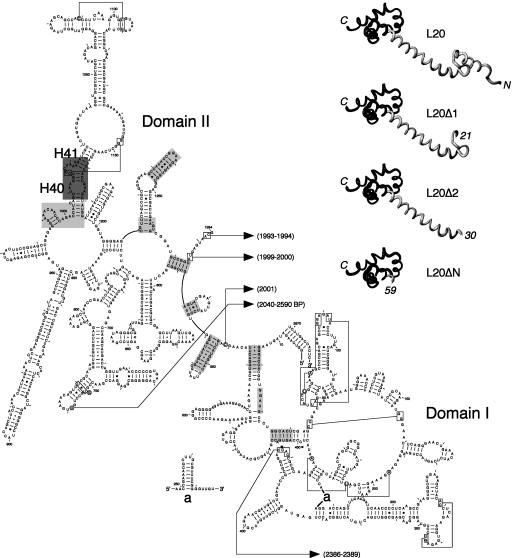
FIGURE 1.
Structure of the ribosome-bound form of L20 and its interactions with domains I and II of 23S rRNA. (Left) Secondary structure diagram of domains I and II of Deinococcus radiodurans 23S rRNA (available online at http://www.rna.icmb.utexas.edu). Domains I (nucleotides 15–535) and II (588–1274) were defined according to the secondary structure model for E. coli 23S rRNA (Noller et al. 1981). Helices are numbered according to the helix numbering of E. coli 23S rRNA (Yusupov et al. 2001). The regions interacting with the N-terminal and C-terminal domains of L20 are shown in light and dark gray backgrounds, respectively. The L20-contacting regions have been determined using the RasMol program (Sayle and Milner-White 1995) for rendering of the coordinates for the molecular structure of the D. radiodurans large ribosomal subunit (Harms et al. 2001) deposited at the Protein Data Bank under PDB accession number 1NKW. L20-contacting regions have been determined using a probing distance of 10 Å around the Cα backbone of the protein. Helices H40 and H41 containing the L20 C-terminal domain-binding site in 23S rRNA are indicated. (Upper right) Structure of the ribosome-bound D. radiodurans L20 (Harms et al. 2001) and structural models of the truncated forms of L20 (L20Δ1, L20Δ2, and L20ΔN) used in this study. The L20Δ1, L20Δ2 and L20ΔN truncations were constructed by deleting amino acid residues 6–20, 6–29, and 6–58, respectively. Amino acid residues 1–5 of L20 are present in the three deleted forms, so that the translation initiation efficiency is assumed to be the same as that of the wild-type L20. The structural models were drawn using the MolMol program (Koradi et al. 1996). The N- and C-terminal domains of L20 are in gray and black, respectively.