20 and pBL20ecΔ6–29 transformants were cotransformed with the compatible plasmids pSGL6 expressing wild-type L20 (L20Δ1 + L20 and L20Δ2 + L20 sectors) or pSUrplTW61Stop expressing the N-terminal domain of L20 (L20Δ1 + L20N and L20Δ2 + L20N sectors). Growth was followed at 37°C upon induction of the expression of the different rplT alleles with 1 mM IPTG (A) and in noninducing conditions (B). (C) Overproduction of L20ΔN results in a dominant-lethal phenotype. The E. coli IBPC5311[λMQ21ΔNB] lysogen was transformed with either pBR322 (empty vector sector) or with plasmids pBL20ec (L20 sector) or its derivative pBL20ecΔN expressing the L20ΔN truncation (L20ΔN sector). The pBL20ecΔN transformant was cotransformed with the compatible plasmids pSGL6 expressing wild-type L20 (L20ΔN + L20 sector) or pSUrplTW61Stop expressing the N-terminal domain of L20 (L20ΔN + L20N sector). Induction was carried out with 1 mM IPTG.
Growth of the E. coli wild-type strain IBPC5421 and its ΔrplT derivative IBPC6801 transformed with the tester plasmids pBL20ec, pBL20ecΔ6–20, and pBL20ecΔ6–29 was compared in LB liquid medium at 30°C, under noninducing and inducing conditions. The doubling time of the wild-type strain was 55 min under both conditions. In contrast, the ΔrplT strain transformed with either of the tester plasmids displayed much slower growth rates under noninducing conditions; doubling times were 150 min, 215 min, and 235 min for the pBL20ec, pBL20ecΔ6–20, and pBL20ecΔ6–29 transformants, respectively. It is likely that the pBL20ecΔ6–20 and pBL20ecΔ6–29 transformants display some growth in liquid medium under noninducing conditions due to leakiness of the lac promoter, allowing residual synthesis of the truncated proteins. Indeed, when we analyzed the accumulation of both the L20Δ1 and L20Δ2 truncations and compared it to that of the wild-type L20 by SDS-PAGE followed by Western blotting analysis using polyclonal antibodies to the L20ΔN truncation (Fig. 3 ▶), we found that nonnegligible amounts of the L20 truncations were produced under noninducing conditions. In addition, in contrast to the phenotype afforded by plasmid pBL20ec (Fig. 2B ▶, L20 sector), no growth was observed on LB plates under noninducing conditions when we used a derivative of plasmid pBAD30 (Guzman et al. 1995) carrying wild-type rplT under control of the tightly regulated arabinose pBAD promoter (data not shown). Under inducing conditions, the pBL20ec-transformed ΔrplT strain had a doubling time of 65 min, which is quite similar to that of the wild-type strain. In contrast to the pBL20ec transformant, the pBL20ecΔ6–20 and pBL20ecΔ6–29 transformants displayed a growth defect under inducing conditions; their doubling times were 90 min and 100 min, respectively. To rule out the possibility that the slow-growth phenotype exhibited by the pBL20ecΔ6–20 and pBL20Δ6–29 transformants is due to instability instead of lack of activity of the L20Δ1 and L20Δ2 truncations, we analyzed the accumulation of both truncations under noninducing and inducing conditions and compared it to that of the wild-type L20 as described above (Fig. 3 ▶). As an internal standard, we also used polyclonal antibodies to the PTH protein, the level of which should remain unchanged in uninduced and induced cells. Our results showed that, under both conditions, the L20 truncations accumulate to levels similar to that of the wild-type L20. Therefore, the slow-growth phenotype observed in liquid medium concurs with the results obtained on LB plates. Furthermore, our results from Western blot analysis indicate that the slow-growth phenotype caused by the L20Δ1 and L20Δ2 truncations is not due to some protein instability but most likely to the fact that the N-terminal domain of L20 by itself is crucial for cell growth.
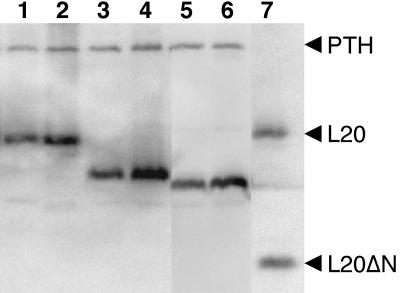
FIGURE 3.
Accumulation of wild-type L20 and of L20Δ1 and L20Δ2 truncations. Total extracts from uninduced (lanes 1,3,5) and induced (lanes 2,4,6) E. coli strain IBPC6801 transformed either with plasmid pBL20ec, expressing wild-type L20 (lanes 1,2) or with plasmids pBL20ecΔ6–20 (lanes 3,4) and pBL20ecΔ6–29 (lanes 5,6), expressing the L20Δ1 and L20Δ2 truncations, respectively, were fractionated on a 17.5% SDS-PAGE gel and subjected to Western blotting analysis using polyclonal antibodies raised against the L20ΔN truncation and PTH protein. L20 and L20ΔN truncation (lane 7) were used as size markers.
Truncations in the N-terminal domain of L20 produce severe defects in ribosome biogenesis
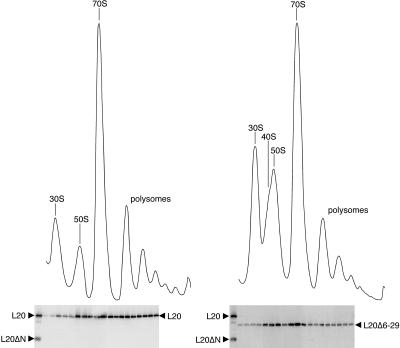
We next investigated the effect of the L20 N-terminal truncations on ribosome biogenesis. The wild-type E. coli strain IBPC5421 and its ΔrplT derivative IBPC6801 transformed with the three tester plasmids (see above) were grown in LB liquid medium at 30°C, under noninducing and inducing conditions. Polysomes were produced from whole-cell lysates and ribosomal particles were then separated on sucrose gradients under conditions ensuring ribosome association. Under both noninducing and inducing conditions, the wild-type strain produced ribosomal particles sedimenting essentially as polysomes or 70S ribosomes with only small amounts of free 30S and 50S particles (Fig. 4 ▶). Under noninducing conditions, the polysome profile was substantially modified with the pBL20ec transformant and even more dramatically altered with the pBL20ecΔ6–20 and pBL20ecΔ6–29 transformants (Fig. 4 ▶). The pBL20ec transformant polysome profile showed an increased proportion of free 30S ribosomal subunits and a relatively reduced amount of polysomes. A significant reduction in the ratio of the 50S to the 30S absorption peaks was correlated with the appearance of a new absorption peak between the 30S and 50S peaks, corresponding to particles sedimenting with an intermediate coefficient of about 40S. This indicates that limiting the amount of L20 inhibits the biogenesis of the 50S ribosomal subunit per se. Interestingly, the defects observed in the polysome profile of the pBL20ec transformant were enhanced in the profiles of the pBL20ecΔ6–20 and pBL20ecΔ6–29 transformants. The proportions of free 30S particles and 70S ribosomes were similar and the polysome absorption peaks were greatly reduced for the pBL20ecΔ6–20 transformant and practically disappeared for the pBL20ecΔ6–29 transformant. With both of these mutant transformants, the ratio of the 50S to the 30S absorption peaks was further reduced and the proportion of the 40S particles increased. In contrast, under inducing conditions, the pBL20ec transformant polysome profile was quite similar to that of the wild-type strain whereas the profiles of the pBL20ecΔ6–20 and pBL20ecΔ6–29 transformants showed increased ratios of the 70S and polysome peaks to the 30S absorption peak compared to that observed under noninducing conditions (Fig. 4 ▶). Nevertheless, it is worth noting that under inducing conditions the proportions of free 30S and 50S particles with both mutant transformants were significantly higher than that observed with the wild-type strain or the pBL20ec transformant. Furthermore, the 40S particles were still present, seen as a shoulder on the left side of the peak containing particles that display an apparent sedimentation coefficient of 50S. Taken together, our data reveal a close correlation between the accumulation of incomplete 40S ribosomal particles due to either limited cellular amounts of L20 or the L20 truncations and the slow-growth phenotype observed on both LB plates and liquid LB medium.
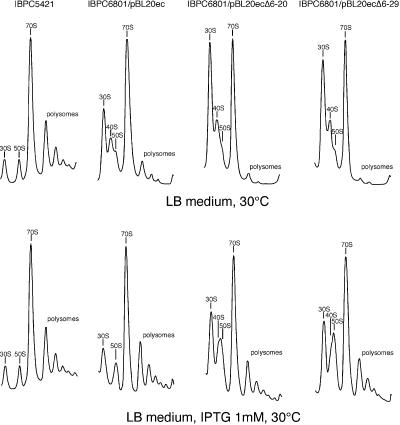
FIGURE 4.
The defects in 50S ribosomal subunit assembly caused by limited amounts of L20 are enhanced by truncations in its N-terminal tail. Sucrose gradient analysis of ribosomal particles produced from E. coli wild-type strain IBPC5421 and ΔrplT strain IBPC6801 (IBPC5421 background) transformed with the indicated plasmids. Cells were grown in LB medium at 30°C without or with 1 mM IPTG. Absorption profiles were recorded at 254 nm. The absorption peaks of free 30S and 50S ribosomal subunits, 40S ribosomal particles, 70S ribosomes including free couples and monosomes, and polysomes are indicated.
The L20Δ2 truncation is assembled into 50S ribosomal particles and is found in 70S ribosomes and in polysomes
Under inducing conditions, the L20Δ2 truncation caused an accumulation of incomplete 40S ribosomal particles. We investigated whether this truncated L20 could be found in the 50S ribosomal particles and 70S ribosomal couples and in the polysomes. After growth of the pBL20ec and pBL20ecΔ6–29 transformants expressing the wild-type L20 and the L20Δ2 truncation in inducing conditions in LB medium at 30°C, crude ribosomes were prepared and sedimented through high-magnesium (10 mM) sucrose gradients. Proteins from individual fractions spanning the gradient region extending from the peak of 30S ribosomal particles to the polysome peaks were separated by SDS-PAGE and their L20 content was analyzed by Western blotting using polyclonal antibodies raised against the L20ΔN truncation (Fig. 5 ▶). The L20Δ2 truncation was readily detected in the fractions containing the incomplete 40S and the 50S ribosomal particles as well as in the fractions containing the 70S ribosomal couples. These results indicate (1) that, under inducing conditions, a deletion removing part of the N-terminal tail of L20 still allows the truncated L20 to be assembled into 50S ribosomal particles having an apparent 50S sedimentation coefficient and (2) that these truncated L20-containing 50S particles are still capable of interacting with 30S particles to form 70S ribosomal couples. A detectable amount of L20Δ2 truncation was also found in the polysome fractions of the gradient. Compared with the distribution of the wild-type L20 (Fig. 5 ▶), however, the L20Δ2 truncation appeared to enrich the 50S and 70S particle-containing fractions of the gradient relative to the polysome fractions. These results suggest that the ability of the truncated L20 to stay assembled in the 50S particles in a polysome context is partially impaired by deletion of part of the N-terminal tail of the protein.
FIGURE 5.
L20 content of the ribosomal particles prepared from the IBPB6801/pBL20ec (left) and IBPC6801/pBL20ecΔ6–29 (right) strains, expressing wild-type L20 and the L20Δ2 truncation, respectively. Cells were grown in LB medium in the presence of 1 mM IPTG at 30°C. Ribosomal particle profiles were analyzed on sucrose gradients by monitoring the absorbance at 254 nm. Proteins in the collected fractions were fractionated on 17.5% SDS-PAGE gels and the L20 content was analyzed by Western blotting using polyclonal antibodies to the L20ΔN truncation. As an internal standard, the PTH protein was detected using the corresponding antibodies. Purified wild-type L20 and the L20ΔN truncation were used as size markers alongside the content of the collected fractions.
L20 N-terminal domain truncations do not affect autogenous control
We next compared the ability of the wild-type L20 and L20Δ1 and L20Δ2 truncations to repress the expression of an rpmI’-‘lacZ translational fusion integrated into the E. coli chromosome. The E. coli lysogenic strain IBPC5311[λMQ21ΔNB] was transformed either with plasmid pBR322 or with plasmid pBL6ec (see Materials and Methods) expressing wild-type L20 and its two derivatives pBL6ecΔ6–20 and pBL6ecΔ6–29 expressing the L20Δ1 and L20Δ2 truncations, respectively. The repression factors associated with wild-type L20, L20Δ1, and L20Δ2 were 62, 75, and 88, respectively (Table 1 ▶). This result clearly indicates that the proximal half of the N-terminal domain, at least up to amino acid residue 29, is dispensable for autogenous control.
TABLE 1.
L20Δ1 and L20Δ2 repress rpmI expression
| Overproduced L20 | β-Galactosidase activity (Miller units) | Repression factor |
| None | 102.8 | |
| Wild-type | 1.67 | 61.6 |
| L20Δ1 | 1.37 | 75.0 |
| L20Δ2 | 1.17 | 87.9 |
The E. coli IBPC5311[λMQ21ΔNB] lysogen, carrying an rpml’-‘lacZ translational fusion, was transformed either with plasmid pBR322 or with plasmid pBL6ec expressing wild-type L20 and its two derivatives pBL6ecΔ6-20 and pBL6ecΔ6-29 expressing the L20Δ1 and L20Δ2 truncations, respectively. pBL6ec is a pBR322 derivative carrying rplT, which is constitutively expressed from a lac promoter and is not subjected to autogenous control. The repression factor is defined as the ratio of the β-galactosidase activity assayed in the presence of control plasmid pBR322 to the activity assayed in the presence of wild-type L20- or truncated L20-overproducing plasmid.
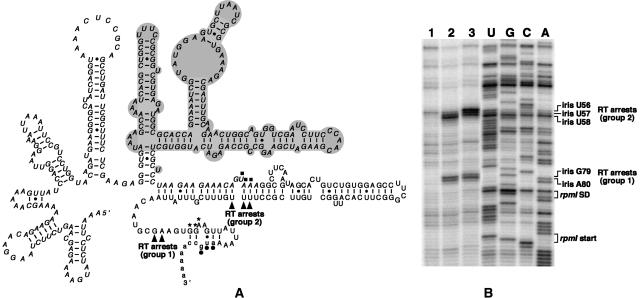
L20 and the L20ΔN truncation induce the same RT stops in the rpmI translational operator in vitro
As mentioned before, steady-state expression of the L20ΔN truncation is lethal to the cell, thus precluding any measure of autogenous control by assaying the β-galactosidase activity of an rpmI’-‘lacZ translational fusion in vivo. We thus decided to study the interaction between the rpmI translational operator and either wild-type L20 or the L20ΔN truncation by primer-extension assay of an in vitro synthesized transcript. Both proteins caused reverse transcriptase (RT) arrests at two regions within the infC-rpmI intergenic sequence (iris). A first group of RT arrests is located at nucleotides iris G79 and iris A80 (group 1 in Fig. 6 ▶) and a second group at nucleotides iris U57 and iris U58 (group 2 in Fig. 6 ▶). Although L20ΔN caused an additional RT arrest at the adjacent nucleotide, iris U56, both proteins clearly recognize the rpmI translational operator similarly in vitro. Our data are consistent with those provided by a previous pulse-chase labeling experiment performed in vivo, showing that L20ΔN is sufficient to repress the expression of rpmI (Raibaud et al. 2002).
FIGURE 6.
L20 and L20ΔN-induced RT arrests are located at similar positions on rpmI mRNA. (A) Secondary structure of the rpmI translational operator, i.e., the region upstream of rpmI mRNA containing the two L20-binding sites required for control (Chiaruttini et al. 1996). Nucleotide residues forming the infC stop codon and the rpmI Shine–Dalgarno (SD) sequence and start codon are indicated by filled squares, asterisks, and dots, respectively. The nucleotide sequences of infC, iris (iris is an acronym for the infC-rpmI intergenic sequence) and rpmI are in italics, uppercase and lowercase, respectively. The ΔApaLI deletion which does not affect control (Chiaruttini et al. 1996) is in gray background. Arrowheads indicate the two groups (groups 1 and 2) of RT arrests caused by L20 and L20ΔN. (B) Primer-extension assay. A shortened form of the operator in which nucleotide residues corresponding to the ΔApaLI deletion have been removed was used in this experiment. (Lane 1) rpmI operator alone; (lane 2) rpmI operator with a 30-fold molar excess of wild-type L20; (lane 3) rpmI operator with a 90-fold molar excess of L20ΔN. The positions of the L20- and L20ΔN-induced RT arrests and those of the nucleotide residues forming the rpmI SD sequence and start codon are indicated to the right of the autoradiogram.
DISCUSSION
More than one-third of the bacterial r-proteins possess loops or tails, often protruding from globular domains located on the ribosomal surface and penetrating deep into the ribosome by making extensive contacts with rRNA (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000; Harms et al. 2001; Brodersen et al. 2002). In most cases, the precise role of these RNA-binding extensions remains unknown. Because they are intertwined with rRNA, interacting with several regions of rRNA and, in some cases, with several of its domains, such extensions have been proposed to stabilize the correct tertiary structure of rRNA (Ban et al. 2000; Wimberly et al. 2000; Brodersen et al. 2002). In addition to playing a scaffolding role in building up and stabilizing rRNA structure, r-protein extensions probably also play an active role in ribosome function since mutations in some of them were shown to alter the structure and function of ribosomes (Gregory and Dahlberg 1999; Gabashvili et al. 2001; Nakatogawa and Ito 2002). A number of r-protein mutations lying in these RNA-binding extensions are known to confer resistance to antibiotics (Ban et al. 2000; Carter et al. 2000). For example, erythromycin resistance mutations mapping in some of the extended loops of r-proteins L4 and L22 have been shown to modulate the gating system of the ribosome polypeptide tunnel. Not surprisingly, r-protein extensions were shown to be crucial for biogenesis of the eukaryotic ribosome also, as mutations in the C-terminal extension of yeast r-protein S14 were reported to block the maturation of preribosomal 43S particles (Jakovljevic et al. 2004).
Here we analyzed the effects of L20 truncations on ribosome assembly and autogenous regulation in E. coli. Truncation is often a source of protein instability, thus making it difficult to assess the precise function that is impaired. Indeed, the cellular level of a truncated protein could be so low that it necessarily produces deleterious effects on the cell. Therefore, a slow-growth phenotype caused by a truncated r-protein could be due to instability rather than to an inability to assemble into a ribosomal particle. The effects of the L20 truncations seen here are unlikely to be attributable to instability. First, both the L20Δ1 and L20Δ2 truncations repress the expression of a rpmI’-‘lacZ fusion as efficiently as the wild-type L20 protein in vivo. Second, both truncations are expressed to levels similar to that of the wild-type L20 as they can be readily detected on protein gels.
Does the complementation defect observed with the N-terminally truncated forms of L20 reflect an alteration in the function or the assembly of the ribosome? We believe that the complementation defect is explained by altered assembly since there is a perfect correlation between phenotype and assembly defect: The slower the growth rate, the stronger the assembly defect. For instance, the relatively slow growth rate observed with the pBL20ecΔ6–20 or pBL20ecΔ6–29 transformants compared to that of the pBL20ec transformant under induced conditions correlates with a moderate assembly defect, whereas the much slower growth with the truncated versions of L20 under noninducing conditions reflects a strong assembly defect. In addition, in vitro data from Nierhaus’s laboratory has shown directly that L20 is not involved in ribosome function (Nowotny and Nierhaus 1980). Is the extended N-terminal domain of L20 crucial for ribosome assembly? Despite the deleterious effect of the L20Δ2 truncation on cell growth, it can assemble into ribosomal particles having an apparent sedimentation coefficient of 50S. This is probably due to the fact that a sufficient number of contacts with 23S rRNA have been maintained, thus permitting assembly to go further, albeit at a reduced rate. However, it should be noted that the L20ΔN truncation that is devoid of almost all the L20 N-terminal domain cannot substitute for L20 expressed from a plasmid carrying rplT in an E. coli strain in which rplT has been deleted. Moreover, the L20ΔN truncation produces a dominant-lethal phenotype in a merodiploid strain, which could be due to its inability to ensure the assembly of functional ribosomes in combination with its ability to repress the expression of the chromosomal wild-type rplT. We thus hypothesize that the N-terminal extension of E. coli r-protein L20 could be crucial for ribosome assembly, while it is clearly not required for autogenous feedback regulation. We indeed demonstrated that the N-terminal domain at least up to amino acid residue 29 is not required for autogenous control. This conclusion concurs with a pulse-chase labeling experiment showing that the L20ΔN truncation, corresponding to the deletion of amino acid residues 6–58, was able to repress the expression of an rpmI’-‘lacZ translational fusion in vivo (Raibaud et al. 2002).
Our results differ from those obtained with truncations of r-proteins L4 and L22, where extended loops are apparently required neither for ribosome assembly nor for L4-mediated translational control (Zengel et al. 2003). This was a quite surprising result since prior binding of “early-binding” r-proteins such as L4 and L22 to 23S rRNA was shown to be a prerequisite for binding of other r-proteins during ribosome assembly in reconstitution experiments in vitro (Herold and Nierhaus 1987; Stelzl et al. 2000). It is likely that L4 and L22 have a scaffolding function in the assembly of the particle as they make extensive contacts with multiple domains in 23S rRNA as revealed from the atomic structure of the prokaryotic ribosome (Ban et al. 2000; Harms et al. 2001). Besides their purely architectural role in the folding of 23S rRNA and thus in ribosome assembly, these extensions have also been shown to play a more functional role as they contribute to a constriction of the peptide exit channel and are thus ideally located to participate in the regulation of the translation rate and in the gating of the ribosome polypeptide tunnel (Nissen et al. 2000; Gabashvili et al. 2001). In addition, truncations in the extended loops of both L4 and L22 included the tip portions, carrying erythromycin-resistance mutations that produce significant changes in the morphology of the 50S ribosomal subunit (Gabashvili et al. 2001). Recently, other examples of r-protein extensions that are apparently not required for assembly and/or function have been reported also in the cases of S9 and S13 (Hoang et al. 2004).
In conclusion, our results indicate that, at least in some cases, r-protein extensions are important for ribosome assembly. The role played by L20 is not surprising as the atomic structure of the mature 50S particle revealed that the N-terminal tail of the protein makes extensive contacts with domains I and II of 23S rRNA (Harms et al. 2001). The deleterious effect afforded by truncations within its extended N-terminal domain is most likely due to the loss of certain rRNA–L20 contacts, which makes it difficult for 23S rRNA to fold at the proper rate into the tertiary structure required for 50S subunit assembly, i.e., assembly of the “late-binding” r-proteins. Further analysis of the protein content of the intermediate 40S particle that accumulates with the L20Δ1 and L20Δ2 truncations will tell us more about which assembly step requires the specific contacts provided by the N-terminal tail of L20.
MATERIALS AND METHODS
Strains and plasmids
The E. coli strains IBPC5311 Δlac, IBPC5421 lacZ (non polar), and IBPC5311[λMQ21ΔNB] have been described previously (Springer et al. 1985; Plumbridge and Söll 1989; Lesage et al. 1992). Unless otherwise stated, all the mutations and deletions introduced into the plasmids used in this study were engineered by PCR using the QuickChange site-directed mutagenesis protocol from Stratagene.
Plasmid pNIBLE was used to construct the E. coli strain IBPC6801 ΔrplT. This plasmid was constructed as follows. First, plasmid pTI5.1 was constructed by cloning the 2.98-kb DraI-DraI fragment of pB5–57 (Plumbridge et al. 1980) containing the 3′ region of thrS, infC, rpmI, rplT, and the 5′ region of pheS into the EcoRV site of pBlueScript SK(+) (Stratagene). The 1.44-kb HincII–HincII fragment of plasmid pUC-4K (Amersham Biosciences) containing a kanamycin cassette was then cloned between the PmlI and EcoRV sites of pTI5.1, thus giving plasmid pTI5.1ΔrplT.1 in which about 70% of the rplT sequences have been deleted and replaced with a DNA fragment containing the kanamycin cassette. Finally, the 2.02-kb SstII/NheI fragment of pTI5.1ΔrplT.1 containing the 3′ region of infC, rpmI, the disrupted allele of rplT gene, and the 5′ region of pheS was used to replace the corresponding SstII/NheI fragment in plasmid pMAK5–57 (Sacerdot et al. 1999), a derivative of plasmid pMAK705 (Hamilton et al. 1989) that contains the 3′ region of thrS, infC, rpmI, rplT, and the 5′ region of pheS. The resulting plasmid, named pNIBLE, carries a temperature-sensitive pSC101 origin of replication, the choramphenicol and the kanamycin resistance markers, the 3′ region of thrS, infC, rpmI, the disrupted rplT gene, and the 5′ region of pheS. Plasmid pSGL6 was used as a resident plasmid to complement the chromosomal disrupted rplT gene. It was constructed by cloning rplT, borne by the 0.79-kb NheI–XhoI fragment of plasmid pBL6 (Lesage et al. 1992) between the XbaI-XhoI sites of the counterselectable plasmid pSG335 (Geissler and Drummond 1993). It carries rplT under the control of a lac promoter, the resistance gene for chloramphenicol, and the sacB marker from Bacillus subtilis, which is lethal to E. coli when grown in the presence of sucrose. Plasmids pBL20ec, pBL20ecΔ6–20, pBL20ec6–29, pBL20ecΔN, and pSUrplTW61Stop used in analysis of growth phenotype were constructed as follows. Plasmid pBL20ec expressing E. coli L20 was constructed in two steps. First, pBL6lacIqΔ was constructed by inserting the 1.4-kb DraI–AseI fragment of pAIQ7 (M. Springer, unpubl.) carrying most of the lacI gene between the same sites in pBL6. Although it carries a truncated form of lacI, pBL6lacIqΔ expresses a lac repressor lacking the 13 C-terminal residues that is still able to repress E. coli rplT expression and therefore expresses E. coli L20 upon induction by IPTG. Plasmid pBL20ec was then constructed by creating a unique BsrGI site immediately downstream of the stop codon of E. coli rplT carried by pBL6lacIqΔ. Plasmids pBL20ecΔ6–20, pBL20ec6–29, and pBL20ecΔN were constructed by deleting nucleotides 16–60, 16–87, and 16–174, respectively, of rplT carried by pBL20ec. Plasmid pSUrplTW61Stop was constructed by cloning the XbaI–SmaI fragment of plasmid pBL6W61Stop, carrying an rplT gene harboring an amber codon from positions 181 to 183 into the same sites of the pACYC184-derived pSU2719 vector (Martinez et al. 1988). Plasmid pBL6W61Stop resulted from hydroxylamine mutagenesis of pBL6 and subsequent screening for derepressed expression of a rpmI’-‘lacZ translational fusion present in the IBPC5311[λMQ21ΔNB] lysogen transformed with the mutagenized plasmid.
Plasmids pBL6ec, pBL6ecΔ6–20, and pBL6ecΔ6–29 used for in vivo regulatory studies were constructed as follows. Plasmid pBL6ec was obtained by creating a unique BsrGI site immediately downstream of rplT carried by plasmid pBL6. Plasmids pBL6ecΔ6–20 and pBL6ecΔ6–29 were constructed by cloning the XbaI–BsrGI fragment carrying the truncated versions of rplT from pBL20ecΔ6–20 and pBL20ec6–29 into the same sites of pBL6ec.
The E. coli strain IBPC6801 ΔrplT was constructed according to a temperature-induced recombination method (Hamilton et al. 1989). Recombination was carried out between homologous sequences present in both the chromosome of strain IBPC5421 and plasmid pNIBLE. IBPC5421 was first transformed with pNIBLE and insertion of the plasmid into the chromosome was carried out at 44°C. Either a recombinant rplT-carrying pNIBLE derivative or pNIBLE was then excised from the chromosome by growing the cells at 30°C. Recombination of the deleted allele of rplT into the chromosome was verified by analyzing the structure of the excised plasmid with the appropriate restriction enzymes and by comparing the PCR products generated from the selected candidates with those generated from the starting strain using the appropriate oligodeoxyribonucleotide primers. The selected strain IBPC6801 ΔrplT containing the excised rplT-carrying pNIBLE derivative was then transformed with plasmid pSGL6 and the transformants were grown at nonpermissive temperature to cure the excised pNIBLE derivative carrying the wild-type allele of rplT. Loss of the excised plasmid was verified by comparing the PCR products generated from the selected transformants with those generated from the transformant containing both pSGL6 and the excised pNIBLE derivative using the appropriate oligodeoxyribonucleotide primers.
Replacement of pSGL6 in strain IBPC6801 ΔrplT with each of the plasmids, pBL20ec, pBL20ecΔN, pBL20ecΔ6–20, and pBL20ecΔ6–29, was carried out by first growing the double transformants in liquid LB medium containing the appropriate antibiotic and IPTG at various concentrations and then plating on LB plates containing sucrose, the appropriate antibiotic, and IPTG at the same concentrations.
Analysis of growth phenotype
The E. coli strain IBPC6801 ΔrplT transformed with the plasmids indicated (see legend to Fig. 2 ▶) was grown at 37°C on LB plates containing 1 mM or lacking IPTG. Growth was checked after 24 and 48 h incubation.
Polysome profile analysis
The E. coli wild-type strain IBPC5421 and strain IBPC6801 ΔrplT transformed with either pBL20ec or pBL20ecΔ6–20 or pBL20ecΔ6–29 were cultured in LB medium at 30°C without or with 1 mM IPTG. At OD650 = 0.5, chloramphenicol was added to a final concentration of 100 μg/mL and incubation was allowed to proceed for various times as a function of the doubling time of the strain. Chloramphenicol treatment was for 5 min with the wild-type strain and scaled up for the transformants according to the ratio of their doubling times to that of the wild-type strain. Polysomes were prepared from whole-cell lysates and analyzed essentially as described (Charollais et al. 2003). For each strain, eight OD260 units were layered on top of 10%–40% sucrose gradients and centrifuged at 35,000 rpm for 4 h at 4°C in a Beckman SW41 rotor.
Protein gel electrophoresis and Western blotting analysis
Crude cell extracts or proteins from collected fractions of the sucrose gradients were fractionated by 17.5% SDS-PAGE (Laemmli 1970) and then electroblotted to a Hybond-C super nitrocellulose membrane (Amersham Biosciences) according to standard procedures. In both cases, purified wild-type L20 and L20ΔN truncation were used as size markers. For probing wild-type L20 and the L20Δ2 truncation, the membrane was incubated with a polyclonal rabbit antiserum specific for the L20ΔN truncation (diluted 1:5000). When crude cell extracts were analyzed, we also added a polyclonal rabbit antiserum raised against the PTH protein (diluted 1:5000). Antigen–antibody complexes were detected using 125I-labeled protein A (Amersham Biosciences) and scanning the membrane on a PhosphorImager (Molecular Dynamics).
In vivo regulatory studies
Plasmid pBL6ec expressing wild-type L20 and plasmids pBL6ecΔ6–20 and pBL6ecΔ6–29 expressing the L20Δ1 or L20Δ2 truncations, respectively, were transformed into the E. coli lysogenic strain IBPC5311[λMQ21ΔNB] carrying an rpmI’-‘lacZ translational fusion on the chromosome. Expression of the fusion in the presence of the different versions of L20 was monitored by measuring the corresponding β-galactosidase activities as described previously (Lesage et al. 1992).
L20-induced primer extension arrest
The interaction between transcript pOTΔApaLI, a shortened but functional version of the translational operator of rpmI mRNA, and L20 or its truncated version was assessed in vitro using a primer-extension assay, as described previously (Guillier et al. 2002).
Acknowledgments
We are indebted to C. Condon and I. Iost for critical reading of the manuscript. We are grateful to Y. Timsit for fruitful discussions. We thank V. Heurgué-Hamard (Institut de Biologie Physico-Chimique, UPR 9073 du CNRS) for the kind gift of antibodies to the PTH protein. This work was supported by the CNRS (UPR 9073), the University of Paris VII, and the French Ministry of Research (ACI Dynamique et Réactivité des Assemblages Biologiques). M.G. and S.R. were recipients of studentships from the French Ministry of Research.
Abbreviations
IF3, translation initiation factor 3
IPTG, isopropyl-β-D-thiogalactopyranoside
PTH, peptidyl-tRNA hydrolase
RT, reverse transcriptase
SDS-PAGE, SDS-polyacrylamide gel electrophoresis.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7134305.
REFERENCES
- Agalarov, S.C., Sridhar Prasad, G., Funke, P.M., Stout, C.D., and Williamson, J.R. 2000. Structure of the S15,S6,S18-rRNA complex: Assembly of the 30S ribosome central domain. Science 288: 107–113. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Brodersen, D.E., Clemons Jr., W.M., Carter, A.P., Wimberly, B.T., and Ramakrishnan, V. 2002. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 316: 725–768. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., and Ramakrishnan, V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348. [DOI] [PubMed] [Google Scholar]
- Charollais, J., Pflieger, D., Vinh, J., Dreyfus, M., and Iost, I. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 48: 1253–1265. [DOI] [PubMed] [Google Scholar]
- Chiaruttini, C., Milet, M., and Springer, M. 1996. A long-range RNA–RNA interaction forms a pseudoknot required for translational control of the IF3-L35-L20 ribosomal protein operon in Escherichia coli. EMBO J. 15: 4402–4413. [PMC free article] [PubMed] [Google Scholar]
- Franceschi, F.J. and Nierhaus, K.H. 1988. Ribosomal protein L20 can replace the assembly-initiator protein L24 at low temperatures. Biochemistry 27: 7056–7059. [DOI] [PubMed] [Google Scholar]
- Gabashvili, I.S., Gregory, S.T., Valle, M., Grassucci, R., Worbs, M., Wahl, M.C., Dahlberg, A.E., and Frank, J. 2001. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell 8: 181–188. [DOI] [PubMed] [Google Scholar]
- Gao, H., Sengupta, J., Valle, M., Korostelev, A., Eswar, N., Stagg, S.M., Van Roey, P., Agrawal, R.K., Harvey, S.C., Sali, A., et al. 2003. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113: 789–801. [DOI] [PubMed] [Google Scholar]
- Geissler, S. and Drummond, M. 1993. A counterselectable pACYC184-based lacZa-complementing plasmid vector with novel multiple cloning sites; construction of chromosomal deletions in Klebsiella pneumoniae. Gene 136: 253–255. [DOI] [PubMed] [Google Scholar]
- Gregory, S.T. and Dahlberg, A.E. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J. Mol. Biol. 289: 827–834. [DOI] [PubMed] [Google Scholar]
- Guillier, M., Allemand, F., Raibaud, S., Dardel, F., Springer, M., and Chiaruttini, C. 2002. Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: A possible case of ribosomal RNA–messenger RNA molecular mimicry. RNA 8: 878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier, M, Allemand, F., Dardel, F., Royer, C.A., Springer, M, and Chiaruttini, C. 2005. Double molecular mimicry in Escherichia coli: Binding of ribosomal protein L20 to its two sites in mRNA is similar to its binding to 23S rRNA. Mol. Microbiol. (in press). [DOI] [PubMed]
- Guzman, L.M., Belin, D., Carson, M.J., and Beckwith, J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, C.M., Aldea, M., Washburn, B.K., Babitzke, P., and Kushner, S.R. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171: 4617–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688. [DOI] [PubMed] [Google Scholar]
- Herold, M. and Nierhaus, K.H. 1987. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem. 262: 8826–8833. [PubMed] [Google Scholar]
- Hoang, L., Fredrick, K., and Noller, H.F. 2004. Creating ribosomes with an all-RNA 30S subunit P site. Proc. Natl. Acad. Sci. 101: 12439–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic, J., De Mayolo, P.A., Miles, T.D., Nguyen, T.M., Leger-Silvestre, I., Gas, N., Woolford Jr., J.L. 2004. The carboxyterminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell 14: 331–342. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MolMol: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14: 51–55. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lesage, P., Truong, H.-N., Graffe, M., Dondon, J., and Springer, M. 1990. Translated translational operator in Escherichia coli. J. Mol. Biol. 213: 465–475. [DOI] [PubMed] [Google Scholar]
- Lesage, P., Chiaruttini, C., Graffe, M., Dondon, J., Milet, M., and Springer, M. 1992. Messenger RNA secondary structure and translational coupling in the Escherichia coli operon encoding translation initiation factor IF3 and the ribosomal proteins, L35 and L20. J. Mol. Biol. 228: 366–386. [DOI] [PubMed] [Google Scholar]
- Martinez, E., Bartolome, B., and de la Cruz, F. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68: 159–162. [DOI] [PubMed] [Google Scholar]
- Nakatogawa, H. and Ito, K. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108: 629–636. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Noller, H.F., Kop, J., Wheaton, V., Brosius, J., Gutell, R.R., Kopylov, A.M., Dohme, F., Herr, W., Stahl, D.A., Gupta, R., et al. 1981. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 9: 6167–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny, V. and Nierhaus, K.H. 1980. Protein L20 from the large subunit of Escherichia coli ribosomes is an assembly protein. J. Mol. Biol. 137: 391–399. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons Jr., W.M., Tarry, M.J., Carter, A.P., and Ramakrishnan, V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., and Ramakrishnan, V. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732. [DOI] [PubMed] [Google Scholar]
- Plumbridge, J. and Söll, D. 1989. Characterization of cis-acting mutations which increase expression of a glnS-lacZ fusion in E. coli. Mol. Gen. Genet. 216: 113–119. [DOI] [PubMed] [Google Scholar]
- Plumbridge, J.A., Springer, M., Graffe, M., Goursot, R., and Grunberg-Manago, M. 1980. Physical localisation and cloning of the structural gene for E. coli initiation factor IF3 from a group of genes concerned with translation. Gene 11: 33–42. [DOI] [PubMed] [Google Scholar]
- Raibaud, S., Lebars, I., Guillier, M., Chiaruttini, C., Bontems, F., Rak, A., Garber, M., Allemand, F., Springer, M., and Dardel, F. 2002. NMR structure of bacterial ribosomal protein l20: Implications for ribosome assembly and translational control. J. Mol. Biol. 323: 143–151. [DOI] [PubMed] [Google Scholar]
- Sacerdot, C., de Cock, E., Engst, K., Graffe, M., Dardel, F., and Springer, M. 1999. Mutations that alter initiation codon discrimination by Escherichia coli initiation factor IF3. J. Mol. Biol. 288: 803–810. [DOI] [PubMed] [Google Scholar]
- Sayle, R.A. and Milner-White, E.J. 1995. RASMOL: Biomolecular graphics for all. Trends Biochem. Sci. 20: 374. [DOI] [PubMed] [Google Scholar]
- Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F., et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623. [DOI] [PubMed] [Google Scholar]
- Spillmann, S., Dohme, F., and Nierhaus, K.H. 1977. Assembly in vitro of the 50S subunit from Escherichia coli ribosomes: Proteins essential for the first heat-dependent conformational change. J. Mol. Biol. 115: 513–523. [DOI] [PubMed] [Google Scholar]
- Springer, M., Plumbridge, J.A., Butler, J.S., Graffe, M., Dondon, J., Mayaux, J.F., Fayat, G., Lestienne, P., Blanquet, S., and Grunberg-Manago, M. 1985. Autogenous control of Escherichia coli threonyl-tRNA synthetase expression in vivo. J. Mol. Biol. 185: 93–104. [DOI] [PubMed] [Google Scholar]
- Stelzl, U., Spahn, C.M., and Nierhaus, K.H. 2000. Selecting rRNA binding sites for the ribosomal proteins L4 and L6 from randomly fragmented rRNA: Application of a method called SERF. Proc. Natl. Acad. Sci. 97: 4597–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons Jr., W.M., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A, Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]
- Zengel, J.M., Jerauld, A., Walker, A, Wahl, M.C., and Lindahl, L. 2003. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA 9: 1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]