Abstract
Trypanosoma brucei mitochondria possess a unique RNA decay pathway in which rapid degradation of polyadenylated mRNAs is dependent on the addition of UTP, as measured by in organello pulse chase assays. To determine the mechanism by which UTP stimulates the degradation of polyadenylated RNAs, we performed in organello pulse chase assays under different conditions. Treatment of mitochondria with proteinase K revealed that UTP does not act through a receptor on the surface of the mitochondria. To determine if the UTP-stimulated RNA decay pathway is triggered by the mitochondrial energy state or ATP:UTP ratio, increasing ATP was added to a constant amount of UTP during the chase period of the assay. Results indicate that rapid turnover is responsive to UTP and not the ATP:UTP ratio. Experiments using UTP analogs demonstrate that UTP polymerization into RNAs is necessary for UTP-dependent degradation. Furthermore, experiments performed with RNAi cells indicate that the RET1 terminal uridylyl transferase (TUTase) is required for UTP-dependent decay of polyadenylated RNAs. Overall, these results show that degradation of polyadenylated RNAs in T. brucei mitochondria can occur through a unique mechanism that requires the polymerization of UTP into RNAs, presumably by the RET1 TUTase.
Keywords: RNA stability, RNA decay, RNA editing, trypanosomes, polyadenylation
INTRODUCTION
RNA decay is an important aspect of gene expression in all systems. This process is less well understood in mitochondria, but the role of RNA stability in regulating mitochondrial gene expression has recently received increasing scrutiny. Studies encompassing multiple species have revealed interesting contrasts and parallels in mitochondrial mRNA decay (for review, see Gagliardi et al. 2004). First, the enzymes involved in mitochondrial mRNA decay differ across species. The mitochondrial degradosome (or mtEXO) was first discovered in the mitochondria of Saccharomyces cerevisiae and has been shown to play a variety of roles in RNA stability and processing (Min et al. 1993; Golik et al. 1995; Margossian et al. 1996; Dziembowski et al. 1998). The degradosome is a two-protein complex, composed of an exo-ribonuclease (DSS-1) and an RNA helicase (SUV3) (Dziembowski et al. 2003). Interestingly, the degradosome is not preserved across all species, as SUV3 homologs are present in plant and human mitochondria, but DSS-1 homologs are apparently absent in these species (Gagliardi et al. 2004). Second, mitochondrial systems differ dramatically in the role of polyadenylation in RNA stability. Polyadenylation of mRNAs has not been detected in yeast mitochondria. However, in plant mitochondria, polyadenylation acts to destabilize mRNAs (Gagliardi and Leaver 1999; Lupold et al. 1999; Kuhn et al. 2001), similar to its role in bacteria and chloroplasts (Schuster et al. 1999; Steege 2000). Destabilization of RNAs by polyadenylation is consistent with the small proportion of polyadenylated RNAs detected in the steady-state population in plant mitochondria (Gagliardi and Leaver 1999; Lupold et al. 1999; Kuhn et al. 2001). In contrast, the majority of mRNAs in the mitochondria of humans are polyadenylated (Gaines and Attardi 1984; Gaines et al. 1987). Consistent with this observation, recent evidence from Temperley et al. (2003), suggests that poly-adenylation stabilizes mRNAs in human mitochondria.
Trypanosoma brucei is one of the earliest branching eukaryotes (Sogin et al. 1989) and the causative agent of African Sleeping Sickness. The mitochondria of T. brucei and related organisms are of great biological interest because of both the unique arrangement of the mitochondrial DNA and the novel posttranscriptional events that govern gene expression, including uridine insertion/deletion-type RNA editing (Shapiro and Englund 1995; Schneider 2001; Stuart and Panigrahi 2002; Simpson et al. 2003). The cis- and trans-acting factors involved in RNA turnover in this system are just beginning to be elucidated and suggest unique features in this aspect of mitochondrial gene regulation in T. brucei as well. Some components of the RNA degradation machinery are conserved between yeast and trypanosomes. For example, T. brucei possess a homolog of the DSS-1 exoribonuclease (TbDSS-1), whose down-regulation by RNA interference (RNAi) affects the levels of both mRNA and gRNAs (Penschow et al. 2004). Genomic searches have also revealed the presence of an SUV3 RNA helicase homolog in T. brucei (J.L. Penschow and L.K. Read, unpubl.). While this suggests that the degradosome structure is similar among lower eukaryotes, differences also exist since TbDSS-1 is apparently not associated with ribosomes as is the yeast degradosome, but is present in an ~20S complex of unknown composition (Penschow et al. 2004). T. brucei differs even more dramatically from yeast in the role of mitochondrial mRNA polyadenylation. The majority of mRNAs in T. brucei mitochondria are polyadenylated, a state that is more like that of human mitochondria (Bhat et al. 1992; Read et al. 1994a; Militello and Read 1999). Moreover, we have shown that polyadenylation plays a dual role in RNA stability in trypanosome mitochondria. In vitro RNA turnover studies demonstrated that unedited mRNAs with a 20-nt poly(A) tail are rapidly degraded compared to their nonadenylated counterparts (Ryan et al. 2003). Conversely, in the same system, a 20-nt poly(A) tail protects fully or partially edited mRNAs from rapid degradation (Kao and Read 2005). In organello assays that primarily measure unedited RNA turnover also demonstrated that, under certain conditions, polyadenylation stimulates decay of pulse-labeled RNAs (Militello and Read 2000). Intriguingly, rapid RNA decay in organello requires the addition of UTP. In this system, UTP stimulates rapid decay of exclusively polyadenylated RNAs, decreasing their half-life from >3 h to <20 min. The linkage between in vitro and in organello rapid poly(A)+ RNA decay pathways is unclear since in vitro decay of polyadenylated RNAs does not require UTP addition (C.M. Ryan and L.K. Read, unpubl.). The two assays may be measuring separate RNA turnover pathways. Alternatively, it is possible that purification of proteins for in vitro decay assays uncouples the UTP effect from degradation of polyadenylated RNAs.
The requirement for UTP in stimulating rapid turnover of polyadenylated RNAs is particularly interesting in light of the multiple roles played by UTP in T. brucei mitochondria. Transcription requires a large amount of UTP, since never edited mRNAs are composed of >45% uridine residues (Jasmer et al. 1985). The post-transcriptional process of RNA editing, which remodels mRNAs by the precise insertion and deletion of uridine residues to create open reading frames in otherwise untranslatable RNAs, requires UTP for addition editing (Estevez and Simpson 1999). In addition, post-transcriptional addition of 3′-oligo(U) tails to gRNAs and rRNAs uses UTP (Blum and Simpson 1990; Adler et al. 1991). This suggests that UTP levels may be sensed as a signal for coordinated regulation of multiple processes, culminating in alterations in mRNA decay rates. We previously put forth three hypotheses that describe potential mechanisms by which UTP could act to stimulate degradation of polyadenylated RNAs (Militello and Read 2000). First, UTP may directly interact with an enzyme involved in RNA degradation, thereby causing its activation. In this scenario, UTP may act as a protein cofactor or as an energy source for a nuclease or helicase. Also, an enzyme could be activated through the covalent addition of UMP (uridylylation) using UTP as a donor. Uridylylation has been shown to modulate the binding properties of PII proteins in Escherichia coli (Rhee et al. 1989). Second, UTP may stimulate degradation by directly modifying RNAs, thus triggering their rapid turnover. In this case, addition of single or multiple uridine residues or uridine-containing motifs would act to mark polyadenylated RNAs for rapid degradation, just as poly-adenylation marks RNAs for decay in bacteria and chloroplasts (Schuster et al. 1999; Steege 2000). Third, UTP may be required for a process that is coupled to RNA degradation, and thus RNA degradation would indirectly require UTP. We previously showed that UTP-dependent RNA decay is not coupled to transcription (Militello and Read 2000). However, it is possible that the RNA degradation is coupled to a process such as RNA editing. To distinguish between these hypotheses, we performed in organello pulse chase assays under different conditions, in the presence of UTP analogs, or in mitochondria down-regulated for specific mitochondrial proteins. We found that UTP does not act through a receptor on the outer membrane of the mitochondria and that UTP-dependent decay is not tied to RNA editing. Moreover, the rapid turnover pathway is responsive to UTP and not the ATP:UTP ratio. Experiments using nonextendable and noncleavable analogs of UTP, in place of unmodified UTP, show that polymerization of UTP into RNA is required for rapid degradation of polyadenylated RNAs. Finally, through RNAi-mediated depletion, we demonstrate that RNA editing terminal uridylyl transferase (RET1) is required for the UTP-dependent decay pathway.
RESULTS
Basic parameters of the UTP-stimulated RNA turnover pathway
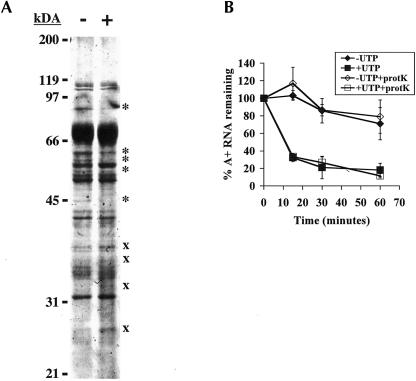
Our hypotheses, that UTP stimulates turnover of polyadenylated RNA by either modifying RNA or interacting with protein components, require that UTP enters the mitochondria to function. Alternatively, it is possible that UTP stimulates turnover through interaction with a protein on the outer mitochondrial membrane. Functional interaction of UTP with membrane proteins is not unprecedented, as a subset of P2Y receptors are activated by extracellular UTP and regulate a wide range of cellular functions (for review, see Lazarowski and Boucher 2001). To examine if UTP is acting through a membrane protein to stimulate RNA turnover, mitochondria were pretreated with proteinase K prior to performing in organello RNA turnover assays. It has previously been shown that treatment of T. brucei mitochondrial vesicles with low concentrations of proteinase K at 4°C inhibits both tRNA and protein import (Yermovsky-Kammerer and Hajduk 1999; Priest and Hajduk 2003). Treatment with five times the concentration of proteinase K used to inhibit protein import (75 μg/mL) led to an alteration in the protein profile of the mitochondria similar to previously observed results (Priest and Hajduk 2003; data not shown). To verify that outer membrane proteins were degraded by proteinase K treatment, the mitochondria were fractionated. Treatment with proteinase K resulted in reduction or loss of at least five high-molecular-weight proteins from fractions enriched in outer mitochondrial membrane and intermembrane space proteins and an increase in at least four low-molecular-weight proteins that are presumably degradation products (Fig. 1A ▶). We do not expect all proteins to degrade since many integral membrane and intermembrane space proteins would be protected from proteinase K treatment. In addition, removal of a small portion of an outer membrane protein might be undetectable on a polyacrylamide gel but still have functional consequences. The results shown in Figure 1A ▶ indicate that proteinase K treatment results in degradation of proteins in a fraction enriched in outer mitochondrial membrane proteins. Pretreatment of mitochondria with proteinase K also caused an average 50% decrease in the labeling of total RNA as compared to mock-treated controls (data not shown). This modest decrease in RNA labeling had no effect on the half-life of total RNA (data not shown). More importantly, proteinase K pretreatment of mitochondrial vesicles had no effect on either the UTP-dependent or basal (-UTP) turnover rate of polyadenylated RNAs (Fig. 1B ▶). While it is possible that proteinase K treatment does not degrade all outer mitochondrial membrane proteins, these results suggest that UTP enters the mitochondria to stimulate decay of polyadenylated RNAs.
FIGURE 1.
UTP acts inside the mitochondria. (A) Isolated mitochondria were mock-treated (−) or treated with 75 μg/mL proteinase K for 15 min at 4°C (+). Mitochondria were fractionated as described in Materials and Methods, and fractions enriched in outer membrane and intermembrane space proteins were separated on a 12.5% acryl-amide gel and visualized by Coomassie staining. The migration positions of molecular mass standards are indicated on the left. (*) Indicates proteins that decrease upon treatment with proteinase K, (x) indicates proteins that increase with proteinase K treatment. (B) Isolated mitochondria were mock-treated (closed symbols) or treated with 75 μg/mL proteinase K for 15 min at 4°C (+prot K, open symbols). Mitochondria were then labeled with [α-32P]CTP and subsequently incubated in transcription buffer containing 2 mM CTP (-UTP, diamonds) or 2 mM CTP and 2 mM UTP (+UTP, squares). At indicated time points, incorporation of [α-32P]CTP into polyadenylated RNAs was measured. Percent polyadenylated RNA remaining as compared to pulse-labeled RNA is plotted. Each point is the average of three experiments with error bars representing one standard deviation.
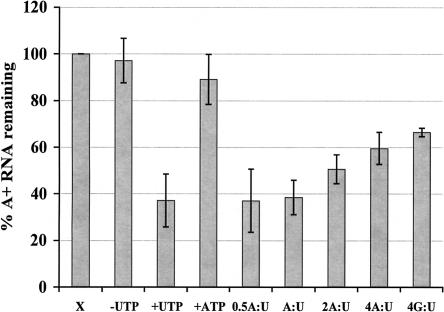
UTP-stimulated decay may be a response to low energy conditions in the cell. High concentrations of UTP or a low ATP:UTP ratio may be sensed as a decreased abundance of available energy sources, which could result in increased RNA decay to conserve energy. In this assay, the addition of 2 mM UTP during the chase period coupled with the negligible amount of ATP present in the mitochondrial vesicles (Harris et al. 1990) could indicate low energy abundance. If low energy conditions or a low ATP:UTP ratio allow UTP to stimulate decay of polyadenylated RNAs, increasing ATP concentrations would be expected to inhibit UTP-stimulated decay. To test this hypothesis, mitochondrial RNA was labeled with [α-32P]CTP, and RNA decay was measured during a chase period in the presence of 2 mM CTP, 2 mM UTP, and between 0.5-fold and fourfold molar excess of ATP. GTP, at a ratio of 4:1 (GTP:UTP), served as a control for nucleotide specificity. While lower concentrations of ATP had no effect, the addition of fourfold molar excess ATP inhibited UTP-stimulated decay ~20% (Fig. 2 ▶). However, this effect was not nucleotide-specific as GTP caused a similar inhibition (Fig. 2 ▶). Since in organello UTP-dependent decay is absolutely specific for UTP, it is unlikely that ATP or GTP directly inhibits the UTP-dependent decay mechanism (Fig. 2 ▶; Militello and Read 2000). More likely, such a large excess of nucleotides inhibited import of UTP into the mitochondria, thus inhibiting decay. These results indicate that the turnover rate of polyadenylated RNAs is controlled by absolute UTP levels rather than the ATP:UTP ratio.
FIGURE 2.
Effect of the ATP:UTP ratio on UTP-dependent RNA turnover. Mitochondrial vesicles were labeled with [α-32P]CTP and subsequently incubated in transcription buffer containing 2 mM CTP (-UTP); 2 mM CTP and 2 mM UTP (+UTP); 2 mM CTP and 2 mM ATP (ATP); or 2 mM CTP, 2 mM UTP, and either 1 mM ATP (0.5 A:U); 2 mM ATP (A:U); 4 mM ATP (2A:U); 8 mM ATP (4A:U); or 8 mM GTP (4G:U). After 15 min, incorporation of [α-32P]CTP into polyadenylated RNAs was measured. Percent polyadenylated RNA remaining as compared to pulse-labeled RNA (X) is plotted. Each point is the average of three experiments, and the error bars represent one standard deviation.
Polymerization of UTP into RNA is required for UTP-dependent RNA turnover
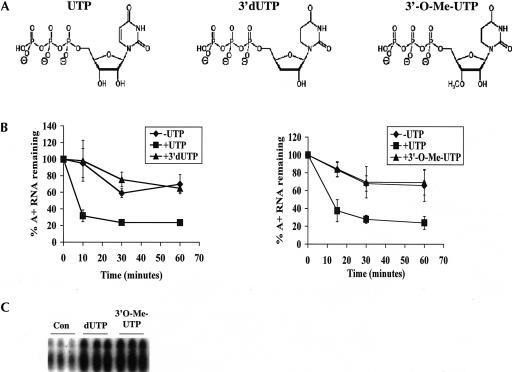
To distinguish between models in which UTP is used to directly modify RNAs from those in which UTP acts as a cofactor or energy source, we used a bank of UTP analogs. Two nonextendable analogs of UTP (3′-dUTP and 3′-O-Me-UTP; Fig. 3A ▶) were added individually to the chase period of in organello RNA degradation experiments in the absence of UTP. The rate of turnover of polyadenylated RNAs was compared in assays using UTP or UTP analogs. Neither 3′-dUTP nor 3′-O-Me-UTP was able to stimulate rapid decay of polyadenylated RNAs (Fig. 3B ▶). Furthermore, neither of these analogs had any effect on degradation of the total RNA population (data not shown). The inability of either 3′-dUTP or 3′-O-Me-UTP to support decay of polyadenylated RNAs could result from their failure to enter the mitochondria. To establish that the 3′-modified UTP analogs enter the mitochondria, we used a transcription assay. In this assay, isolated mitochondria were pulsed with [α-32P]CTP in the presence of 0.1 mM GTP, UTP, and ATP and either 1 mM or 2 mM of one of the UTP analogs. After purification, total RNA was resolved on a denaturing acrylamide gel and visualized by autoradiography. RNA migration patterns, but not relative RNA abundance, were similar to previously published results (Harris et al. 1990; Militello and Read 2000). RNAs corresponding to transcripts encoded by the maxicircle (mRNAs and pre-mRNAs) (Harris et al. 1990; Militello and Read 2000), as well as transcripts that are presumably precursor and mature tRNAs (Hancock et al. 1992), were observed. Upon addition of either 3′-modified analog, we observed a reduction in mRNA labeling and a corresponding increase in labeling of mature tRNAs in a concentration-dependent manner (Fig. 3C ▶; data not shown). tRNA labeling is presumably due to 3′-CCA addition to tRNAs (Hancock et al. 1992). The increase we observed is likely a result of the presence of increased free [α-32P]CTP that is unable to be incorporated by transcription. The effects of both 3′-dUTP and 3′-O-Me-UTP on mitochondrial RNA labeling demonstrate that both analogs enter the mitochondria. Thus, the inability of 3′-dUTP or 3′-O-Me-UTP to stimulate rapid decay of polyadenylated RNAs is due to intrinsic properties of the nucleotides rather than their inability to enter the mitochondria. Taken together, these results suggest that polymerization of UTP is required to stimulate rapid degradation of polyadenylated RNAs. In addition, these experiments rule out uridylylation of a protein as the mechanism of UTP-dependent decay, since uridylylation requires covalent addition of UMP with no restraints on the 3′-moiety (Rhee et al. 1989). Although data with 3′-dUTP and 3′-O-Me-UTP suggest that UTP polymerization into RNA is required for rapid decay of polyadenylated RNAs, these experiments do not rule out that UTP acts as a protein cofactor and that the presence of a 3′-hydroxyl group (see Fig. 3A ▶) is necessary for proper interaction of UTP with the proteins of the decay pathway.
FIGURE 3.
Analogs modified at their 3′- positions fail to stimulate rapid decay. (A) Structures of UTP analogs used in these experiments. (From left to right) UTP (uridine-5′-triphosphate), 3′-dUTP (3′-deoxyuridine-5′-triphosphate), and 3′-O-Me-UTP (3′-O-methyluridine-5′-triphosphate). (B) Mitochondria were labeled with [α-32P]CTP and subsequently incubated in transcription buffer containing 2 mM CTP (-UTP, diamonds), 2 mM CTP and 2 mM UTP (+UTP, squares), or 2 mM CTP and 2 mM 3′-dUTP (+3′-dUTP, triangles, left panel) or 2 mM 3′-O-Me-UTP (+3′-O-Me-UTP, triangles, right panel). At indicated time points, incorporation of [α-32P]CTP into polyadenylated RNAs was measured. Percent polyadenylated RNA remaining as compared to pulse-labeled RNA is plotted. Each point is the average of three experiments with error bars representing one standard deviation. (C) Isolated mitochondria were pulsed in triplicate with [α-32P]CTP in the presence of 0.1 mM GTP, UTP, and ATP (Con) or these nucleotides and 2 mM dUTP or 2 mM 3′-O-Me-UTP. After purification, total RNA was resolved on a 7 M urea and 6% acrylamide gel and visualized by autoradiography. The region of the gel at which tRNAs migrate is shown.
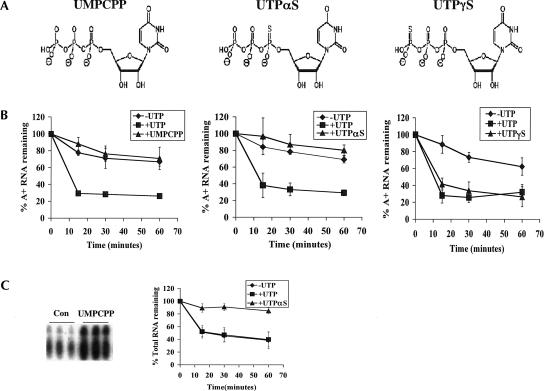
To further address the requirement for UTP polymerization, we examined whether UTP analogs with noncleavable α–β bonds (UMPCPP and UTPαS) (Fig. 4A ▶) or noncleavable β–γ bonds (UTPγS) (Fig. 4A ▶) could stimulate rapid degradation of polyadenylated RNAs. As with 3′-dUTP and 3′-O-Me-UTP, analogs with noncleavable α–β bonds would not support rapid decay of polyadenylated RNAs if this effect requires UTP polymerization. Conversely, UTPγS should support rapid turnover in this case as hydrolysis of the β–γ bond is not required for UTP polymerization. Neither UMPCPP nor UTPαS was able to support rapid degradation of polyadenylated RNAs (Fig. 4B ▶). In addition, UMPCPP had no effect on decay of the total RNA population, whereas UTPαS significantly stabilized the total RNA population by an unknown mechanism (Fig. 4C ▶). In contrast to α–β noncleavable UTP analogs, UTPγS effectively stimulated rapid degradation of polyadenylated RNAs (Fig. 4B ▶) and had no effect on turnover of the total RNA population (data not shown). Since UTPγS and UTPαS affect polyadenylated and total RNA stability, respectively, there is no question that these nucleotides enter the mitochondria. To determine if UMPCPP enters the mitochondria, a transcription assay was performed as described above. Addition of 2 mM UMPCPP to transcription assays caused a similar increase in tRNA labeling as addition of either 3′-dUTP or 3′-O-Me-UTP (Fig. 4C ▶), indicating that UMPCPP enters the mitochondria. These results demonstrate that hydrolysis of the UTP α–β bond and not the β–γ bond is necessary for UTP-dependent degradation of polyadenylated RNAs. This shows that UTP is not used as an energy source for decay, since hydrolysis of the β–γ bond is not required to stimulate decay. Furthermore, these results, along with our previous results with 3′-modified analogs, indicate that rapid turnover of polyadenylated RNAs requires polymerization of UTP into RNA.
FIGURE 4.
The effect of nonhydrolyzable analogs on degradation of polyadenylated RNAs. (A) Structures of the nonhydrolyzable analogs used in this assay. (From left to right) UMPCPP [uridine-5-O-(α,β-methylene-triphosphate)], UTPαS [uridine-5′-(α-thio)-triphosphate], and UTPγS [uridine-5′-(γ-thio)-triphosphate]. (B) Mitochondria were labeled with [α-32P]CTP and subsequently incubated in transcription buffer containing 2 mM CTP (-UTP, diamonds), 2 mM CTP and 2 mM UTP (+UTP, squares), or 2 mM CTP and 2 mM UTP analog [triangles; +UMPCPP (left), +UTPαS (middle), and +UTPγS (right)]. At indicated time points, incorporation of [α-32P]CTP into polyadenylated RNAs was measured. Percent polyadenylated RNA remaining as compared to pulse-labeled RNA is plotted. Each point is the average of three experiments with error bars representing one standard deviation. (C, left panel) Mitochondria were pulsed in triplicate with [α-32P]CTP in the presence of 0.1 mM GTP, UTP, and ATP (Con) or these nucleotides and 2 mM UMPCPP. After purification, total RNA was resolved on a 7 M urea and 6% acrylamide gel and visualized by autoradiography. The region of the gel at which tRNAs migrate is shown. (C, right panel) Mitochondria were labeled with [α-32P]CTP and subsequently incubated in transcription buffer containing 2 mM CTP (-UTP, diamonds), 2 mM CTP and 2 mM UTP (+UTP, squares), or 2 mM CTP and 2 mM UTPαS (+UTPαS, triangles). At indicated times, incorporation of [α-32P]CTP into total RNAs was measured. Percent total RNA remaining as compared to pulse-labeled RNA is plotted.
RET1 is involved in UTP-stimulated turnover of polyadenylated RNAs
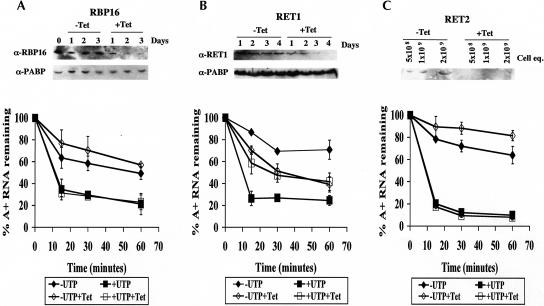
To further examine the mechanism of UTP-dependent decay, we attempted to identify proteins involved in this process by using RNAi. We first determined whether the T. brucei mitochondrial RNA-binding protein RBP16 was involved in this UTP-dependent decay. RBP16 has a strong affinity for poly(U) ribohomopolymers and binds both gRNAs and rRNAs presumably through their oligo(U) tails (Hayman and Read 1999; Pelletier et al. 2000). In addition, disruption of RBP16 by RNAi demonstrated a role for this protein in regulating RNA stability (Pelletier and Read 2003). To determine whether RBP16 functions in the UTP-stimulated turnover pathway, T. brucei cell lines harboring an integrated tetracycline-inducible expression system for induction of RNAi of RBP16 were grown in the presence or absence of tetracycline (Pelletier and Read 2003). After 3 d of induction, protein levels of RBP16 were significantly reduced; however, there was no major growth defect (Fig. 5A ▶; data not shown). Mitochondria isolated on day 3 from either uninduced or induced cells had identical RNA turnover properties in the in organello assay. UTP stimulated rapid degradation of polyadenylated RNAs while CTP had no effect, identical to wild-type cells (Fig. 5A ▶). These data strongly suggest that RBP16 is not involved in UTP-dependent decay of polyadenylated RNAs.
FIGURE 5.
In organello RNA turnover assays with RNAi cells. Mitochondria were isolated from RNAi cell lines: RBP16 (A), RET1 (B), or RET2 (C). Cells were uninduced (closed symbols) or induced with tetracycline (open symbols) for 3 d (RBP16) or 4 d (RET1 and RET2). Isolated mitochondria were labeled with [α-32P]CTP and subsequently incubated in transcription buffer containing 2 mM CTP (-UTP, diamonds) or 2 mM CTP and 2 mM UTP (+UTP, squares). Labeling of polyadenylated RNAs was measured at indicated time points, and percent RNA remaining as compared with pulse-labeled RNA is plotted. Each point is the average of three experiments with error bars representing one standard deviation. Western blots of whole-cell extracts (A,B) or mitochondrial extracts (C) are shown in the top panel of each experiment.
The requirement for UTP polymerization (Figs. 3 ▶, 4 ▶) suggests the involvement of a terminal uridylyl transferase (TUTase) in stimulating RNA turnover. TUTases catalyze the addition of uridylate residues to the 3′-ends of RNA using UTP as a substrate. Two TUTases have been characterized in the mitochondria of T. brucei. RET1 is a processive enzyme that has a role in adding oligo(U) tails to gRNAs (Aphasizhev et al. 2002, 2003). RET2 (RNA editing TUTase 2) is a distributive enzyme and is a stable component of the editosome or L-complex that adds uridines to mRNAs undergoing U-insertional editing (Aphasizhev et al. 2003; Ernst et al. 2003). As with RBP16, the expression of RET1 and RET2 was down-regulated by RNAi to study their role in UTP-stimulated RNA decay. Then, 4 d after induction, mitochondria were isolated from uninduced and induced RET1 or RET2 knock-down cells for use in organello assays. On day 4, we observed a complete loss of RET1 or RET2 protein (Fig. 5B,C ▶). In addition, there was no major growth defect on day 4, thereby allowing us to avoid nonspecific effects due to cell death (data not shown). Mitochondria isolated from cells lacking RET2 behaved identically to uninduced cells, indicating that RET2 has no role in the UTP-stimulated RNA turnover pathway (Fig. 5C ▶). In contrast, in mitochondria isolated from cells deficient in RET1, UTP failed to stimulate rapid turnover of polyadenylated RNAs (Fig. 5B ▶, cf. open squares and open diamonds). In addition, the rate of basal poly(A)+ RNA turnover was somewhat elevated in RET1-deficient mitochondria (Fig. 5B ▶, cf. open and closed diamonds). The inability of UTP to stimulate turnover in RET1-deficient cells is not a secondary effect due to an RNA editing defect since RET2 RNAi cells, which are deficient in RNA editing (Aphasizhev et al. 2003), are fully competent for UTP-stimulated turnover of polyadenylated RNAs (Fig. 5C ▶). These results suggest that RET1 is required for UTP-stimulated turnover of polyadenylated RNAs. However, since induction of RNAi causes a reduction in stability of polyadenylated RNAs in the absence of UTP (Fig. 5B ▶, cf. open and closed diamonds), it is possible that RET1 knock-down causes a general defect in RNA metabolism that also affects UTP-dependent decay of polyadenylated RNAs. Nevertheless, the defect in UTP-dependent decay in RET1-deficient mitochondria is consistent with the UTP analog data, indicating a role for UTP polymerization. Thus, the function of added UTP in in organello assays is presumably as a substrate for RET1.
DISCUSSION
In this study, we used our previously established in organello pulse chase assay to study the mechanism by which UTP stimulates decay of polyadenylated RNAs in T. brucei mitochondria. Our results suggest that UTP acts inside the mitochondria to activate RNA decay. Furthermore, we demonstrate that the turnover rate of polyadenylated RNAs is controlled by absolute UTP levels and not the ATP:UTP ratio. By performing in organello assays with UTP analogs, we show that UTP polymerization into RNA is required for UTP-dependent decay of polyadenylated RNAs. Analysis of decay in mitochondria from RNAi-depleted cells revealed a role for the RET1 TUTase in UTP-stimulated turnover of polyadenylated RNAs.
Pre-treatment of the mitochondria with proteinase K had no effect on basal or UTP-stimulated decay of polyadenylated RNAs, suggesting that UTP must enter the mitochondria to stimulate decay of polyadenylated RNAs. However, some mitochondrial surface proteins may be refractory to proteinase K treatment, as several proteins appear stable after proteinase K treatment in a fraction enriched in outer membrane and intermembrane proteins, as visualized in Coomassie-stained gels. These stable proteins may be inter-membrane proteins or outer membrane proteins that are not degraded or those with such a small portion removed that a shift cannot be observed. We cannot rule out that a few remaining receptor proteins could interact with UTP to stimulate a signal cascade, ultimately resulting in rapid decay of polyadenylated RNAs. Nevertheless, the lack of any effect of proteinase K treatment on turnover of polyadenylated RNAs and the inability of RET1-deficient mitochondria to support decay support the hypothesis that UTP enters the mitochondria to stimulate decay of polyadenylated RNAs. UTP must enter the mitochondria to interact with RET1.
We also demonstrated that rapid turnover of polyadenylated RNAs requires polymerization of UTP into RNA. Three mechanisms by which polymerization of UTP into RNA could stimulate degradation of polyadenylated RNAs can be envisioned. First, oligo(U) at the 3′-end of an RNA may recruit a U-specific nuclease or nuclease complex. U-specific 3′–5′ exoribonuclease activities have been characterized from both Leishmania tarentolae and T. brucei mitochondria (Aphasizhev and Simpson 2001; Igo et al. 2002). The role of these nuclease activities in RNA deletion editing has been investigated, but the possibility that these enzymes have additional functions in mitochondrial RNA metabolism has not been addressed. It is feasible that the editing U-exonuclease can also associate with other ribonucleases, resulting in a nuclease complex that degrades uridylylated RNAs. A different, and yet uncharacterized, U-specific or U-dependent ribonuclease could also be involved. Interestingly, addition of 3′-oligo(U) tails has recently been reported to signal decay of miRNA-directed mRNA cleavage products in both plants and mice (Shen and Goodman 2004). The enzymes that catalyze U addition and degradation of oligo-uridylylated RNA in these organisms are unknown. A second mechanism by which uridine polymeriza-tion may trigger RNA decay in T. brucei mitochondria is through addition of uridines to the poly(A) tail to form an AU-rich element (ARE). AREs are present in the 3′-UTRs of many eukaryotic cytoplasmic mRNAs, where they act as stabilizing or destabilizing elements depending on the RNA and cellular conditions (for review, see Dean et al. 2004). In both Trypanosoma cruzi and T. brucei, AREs are located in the 3′-UTRs of certain nuclear-encoded mRNAs, and these elements have been shown to confer selective destabilization in a stage-specific manner (D’Orso et al. 2003). Proteins involved in this cytoplasmic RNA decay pathway could be imported into the mitochondria and used in a similar decay pathway. In support of this hypothesis, Us are often found in the poly(A) tails of mitochondrial mRNAs in trypanosomes (Decker and Sollner-Webb 1990; Read et al. 1992; Souza et al. 1992, 1993; Read et al. 1994b). In a third potential scenario, addition of a poly(U) tail onto an already polyadenylated RNAs could result in formation of a poly(A)/poly(U) duplex. This duplex could then attract a double-stranded RNase (dsRNase) or a complex containing dsRNase, dsRNA-binding proteins, and/or other ribonucleases that rapidly degrade A/U duplex-containing RNAs. RNAi functions through a dsRNase and is a common mechanism used to regulate gene expression in the cytoplasm of T. brucei (Ullu et al. 2004). In addition, argonaute proteins, which are key components in RNAi, are present in T. brucei and have been shown to bind small interfering RNAs (Shi et al. 2004). This RNAi machinery or the dsRNase itself could be imported into the mitochondria to degrade RNAs with duplexed A/U tails. We have previously used in vitro RNA decay assays to address the roles of 3′-extensions in unedited RNA turnover. In these assays, a poly(U) tail does not promote rapid RNA decay and addition of uridines into the poly(A) tail slows RNA decay (Ryan et al. 2003). A 3′-extension with the potential to form a long duplex was never tested in this system. Thus, our in vitro results seem to suggest that UTP-dependent rapid decay is not due to poly(U) tail formation alone or to creation of an ARE. However, the in vitro and in organello assays may be measuring different pathways, or specific factors such as a U-specific nuclease or AU-binding proteins may be lost in purification of proteins for the in vitro decay assay. In addition, the cis-acting sequences tested may not be in the correct arrangement for RNA decay. Further work is needed to clarify these possibilities.
Depletion of RET1 by RNAi results in a loss of UTP-dependent turnover, indicating that RET1 plays a critical role in UTP-dependent RNA decay. We favor a model in which RET1 uses UTP to polymerize uridine residues on the 3′-end of mRNAs, which then acts in cis as a signal for rapid decay. Alternatively, the loss of function could be a secondary effect of RET1 depletion. Previously, gRNAs in RET1-deficient cells were shown to have shortened 3′-oligo(U) tails (Aphasizhev et al. 2003). Shorter gRNA tails could lead to indirect destabilization of mRNAs by decreasing mRNA–gRNA binding, thus making the mRNA more susceptible to degradation. However, Aphasizhev et al. (2003) showed that in RET1-depleted cells, oligo(U) tails are only shortened, not absent, and that ~40% of the gRNAs retain full-length tails. This modest reduction in gRNA oligo(U) tails is unlikely to substantially affect mRNA stability. Furthermore, we showed that the deficiency in UTP-stimulated decay observed in RET1 RNAi cells is not a secondary effect of an RNA editing defect since RET2-deficient mitochondria, which are defective in editing, are fully competent for UTP-stimulated decay (Aphasizhev et al. 2003). Thus, the simplest explanation is that the effect of RET1 on poly(A)+ RNA turnover is a direct effect, mediated by one of the U-containing modifications discussed above. RET1 is not a stable component of the multi-subunit editosome or L-complex, but exists in mitochondria in a complex with an apparent mass of ~500 or 700 kDa, depending on the organism (Aphasizhev et al. 2002; Ernst et al. 2003). Proteins associated with RET1 as part of a complex may possess or recruit ribonuclease activity for decay of polyadenylated mRNAs. Furthermore, RET1 may be a component of multiple 500- to 700-kDa complexes that are involved in diverse processes including mRNA decay and oligo(U) addition to gRNAs. Identification of RET1-associated proteins may provide insight into the role of this enzyme in turnover of polyadenylated RNAs.
The mechanism by which RET1-mediated RNA decay is regulated is currently unclear. While this pathway is dramatically stimulated by UTP addition in organello, UTP may simply be providing a substrate for RET1 in nucleotide-depleted mitochondrial vesicles, rather than acting as a regulatory signal. Alternatively, it remains possible that mitochondrial UTP levels are sensed as a signal for rapid decay of specific RNA molecules. Future studies to address the function and mechanism of UTP-dependent decay pathway will entail a comprehensive analysis of RNAs specifically stabilized in RET1-deficient cells and comparison of RNA 3′-end modification in uninduced and induced RET1 RNAi cells. In addition, identification of nucleases that act in this pathway may reveal regulatory mechanisms. These studies will provide insight into the role of UTP-stimulated RNA degradation in T. brucei mitochondrial function.
MATERIALS AND METHODS
Trypanosome growth, mitochondrial isolation, and induction of RNAi
Procyclic form T. brucei brucei clone IsTaR1 stock EATRO 164 was grown as described previously (Brun and Schonenberger 1979). Mitochondrial vesicles were isolated on linear 20%–35% Percoll gradients and stored at −80°C (Harris et al. 1990).
Procyclic T. brucei strain 29-13 with integrated genes for the induction of RNAi of RET-1 or RET-2 proteins (generous gifts from Larry Simpson, University of California–Los Angeles) were grown in SDM-79 supplemented with 10% fetal bovine serum, in the presence of G418 (30 mg/L), hygromycin (50 mg/L), and phleomycin (2.5 mg/L) (Aphasizhev et al. 2002, 2003). For induction of double-stranded RNA, cells were cultured in the presence of 1 mg/L tetracycline. After 4 d of induction, mitochondria were isolated by the method of Harris et al. (1990). Cell growth was comparable to previously published results (Aphasizhev et al. 2002, 2003), with very slight growth defects observed at day 4 for both cell lines (data not shown).
Procyclic T. brucei strain 29-13 with integrated genes for the induction of RNAi of RBP16 was grown in SDM-79 supplemented with 15% fetal bovine serum in the presence of G418 (15 mg/L), hygromycin (50 mg/L), and phleomycin (2.5 mg/L) (Pelletier and Read 2003). RNAi was induced by the addition of 1 mg/L tetracycline. After 3 d of induction, mitochondria were isolated as previously described (Harris et al. 1990). Previously published work on RBP16 RNAi cells was performed with nonclonal cultures (Pelletier and Read 2003). The cells used in this study were from a clonal line produced from the previously studied culture. No significant differences in growth or RNA metabolism were noted between the culture and clonal cell line (M. Pelletier and L.K. Read, unpubl.). Using the clonal line, we observed a slight growth defect by day 3 (data not shown) that was comparable to previously published results (Pelletier and Read 2003).
In organello pulse chase assays
In organello pulse chase assays were performed as described previously with minor modifications (Militello and Read 2000). Following collection, mitochondrial vesicles were resuspended at a concentration of 5 mg/mL in transcription buffer (5 mM HEPES at pH 7.6, 3 mM potassium phosphate at pH 7.8, 125 mM sucrose, 6 mM KCl, 10 mM MgCl2, 1 mM EDTA, 2 mM 2-mercaptoethanol) containing 0.1 mM ATP. Vesicles were labeled with [α-32P]CTP (400 Ci/mmol; Amersham) at a concentration of 200 μCi/mL for 10 min at 27°C in the presence of 0.1 mM GTP, ATP, and UTP. Vesicles were subsequently incubated in transcription buffer, containing 2 mM unlabeled CTP, at 27°C for times indicated. Other unlabeled nucleotides were added during the chase period, with CTP at a concentration of 2 mM unless stated otherwise. Pulse reactions were carried out in a volume of 50 μL, and chase reactions were performed in batch (unless there was a single time point) with a volume of 50 μL per time point. At indicated time points, 50 μL was removed from batch chase reactions for termination. Reactions were stopped, and total and polyadenylated RNA was isolated as previously described (Militello and Read 2000).
Treatment of mitochondria with proteinase K
For proteinase K treatment, mitochondria from 5 × 108 cells (~250 μg of protein) were resuspended in transcription buffer either with or without 75 μg/mL proteinase K, in reaction volumes as outlined above. Reactions were incubated at 4°C for 15 min. Mitochondria were then collected by centrifugation for 10 min at 13,000 rpm at 4°C in a Heraeus Instruments Biofuge Pico. Pelleted mitochondrial vesicles were diluted with 9 volumes of transcription buffer containing 1 mM PMSF and then collected by centrifugation for 10 min at 13,000 rpm at 4°C. These vesicles were resuspended at a concentration of 5 mg/mL in transcription buffer containing 0.1 mM ATP, and in organello reactions were carried out as described above. To enrich the outer membrane and intermembrane space proteins, mitochondria were fractionated as previously described (Geenawalt 1974; Williams and Frank 1990). Then 2 mg of isolated mitochondria was resuspended in transcription buffer with or without 75 μg/mL proteinase K. Reactions were incubated at 4°C for 15 min. Mitochondria were then collected by centrifugation for 10 min at 13,000 rpm at 4°C. Pelleted mitochondrial vesicles were diluted with 9 volumes of transcription buffer containing 1 mM PMSF and then collected by centrifugation for 10 min at 13,000 rpm at 4°C. These mitochondria were resuspended in H medium (2.0 mM HEPES, 220 mM mannitol, 70 mM sucrose, 0.5 mg/mL BSA) at a concentration of 70 mg/mL. An equal volume of 12 mg/mL digitonin was added, and the mixture was stirred on ice for 20 min. Inner membranes were collected by centrifugation at 10,000g, the supernatant containing outer membrane and inter-membrane space proteins was removed, resuspended in SDS-PAGE sample buffer, boiled for 10 min, and separated on a 12.5% SDS-polyacrylamide gel. These gels were stained with Coomassie blue to visualize differences in protein profiles. Using the same mitochondrial aliquot, in organello pulse chase experiments were performed in parallel.
UTP analogs
3′-dUTP and 3′-O-Me-UTP were obtained from TriLink Bio-Technologies. UTPαS and UTPγS were purchased from Jena Bioscience. Uridine-5′-O-(α,β-methylene-triphosphate; UMPCPP) was synthesized by the BIOLOG Life Science Institute (Bremen, Germany).
Western blot analysis of RNAi cells
For analysis of RET1 and RBP16 protein levels, aliquots (1 × 107 cells) of uninduced and induced cells were suspended in SDS-PAGE sample buffer, boiled for 10 min, and stored at −20°C. For Western hybridization, the cell aliquots were electrophoresed by SDS-PAGE on 10% polyacrylamide and transferred to nitrocellulose membranes. Blots were probed with 1:2000 anti-RET1 (a generous gift from Larry Simpson, University of California–Los Angeles) (Aphasizhev et al. 2002), 1:1000 anti-RBP16 (Pelletier and Read 2003), or 1:10,000 anti-PABPI (generously provided by Noreen Williams, SUNY Buffalo) (Pitula et al. 2002). Primary antibodies were detected using goat anti-rabbit (PABP, RBP16) or goat anti-mouse (RET1) antibodies (1:10,000 dilution; Pierce) coupled to horseradish peroxidase and detected by enhanced chemiluminescence (Pierce).
For analysis of RET2 protein levels, 1 × 1010 cell equivalents of mitochondria were suspended in SDS-PAGE sample buffer, boiled for 10 min, and stored at −20°C. For Western hybridization, 5 × 108, 1 × 109, or 2 × 109 cell equivalents of mitochondria were electrophoresed by SDS-PAGE on a 12.5% polyacrylamide and transferred to nitrocellulose membranes. Blots were probed with 1:40 anti-RET2 (TbMP57; a generous gift from Kenneth Stuart, Seattle Biomedical Research Institute) (Ernst et al. 2003). Primary antibodies were detected with goat anti-rabbit antibodies (1:10,000 dilution; Pierce) and detected by enhanced chemiluminescence (Pierce).
Acknowledgments
We are grateful to Larry Simpson for providing RET1 and RET2 RNAi cells as well as anti-RET1 antibodies. We thank Kenneth Stuart for supplying anti-RET2 antibodies, and Noreen Williams for providing anti-PABP antibodies. We also thank Ruslan Aphasizhev and Nancy Ernst for technical advice and Michel Pelletier for critical reading of the manuscript. This work was supported by the National Institutes of Health Grant AI47329 (to L.K.R.). C.M.R. was supported in part by NIH Training Grant AI07614.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7248605.
REFERENCES
- Adler, B.K., Harris, M.E., Bertrand, K.I., and Hajduk, S.L. 1991. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol. Cell. Biol. 11: 5878–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev, R. and Simpson, L. 2001. Isolation and characterization of a U-specific 3′-5′-exonuclease from mitochondria of Leishmania tarentolae. J. Biol. Chem. 276: 21280–21284. [DOI] [PubMed] [Google Scholar]
- Aphasizhev, R., Sbicego, S., Peris, M., Jang, S.H., Aphasizheva, I., Simpson, A.M., Rivlin, A., and Simpson, L. 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell 108: 637–648. [DOI] [PubMed] [Google Scholar]
- Aphasizhev, R., Aphasizheva, I., and Simpson, L. 2003. A tale of two TUTases. Proc. Natl. Acad. Sci. 100: 10617–10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, G.J., Souza, A.E., Feagin, J.E., and Stuart, K. 1992. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 52: 231–240. [DOI] [PubMed] [Google Scholar]
- Blum, B. and Simpson, L. 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell 62: 391–397. [DOI] [PubMed] [Google Scholar]
- Brun, R. and Schonenberger, M. 1979. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36: 289–292. [PubMed] [Google Scholar]
- Dean, J.L., Sully, G., Clark, A.R., and Saklatvala, J. 2004. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal 16: 1113–1121. [DOI] [PubMed] [Google Scholar]
- Decker, C.J. and Sollner-Webb, B. 1990. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell 61: 1001–1011. [DOI] [PubMed] [Google Scholar]
- D’Orso, I., De Gaudenzi, J.G., and Frasch, A.C. 2003. RNA-binding proteins and mRNA turnover in trypanosomes. Trends Parasitol 19: 151–155. [DOI] [PubMed] [Google Scholar]
- Dziembowski, A., Malewicz, M., Minczuk, M., Golik, P., Dmochowska, A., and Stepien, P.P. 1998. The yeast nuclear gene DSS1, which codes for a putative RNase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol. Gen. Genet. 260: 108–114. [DOI] [PubMed] [Google Scholar]
- Dziembowski, A., Piwowarski, J., Hoser, R., Minczuk, M., Dmochowska, A., Siep, M., van der Spek, H., Grivell, L., and Stepien, P.P. 2003. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 278: 1603–1611. [DOI] [PubMed] [Google Scholar]
- Ernst, N.L., Panicucci, B., Igo Jr., R.P., Panigrahi, A.K., Salavati, R., and Stuart, K. 2003. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell 11: 1525–1536. [DOI] [PubMed] [Google Scholar]
- Estevez, A.M. and Simpson, L. 1999. Uridine insertion/deletion RNA editing in trypanosome mitochondria—A review. Gene 240: 247–260. [DOI] [PubMed] [Google Scholar]
- Gagliardi, D. and Leaver, C.J. 1999. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 18: 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi, D., Stepien, P.P., Temperley, R.J., Lightowlers, R.N., and Chrzanowska-Lightowlers, Z.M. 2004. Messenger RNA stability in mitochondria: Different means to an end. Trends Genet. 20: 260–267. [DOI] [PubMed] [Google Scholar]
- Gaines, G. and Attardi, G. 1984. Highly efficient RNA-synthesizing system that uses isolated human mitochondria: New initiation events and in vivo-like processing patterns. Mol. Cell. Biol. 4: 1605–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines, G., Rossi, C., and Attardi, G. 1987. The excised leader of human cytochrome c oxidase subunit I mRNA which contains the origin of mitochondrial DNA light-strand synthesis accumulates in mitochondria and is polyadenylated. Mol. Cell. Biol. 7: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geenawalt, J.H. 1974. The isolation of outer and inner mitochondrial membranes. Methods Enzymol. 31: 310–323. [DOI] [PubMed] [Google Scholar]
- Golik, P., Szczepanek, T., Bartnik, E., Stepien, P.P., and Lazowska, J. 1995. The S. cerevisiae nuclear gene SUV3 encoding a putative RNA helicase is necessary for the stability of mitochondrial transcripts containing multiple introns. Curr. Genet. 28: 217–224. [DOI] [PubMed] [Google Scholar]
- Hancock, K., LeBlanc, A.J., Donze, D., and Hajduk, S.L. 1992. Identification of nuclear encoded precursor tRNAs within the mitochondrion of Trypanosoma brucei. J. Biol. Chem. 267: 23963–23971. [PubMed] [Google Scholar]
- Harris, M.E., Moore, D.R., and Hajduk, S.L. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265: 11368–11376. [PubMed] [Google Scholar]
- Hayman, M.L. and Read, L.K. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 274: 12067–12074. [DOI] [PubMed] [Google Scholar]
- Igo Jr., R.P., Weston, D.S., Ernst, N.L., Panigrahi, A.K., Salavati, R., and Stuart, K. 2002. Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukaryot. Cell 1: 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmer, D.P., Feagin, J.E., and Stuart, K. 1985. Diverse patterns of expression of the cytochrome c oxidase subunit I gene and unassigned reading frames 4 and 5 during the life cycle of Trypanosoma brucei. Mol. Cell. Biol. 5: 3041–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, C.Y. and Read, L.K. 2005. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol. Cell. Biol. 25: 1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, J., Tengler, U., and Binder, S. 2001. Transcript lifetime is balanced between stabilizing stem–loop structures and degradation-promoting polyadenylation in plant mitochondria. Mol. Cell. Biol. 21: 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowski, E.R. and Boucher, R.C. 2001. UTP as an extracellular signaling molecule. News Physiol. Sci. 16: 1–5. [DOI] [PubMed] [Google Scholar]
- Lupold, D.S., Caoile, A.G., and Stern, D.B. 1999. Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell 11: 1565–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margossian, S.P., Li, H., Zassenhaus, H.P., and Butow, R.A. 1996. The DExH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell 84: 199–209. [DOI] [PubMed] [Google Scholar]
- Militello, K.T. and Read, L.K. 1999. Coordination of kRNA editing and polyadenylation in Trypanosoma brucei mitochondria: Complete editing is not required for long poly(A) tract addition. Nucleic Acids Res. 27: 1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. UTP-dependent and -independent pathways of mRNA turnover in Trypanosoma brucei mitochondria. Mol. Cell. Biol. 20: 2308–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, J., Heuertz, R.M., and Zassenhaus, H.P. 1993. Isolation and characterization of an NTP-dependent 3′-exoribonuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 268: 7350–7357. [PubMed] [Google Scholar]
- Pelletier, M. and Read, L.K. 2003. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA 9: 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, M., Miller, M.M., and Read, L.K. 2000. RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res. 28: 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penschow, J.L., Sleve, D.A., Ryan, C.M., and Read, L.K. 2004. TbDSS-1, an essential Trypanosoma brucei exoribonuclease homolog that has pleiotropic effects on mitochondrial RNA metabolism. Eukaryot. Cell 3: 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitula, J., Ruyechan, W.T., and Williams, N. 2002. Two novel RNA binding proteins from Trypanosoma brucei are associated with 5S rRNA. Biochem. Biophys. Res. Commun. 290: 569–576. [DOI] [PubMed] [Google Scholar]
- Priest, J.W. and Hajduk, S.L. 2003. Trypanosoma brucei cytochrome c1 is imported into mitochondria along an unusual pathway. J. Biol. Chem. 278: 15084–15094. [DOI] [PubMed] [Google Scholar]
- Read, L.K., Myler, P.J., and Stuart, K. 1992. Extensive editing of both processed and preprocessed maxicircle CR6 transcripts in Trypanosoma brucei. J. Biol. Chem. 267: 1123–1128. [PubMed] [Google Scholar]
- Read, L.K., Stankey, K.A., Fish, W.R., Muthiani, A.M., and Stuart, K. 1994a. Developmental regulation of RNA editing and polyadenylation in four life cycle stages of Trypanosoma congolense. Mol. Biochem. Parasitol. 68: 297–306. [DOI] [PubMed] [Google Scholar]
- Read, L.K., Wilson, K.D., Myler, P.J., and Stuart, K. 1994b. Editing of Trypanosoma brucei maxicircle CR5 mRNA generates variable carboxy terminal predicted protein sequences. Nucleic Acids Res. 22: 1489–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S.G., Chock, P.B., and Stadtman, E.R. 1989. Regulation of Escherichia coli glutamine synthetase. Adv. Enzymol. Relat. Areas Mol. Biol. 62: 37–92. [DOI] [PubMed] [Google Scholar]
- Ryan, C.M., Militello, K.T., and Read, L.K. 2003. Polyadenylation regulates the stability of Trypanosoma brucei mitochondrial RNAs. J. Biol. Chem. 278: 32753–32762. [DOI] [PubMed] [Google Scholar]
- Schneider, A. 2001. Unique aspects of mitochondrial biogenesis in trypanosomatids. Int. J. Parasitol. 31: 1403–1415. [DOI] [PubMed] [Google Scholar]
- Schuster, G., Lisitsky, I., and Klaff, P. 1999. Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol. 120: 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, T.A. and Englund, P.T. 1995. The structure and replication of kinetoplast DNA. Annu. Rev. Microbiol. 49: 117–143. [DOI] [PubMed] [Google Scholar]
- Shen, B. and Goodman, H.M. 2004. Uridine addition after micro-RNA-directed cleavage. Science 306: 997. [DOI] [PubMed] [Google Scholar]
- Shi, H., Ullu, E., and Tschudi, C. 2004. Function of the Trypanosome Argonaute 1 protein in RNA interference requires the N-terminal RGG domain and arginine 735 in the Piwi domain. J. Biol. Chem. 279: 49889–49893. [DOI] [PubMed] [Google Scholar]
- Simpson, L., Sbicego, S., and Aphasizhev, R. 2003. Uridine insertion/ deletion RNA editing in trypanosome mitochondria: A complex business. RNA 9: 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin, M.L., Gunderson, J.H., Elwood, H.J., Alonso, R.A., and Peattie, D.A. 1989. Phylogenetic meaning of the kingdom concept: An unusual ribosomal RNA from Giardia lamblia. Science 243: 75–77. [DOI] [PubMed] [Google Scholar]
- Souza, A.E., Myler, P.J., and Stuart, K. 1992. Maxicircle CR1 transcripts of Trypanosoma brucei are edited and developmentally regulated and encode a putative iron–sulfur protein homologous to an NADH dehydrogenase subunit. Mol. Cell. Biol. 12: 2100–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, A.E., Shu, H.H., Read, L.K., Myler, P.J., and Stuart, K.D. 1993. Extensive editing of CR2 maxicircle transcripts of Trypanosoma brucei predicts a protein with homology to a subunit of NADH dehydrogenase. Mol. Cell. Biol. 13: 6832–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steege, D.A. 2000. Emerging features of mRNA decay in bacteria. RNA 6: 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, K. and Panigrahi, A.K. 2002. RNA editing: Complexity and complications. Mol. Microbiol. 45: 591–596. [DOI] [PubMed] [Google Scholar]
- Temperley, R.J., Seneca, S.H., Tonska, K., Bartnik, E., Bindoff, L.A., Lightowlers, R.N., and Chrzanowska-Lightowlers, Z.M. 2003. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum. Mol. Genet. 12: 2341–2348. [DOI] [PubMed] [Google Scholar]
- Ullu, E., Tschudi, C., and Chakraborty, T. 2004. RNA interference in protozoan parasites. Cell Microbiol. 6: 509–519. [DOI] [PubMed] [Google Scholar]
- Williams, N. and Frank, P.H. 1990. The mitochondrial ATP synthase of Trypanosoma brucei: Isolation and characterization of the intact F1 moiety. Mol. Biochem. Parasitol. 43: 125–132. [DOI] [PubMed] [Google Scholar]
- Yermovsky-Kammerer, A.E. and Hajduk, S.L. 1999. In vitro import of a nuclearly encoded tRNA into the mitochondrion of Trypanosoma brucei. Mol. Cell. Biol. 19: 6253–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]