Abstract
We systematically examined transcription and RNA-processing in mitochondria of the petite-negative fission yeast Schizosaccharomyces pombe. Two presumptive transcription initiation sites at opposite positions on the circular-mapping mtDNA were confirmed by in vitro capping of primary transcripts with guanylyl-transferase. The major promoter (Pma) is located adjacent to the 5′-end of the rnl gene, and a second, minor promoter (Pmi) upstream from cox3. The primary 5′-termini of the mature rnl and cox3 transcripts remain unmodified. A third predicted accessory transcription initiation site is within the group IIA1 intron of the cob gene (cobI1). The consensus promoter motif of S. pombe closely resembles the nonanucleotide promoter motifs of various yeast mtDNAs. We further characterized all mRNAs and the two ribosomal RNAs by Northern hybridization, and precisely mapped their 5′- and 3′-ends. The mRNAs have leader sequences with a length of 38 up to 220 nt and, in most instances, are created by removal of tRNAs from large precursor RNAs. Like cox2 and rnl, cox1 and cox3 are not separated by tRNA genes; instead, transcription initiation from the promoters upstream from rnl and cox3 compensates for the lack of tRNA-mediated 5′-processing. The 3′-termini of mRNAs and of SSU rRNA are processed at distinct, C-rich motifs that are located at a variable distance (1–15 nt) downstream from mRNA and SSU-rRNA coding regions. The accuracy of RNA-processing at these sites is sequence-dependent. Similar 3′-RNA-processing motifs are present in species of the genus Schizosaccharomyces, but not in budding yeasts that have functionally analogous A+T-rich dodecamer processing signals.
Keywords: mitochondrial promoters, tRNA punctuation model, mRNA, processing signals
INTRODUCTION
Mitochondria—the energy-producing organelles present in most eukaryotic cells—contain their own system to express the genetic information encoded in mitochondrial DNA (mtDNA). Since mitochondria derive from an α-proteo-bacterial ancestor, one might expect to find conserved eubacterial features in mitochondrial gene expression. However, while an overwhelming number of DNA motifs and protein factors are known to control nuclear transcription and RNA maturation, comparable mitochondrial elements are either difficult to recognize (because of the accelerated evolutionary rate of mtDNA), or totally missing because of radical changes in mitochondrial gene expression (Gray 1999).
A well-studied example is mitochondrial RNA polymerase. In most eukaryotes, the eubacterial-type RNA polymerase complex was replaced by a T3/T7 phage-like enzyme of uncertain evolutionary origin. This phage-like RNA polymerase promotes RNA synthesis from within short sequence motifs producing polycistronic transcripts (Cermakian et al. 1997; Gray and Lang 1998). The four subunits of a typical eubacterial α2ββ′σ RNA polymerase complex are encoded only in jakobid protist mtDNAs (rpoA–rpoD). However, even in these primitive mtDNAs, the sequences of promoter motifs do not appear to be eubacteria-like, and remain to be determined (Lang et al. 1997).
Mitochondrial promoters are best characterized in Saccharomyces cerevisiae, for which at least 19 transcription units have been demonstrated experimentally (Foury et al. 1998). Despite the high number of promoters, most transcripts are polycistronic, usually composed of two or more coding sequences including various combinations of rRNAs, tRNAs, and mRNAs. Transcription is initiated both in vivo and in vitro at the last A residue of the conserved nonanucleotide motif ATATAAGTA (Christianson et al. 1982; Osinga and Tabak 1982; Christianson and Rabinowitz 1983; Osinga et al. 1984b). A slightly different promoter motif (TTATAAGTA) is characteristic for tRNA gene transcription (Edwards et al. 1983; Tabak et al. 1983; Burke et al. 1987). Identical or very similar promoter sequences have been identified in mitochondria of Kluyveromyces lactis (Ragnini and Frontali 1994), Torulopsis glabrata (Clark-Walker et al. 1985; Koszul et al. 2003), Williopsis (Hansenula) mrakii (Drissi et al. 1993, 1994), and Yarrowia lipolytica (Kerscher et al. 2001). In contrast, mitochondrial promoters of the distantly related ascomycete fungus Neurospora crassa are only marginally similar to the nona-nucleotide motif of S. cerevisiae (Kubelik et al. 1990). Although several studies in S. cerevisiae have shown that the consensus sequence alone is sufficient to promote initiation of transcription in vitro (Biswas et al. 1985; Schinkel et al. 1986), the relative strength of these promoters is affected by the context of flanking sequences (Biswas and Getz 1986).
The function of the mitochondrial promoter region is to recruit not only RNA polymerase but also additional specificity factors that initiate, enhance, and control transcription rates. In S. cerevisiae, in vitro and in vivo experimental data suggest initial binding of the specificity factor (Mtf1p) to the core RNA polymerase (Rpo41p), a mechanism that resembles activation by the eubacterial σ factor. This preinitiation complex associates nonspecifically with mtDNA and walks along the DNA template scanning for a promoter. After binding to this target, the preinitiation complex induces conformational changes in the DNA at the promoter site, changes required for initiating RNA synthesis. Immediately following early stages of elongation by RNA polymerase, Mtf1p is released from the transcription complex (Shadel and Clayton 1993; Mangus et al. 1994; Cliften et al. 1997). Processing of the growing polycistronic messengers occurs simultaneously with transcription (Dieckmann and Staples 1994). In the absence of mechanisms for transcription modulation in mitochondria, RNA maturation (e.g., endonucleolytic cleavage, trimming of 5′-and 3′-ends, splicing) and degradation are key steps in the control of mitochondrial gene expression. In addition, in animals, plants, and in certain protist mitochondria, polyadenylation regulates mRNA stability (for review, see Gagliardi et al. 2004). Apparently, mitochondrial polyadenylation is absent in S. cerevisiae (Gagliardi and Leaver 1999; Gagliardi et al. 2001).
The tRNA sequences are among the most general and important mitochondrial RNA- processing signals. In polycistronic precursor RNAs, exonucleolytic cleavage of their 5′- and 3′-ends liberates flanking mRNA and rRNA molecules in a wide range of eukaryotes including S. cerevisiae, N. crassa, Aspergillus nidulans, and green and red algae (Burger et al. 1985; Dyson et al. 1989; Wolff and Kück 1996; Richard et al. 1998). This mechanism of RNA-processing is known as the “tRNA punctuation model.” The most advanced example was found in animal mitochondria, where this mechanism directly produces both 5′- and 3′-ends of most mRNA sequences (Ojala et al. 1980). In contrast, the 3′-termini of yeast mitochondrial mRNAs are specified by a dodecamer motif 5′-AAUAA(U/C)AUUCUU-3′ in the 3′-UTR region, which serves as a target for the mitochondrial exosome (mtExo), also known as the yeast mitochondrial degradosome (Osinga et al. 1984a; Hofmann et al. 1993).
In general, trans-acting mitochondrial RNA-processing factors are nuclear-encoded, except for the mtDNA-encoded intron maturases. In yeast, only three mitochondrial exoribonucleases are involved in RNA metabolism: the oligoribonuclease Ynt20p (Hanekamp and Thorsness 1999), the nonspecific ribonuclease Nuc1p (Vincent et al. 1988), and the yeast mitochondrial degradosome (Min et al. 1993). The degradosome is a protein complex with both the 3′–5′ exoribonuclease Dss1p and the RNA helicase Suv3p (Min et al. 1993; Dziembowski et al. 1998, 2003). RNA maturation and turnover are also controlled by several RNA-stabilizing proteins and translation activator proteins that bind to the 5′-UTL of mitochondrial RNAs (Wiesenberger et al. 1995; Wiesenberger and Fox 1997; Green-Willms et al. 1998; Chen et al. 1999; Islas-Osuna et al. 2002, 2003). Likewise, the 55-kDa RNA dodecamer sequence-binding protein (DBP) recognizes a dodecamer sequence in 3′-UTRs (Li and Zassenhaus 1999).
Taken together, current knowledge of mitochondrial transcription and RNA metabolism in S. cerevisiae and vertebrates is extensive, but little is known regarding the mechanisms in alternative model organisms. Here, we have chosen the petite-negative “fission yeast” Schizosaccharomyces pombe (S. pombe) as a model. Contrary to baker’s yeast, its circular-mapping mitochondrial genome is very stable, relatively small, and devoid of long repetitive sequences in intergenic regions (for recent reviews, see Bullerwell et al. 2003; Schäfer 2003). We demonstrate that fission yeast mitochondrial transcription and RNA-processing and regulation differ significantly from the budding yeast S. cerevisiae.
RESULTS
Mitochondrial transcription in S. pombe initiates from yeast-like nonanucleotide motifs
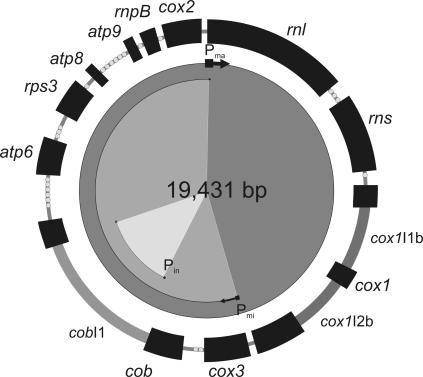
The mtDNA of S. pombe (Fig. 1 ▶) carries genes for two ribosomal RNAs (rnl, rns), a complete set of 25 tRNAs, the RNA-encoding RNase P RNA (rnpB) (Seif et al. 2003), and a total of eight proteins (Paquin et al. 1997; for review, see Schäfer 2003). In the strain used here, the cox1 gene contains two group I introns, whereas the cob gene is endowed with a single group IIA1 intron; all introns are not essential (Schäfer et al. 1991). Every one of the coding regions is on the same DNA strand. Since mitochondrial promoter motifs exhibit conservation among distantly related yeast species (see Introduction), we attempted to identify similar motifs in the S. pombe mtDNA.
FIGURE 1.
Genetic map of S. pombe mtDNA (strain 972h-). The nomenclature for the genes is as follows: rnl and rns: genes encoding large and small rRNAs; the 25 tRNA genes are indicated as dots; cox1, cox2, and cox3: genes encoding subunits 1, 2, and 3 of cytochrome c oxidase; atp6, atp8, and atp9: genes encoding subunits 6, 8, and 9 of ATPase; rps3: ribosomal protein of the small subunit of mitochondrial ribosomes (formerly urf a); rnpB: gene encoding RNA component of the mitochondrial RNase P; group I introns in cox1: (cox1I1b and cox1I2b) and group II intron in cob (cobI1) in dark gray bars. The location of the identified promoters (Pma, Pmi, and Pin) and the corresponding transcription units are indicated in the inner circles.
The analysis of S. pombe mtDNA identified four yeast-like promoter motifs in the unique direction of transcription: (1) the motif ATATATGTA (Pma) immediately adjacent to the rnl coding region; (2) TTATATGTG (Pmi) upstream from the cox3 gene; (3) ATATATGTG (Pin) 15 bp downstream of the cob group II intron 5′-splice junction; and finally (4) TTATATGTA at a position within the rps3 gene. These A+T-rich motifs may possibly occur by chance in the A+T-rich S. pombe mtDNA; therefore, experimental evidence is mandatory to pinpoint potential promoters with confidence.
To identify transcription initiation sites, we used a method that is widely used in yeast and plant mitochondria (Christianson and Rabinowitz 1983; Auchincloss and Brown 1989). The 5′-termini of primary transcripts (not capped in vivo) are labeled in vitro with the Vaccinia virus capping enzyme (guanylyltransferase) in the presence of [α-32P]GTP. Primary, strong initiation occurred at the last A of Pma, which corresponds to the first nucleotide position of the rnl-coding sequence (Fig. 2A,B ▶). Less efficient RNA synthesis initiates at Pmi corresponding to a position 38 nt upstream of cox3 (Fig. 2C ▶), and no signal was detected for the other two predicted promoters. However, primer extension analyses identified a minute fraction of RNA molecules starting at Pin, close to the 5′-end of the group II intron in cob (Fig. 2D ▶). Finally, primer extension analyses indicated that further 5′-processing of both rnl and cox3 RNAs does not take place. A small fraction of a stable cox1–cox3 transcript is likely initialized from Pma (see below). In conclusion, two of four predicted promoters exhibit significant transcriptional activity.
FIGURE 2.
(A) Mapping of transcription initiation sites by in vitro capping with guanylyl-transferase. Shown here are primary RNA molecules as products of transcriptional initiation that are specifically labeled by in vitro capping of RNA purified from intact, purified mitochondria with Vaccinia virus-derived guanylyltransferase in the presence of [α-32P]GTP. (B) Sequencing of in vitro capped rnl RNA. The labeled RNA (identified by direct exposure of an X-ray film) has been excised and purified by preparative PAGE, prior to sequencing. (C,D) Confirmation of the 5′-termini of cox3 and cobI1 RNAs by primer extension analysis (gel views from A.L.F. runs of reactions with primers specific for cox3 and cobI1).
Mapping of mitochondrial transcripts by Northern hybridization
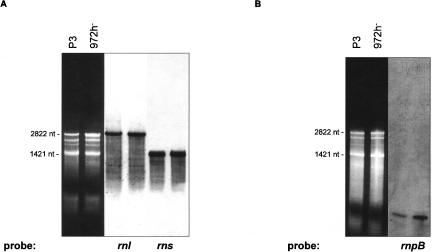
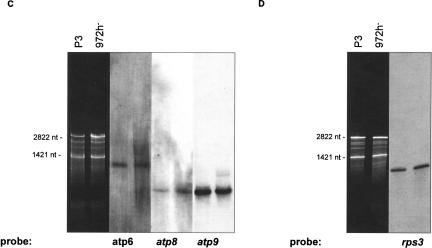
The S. pombe mitochondrial genome (unlike S. cerevisiae) is transcribed in two major units each producing a large precursor RNA. The excision of tRNAs, rRNAs, and mRNAs from the large precursor RNAs likely involves several RNA-processing mechanisms. To identify mRNAs and mature rRNA as well as processing intermediates, Northern hybridizations were performed using comprehensive sets of labeled oligonucleotides covering both DNA strands. As expected, (1) significant transcription occurred only from the predicted sense strand (data not shown). (2) The most abundant sense-strand RNAs are the two rRNAs with sizes of 2822 and 1422 nt (rnl, rns) (Fig. 3A ▶), as predicted from modeling of rRNA secondary structures (Lang et al. 1987; Trinkl et al. 1989). (3) The rnpB RNA was detected at a size of 274 nt (Fig. 3B ▶), confirming an earlier prediction using sequence and structure similarity to other mitochondrial RNase P RNAs, and precise mapping of 5′- and 3′-ends (Seif et al. 2003).
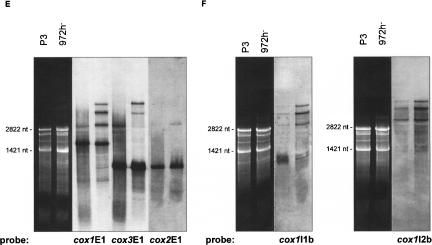
FIGURE 3.
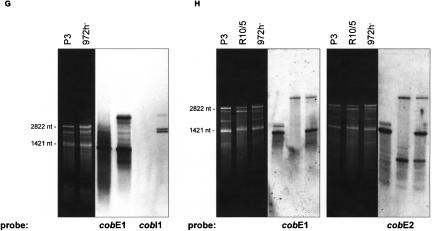
(A–H) Northern hybridizations with comprehensive sets of labeled oligonucleotides covering the sense DNA strand. (A) rnl, rns; (B) rnpB; (C) atp6, atp8, atp9; (D) rps3; (E) cox1, cox2, cox3; (F) cox1I1b/I2b; (G,H) cobE1/E2/I1. In cells grown under standard conditions (3% glucose), only small concentrations of RNA-processing intermediates are present (E–H). The most prominent type of RNA intermediates represent pre-mRNAs and splicing intermediates of the mosaic genes cox1 (E,F) and cob (G,H). Mitochondrial RNA prepared from an intron-less strain (P3) was used to identify mRNA (A–H, first lanes), and the mutant R10/5 defective in cobI1 splicing was used to identify cobI1 precursors (G,H, second lanes).
However, the size of some RNAs seemed to differ from their respective coding regions. One reason was the occurrence of a stable cox1–cox3 transcript, which is not further processed because of the lack of an RNA-processing signal in the intergenic region between cox1 and cox3. Our results imply that the cox1–cox3 cotranscripts are transcribed from the main promoter (upstream rnl), whereas the cox3 mRNA is initiated at the second, minor promoter Pmi upstream of cox3. While small concentrations of RNA-processing intermediates are present in stationary phase, glucose-grown cells (standard experiments with 3% glucose) (Fig. 3E–H ▶), these become more abundant under glucose-repression, but are virtually absent in glycerol or raffinose-grown cells (both carbon sources are known to induce no or only modest glucose repression in fission yeast) (data not shown).
Splicing of group I and group II introns
The cox1 gene in the wild-type strain of S. pombe contains two group I introns (cox1I1b, cox1I2b). Northern hybridization analysis indicates a highly complex RNA-processing pattern (Fig. 3E,F ▶). The smallest of the RNA molecules (~1800 nt) is cox1 mRNA. Larger RNA species are partially and fully spliced cox1 RNA precursors (Fig. 3F ▶), and some are cotranscripts with cox3 as already mentioned (~2800 nt: cox1+cox3 mRNAs; two species at ~4000 nt: cox1–cox3 plus cox1I1b, and cox1–cox3 plus cox1I2b; ~5000 nt: cox1–cox3 plus cox1I1b and cox1I2b). To enable identification of the mature cox1 RNA, mitochondrial RNA prepared from an intron-less strain (P3) was used in addition to RNA from strain 972h- (Fig. 3E ▶, lane 1; Fig. 3F ▶, lane 1).
With cob-specific probes, five RNA species were identified (Fig. 3G,H ▶). The cob mRNA correlates to the band at ~1400 nt while the smaller ~500-nt band hybridizes exclusively to a cob exon 2 probe. Two observations lead us to assume that this RNA might be either a splicing intermediate, or (less likely, because of the low promoter strength) arises via transcription initiation at Pin (within cobI1) and subsequent RNA-processing: (1) The band appears only in intron-containing strains, as seen in comparisons between an intron-less strain (P3: identification of mature cob RNA) (Fig. 3G ▶, lane 1; Fig. 3H ▶, lane 1), a mutant defective in cobI1 splicing (R10/5: identification of cobI1 precursors) (Fig. 3G ▶, lane 2; Fig. 3H ▶, lane 2) and the wild-type strain 972h- (Fig. 3G ▶, lane 3; Fig. 3H ▶, lane 3). (2) The size of the band in question (~500 nt) is close to that of cob exon 2 (480 nt).
The three larger bands (a double band of ~2500 nt and a single band at ~4000 nt) represent two species of the spliced group II intron RNA and the unspliced cob precursor RNA (Fig. 3G,H ▶). To identify the two intron RNAs as lariat, broken lariat, or circular RNA, we amplified cDNAs from the gel-purified RNAs, covering the 5′- and the 3′-end of the intron. The resulting cDNAs were cloned, and >20 independent clones were sequenced. The analysis of the cloned cDNAs obtained with both intron RNA species as templates verified that both types contained the entire 5′-and 3′-junctions (data not shown). Furthermore, primer extension experiments showed linear intron RNAs that end either exactly at the 5′-exon/intron junction or adjacent to Pin in the cobI1 (Fig. 2D ▶).
Transcription initiation and tRNA-processing generate 5′-ends of transcripts
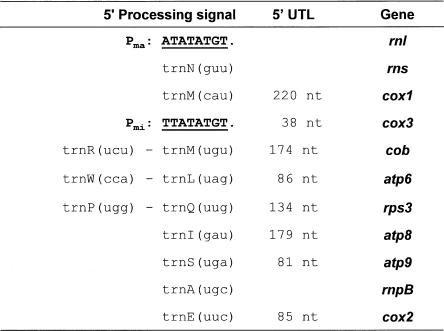
To identify mitochondrial 5′-RNA-processing mechanisms, we mapped the 5′-termini of all transcripts except tRNAs. As already noted, the 5′-ends of the large subunit rRNA and cox3 mRNA coincided with the transcription initiation sites Pma and Pmi, respectively. In numerous experiments using different RNA preparations and primers, we did not detect any cox3 RNA with longer 5′-UTLs. Thus, 5′-ends of rnl and cox3 RNAs are defined by the site of transcription initiation in promoters upstream of each coding region (Fig. 4 ▶).
FIGURE 4.
Mapping of the 5′-termini of mitochondrial transcripts by primer extension. The 5′-ends of mature RNAs coincide either with transcription initiation sites or with 3′-ends of upstream tRNAs.
In contrast, the 5′-termini of most RNA molecules (rns, rps3, cox1, cox2, cob, atp6, atp8, atp9) have leader sequences of 81 to 220 nt that precisely coincide with the 3′-ends of upstream tRNAs (Fig. 4 ▶). Thus, tRNA punctuation creates the majority of RNA 5′-ends. Only a small fraction of 5′-termini remains unprocessed under standard growth conditions. However, an increase in growth temperature from 30°C to 37°C or growth at high glucose concentrations (10% w/v) results in an accumulation of precursors that retained the upstream (e.g., atp9 transcripts with a tRNA cluster) or downstream tRNAs (rnpB RNA precursors).
The 3′-termini of mature mitochondrial transcripts are - characterized by a C-rich motif
Previously, we used S1 Nuclease protection assays to locate the 3′-ends of S. pombe rRNAs and mRNAs (Lang et al. 1983; Trinkl et al. 1989). In many cases, this assay failed to recover precise nucleotide positions because of ”breezing” of AT-rich terminal sequences. An alternative strategy is to identify the 3′-ends in cDNAs produced from circular transcripts obtained after intramolecular RNA ligation of 5′ and 3′ RNA termini with T4 RNA ligase.
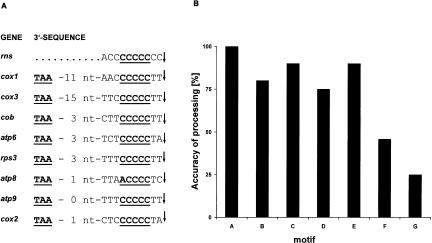
Neither the mtP-RNA nor the LSU rRNA contained an obvious conserved signal motif at its 3′-end. The processing of mtP-RNA is mediated by tRNA punctuation at both ends (Seif et al. 2003). Yet, the specific signal responsible for 3′-processing of LSU rRNA is unknown. In contrast, all mRNAs and the SSU rRNA end 2 nt downstream of a five-C-residue motif (C-core motif) that occurs at a variable distance (1–15 nt) from the end of coding regions (Fig. 5A ▶). Processing at the motif 5′-(CC)-CCCCC-(CC)-3′ (SSU rRNA 3′-end) is precise. In other cases, the accuracy of 3′-RNA-processing is sequence-dependant with ragged ends in ~20%–75% of analyzed RNA molecules (Fig. 5B ▶). The C-core motif is not present at the 3′-end of mobile group I and group II intron ORFs. Despite the sequence-dependent 3′ extensions that are the result of inaccurate 3′-RNA-processing, we found no evidence for nontemplate-specific poly(A) extensions in S. pombe mitochondrial mRNAs. It is interesting that similar 3′-terminal processing signals exist in mitochondrial genomes throughout the Schizosaccharomycetales, that is, including species such as Schizosaccharomyces octosporus and Schizosaccharomyces ja-ponicus var. japonicus (Bullerwell et al. 2003). As in S. pombe, the motif is absent at the end of the rnl-coding region in both yeasts. Also, the atp6 gene in S. japonicus var. japonicus lacks this motif.
FIGURE 5.
(A) Mapping of the 3′-termini of mitochondrial transcripts. The 3′-UTR region of almost all mitochondrial transcripts end 2 nt downstream to the conserved C-core motif. (B) Accuracy of 3′-processing at various C-core motifs. The accuracy of the 3′-processing in vivo was determined by measuring the frequency of clones with imprecise downstream processing among at least 10 clones from three independent reactions. The motifs are: A = CC+CC CCC+CC (rns); B = AC+CCCCC+TT (cox1); C = TC+CCCCC+TT (cox3); D = TC+CCC CC+TA (cox2); E = CT+CCCCC+TA (atp6); F = TT+CCCCC+TT (cob, rps3, and atp9); G = TA+ACCCC+TC (atp8). The C-core motif is lacking at the 3′-ends of rnl and rnpB.
DISCUSSION
This work presents precise mapping of the S. pombe mitochondrial transcriptome, and shows by in vitro capping and primer extension experiments that this mitochondrial genome is transcribed in two transcription units. The results further uncover the mechanisms that are responsible for processing of the large RNA precursors into mature, stable RNAs.
Nonanucleotide motif promoters mark transcription units on S. pombe mtDNA
The S. pombe mitochondrial promoters are similar to those of S. cerevisiae, both in sequence, and in using transcription initiation sites from within the same position of the conserved nonanucleotide motif. There is no evidence of transcription initiation at a motif located within the rps3 gene, despite its sequence identity to tRNA-specific promoters in S. cerevisiae. This indicates high promoter specificity for the mitochondrial RNA polymerase of S. pombe, although the four nucleotide positions in the nonanucleotide motif required for efficient initiation of transcription in S. cerevisiae (i.e., -2G, -4A, -6A, and -7T) are also conserved in fission yeast. In addition, the strongest S. pombe promoter (Pma, required for transcription of high levels of the two rRNA subunits) also contains a purine at position +2 followed by a pyrimidine residue at +3, both known to enhance in vitro transcription rates in S. cerevisiae (Biswas and Getz 1986). In accordance with this rule, the second minor promoter (Pmi, upstream cox3) has instead two purine residues at these two positions, and the potential, very weak cobI1 promoter (Pin) has mismatches in both positions. Pin is not likely used for transcription of structural genes. Instead, transcription from this promoter might be instrumental for intron propagation (Schäfer et al. 2003).
The low density of promoters in S. pombe is unexpected, since substantially higher densities are observed in budding yeasts such as S. cerevisiae, Y. lipolytica, K. lactis and Candida (Torulopsis) glabrata. In this respect, S. pombe more resembles animals (Tracy and Stern 1995), Candida parapsilosis (Nosek et al. 2004), Aspergillus nidulans (Dyson et al. 1989), N. crassa (Burger et al. 1985; Kubelik et al. 1990), the chlorophytic unicellular alga Prototheca wickerhamii (Wolff and Kück 1996), and the red alga Chondrus crispus (Richard et al. 1998). This broad evolutionary distribution of species with only a few mitochondrial promoters suggests that S. pombe retained a more ancestral mode of transcription and RNA-processing, and that high mitochondrial promoter density is a secondary adaptation.
Maturation at the 5′-termini of transcripts follows the tRNA punctuation model
The fission yeast mitochondrial genome gives rise to two long primary transcripts. Initiation at the major promoter (Pma) likely covers the entire 19.4-kb genome because of an apparent lack of transcription termination (i.e., the existence of cox1–cox3 cotranscripts). The minor promoter (Pmi) yields a transcript that is responsible for synthesis of mature cox3 mRNA and presumably other RNAs following down-stream to Pmi on the genome map. Based on these long precursor RNA molecules (presumably 19.4 and 10.5 kb as predicted from the mtDNA map), the 5′-ends of most mature RNAs are generated by tRNA-mediated processing of precursor RNAs. Consequently, the protein-coding genes of S. pombe have 5′-UTL sequences of 38 up to 220 nt length. However, the 5′-processing of RNAs is strongly affected by growth conditions.
The intergenic region between cox1 and cox3 is an exception, because the intergenic region does not contain a tRNA gene or a known RNA-processing or transcription termination signal. We interpret the observed cox1–cox3 precursor RNA as a functionally inactive form, and assume that the biologically active cox3 mRNA is synthesized by initiation at the Pmi promoter, followed by 3′-processing at a C-core motif, as discussed below. In other words, the Pmi promoter is essential for the formation of a unique cox3 mRNA, and compensates for the lack of a tRNA-processing signal between cox1 and cox3.
3′-processing involves a conserved processing motif
Recognition and processing at the 3′-end of transcripts is similar in S. pombe and S. cerevisiae: the 3′-UTR region of nearly all mitochondrial transcripts in fission yeast contains a conserved motif (a distinctive run of several Cs, the C-core motif) that is analogous to the dodecamer motif in S. cerevisiae mtDNA. In S. pombe, this motif occurs at a variable distance 1–15 nt downstream of all protein-coding regions and the small subunit rRNA sequence. We have not succeeded to identify conserved RNA secondary structure motifs including the C-core motif, excluding the interpretation of a bacteria-like transcription termination structure. As shown in Figure 5B ▶, the accuracy of the 3′-processing in vivo decreases with the overall GC-content within the 3′-terminal sequence. Additional support for the importance of this sequence motif in 3′-processing comes from its conservation in all members of the Schizosaccharomycetales (Bullerwell et al. 2003).
Intron splicing
In previous studies, we could show that splicing of both group I introns in the cox1 gene (cox1I1b, cox1I2b) and the group IIA1 intron in the cob gene (cobI1) are maturase-dependent (Schäfer et al. 1991, 1994). Because translation of the cox1I2b maturase might depend on the removal of the cox1I1b intron sequence (i.e., translation of a joint exon1–exon2-I1b ORF), splicing of cox1I2b might occur subsequently to cox1I1b. However, we did not find a dependence on the intron splicing pathway, indicating that translation might be initiated from independent sites.
Lack of mRNA polyadenylation in S. pombe mitochondria
We did not find any evidence for transcripts with poly(A) extensions in fission yeast mitochondria. Polyadenylation of mRNAs was reported in prokaryotes, the cytoplasm of eukaryotes, and some of their organelles. While polyadenylation in the cytoplasm of eukaryotes and in human mitochondria stabilizes mRNA, steady-state levels of polyadenylated mRNAs are low in prokaryotes and in plant organelles, serving to tag mRNAs for degradation (Coburn and Mackie 1999; Gagliardi and Leaver 1999). We believe that mRNA concentrations in S. pombe mitochondria are not controlled by degradation as in plants, but are stabilized as in metazoa, although via a C-core motif rather than by polyadenylation.
Conclusion
The organization of the transcription system in fission yeast mitochondria is simple but efficient. Only a small subset of mechanisms involved in RNA maturation and control of transcript concentrations is present, thereby limiting the necessity to import nuclear-encoded factors. The S. pombe mitochondrial RNA-processing mechanisms have changed little with reference to a postulated ancestral state, similarly found in plants, protists, several non-yeast fungi, and animals. This system is characterized by the synthesis of large RNA precursors from few phage-like promoters, and RNA-processing by tRNA punctuation. It will be interesting to trace its evolutionary pathway within all fungal lineages, and to identify the origin of the dramatic differences in transcript maturation seen in budding yeasts.
MATERIALS AND METHODS
Strains
The fission yeast strain 972h- is a wild-type strain from the Leupold collection. Strain R10/5 is a cob intron mutant of a strain that was constructed by elimination of all group I introns from the mtDNA of strain 972h-. A point mutation at nucleotide position 2734 blocks splicing (presumably as a consequence of an exchange of the proline to a threonine residue close to the end of domain X of the cobI1 ORF) (Schäfer et al. 1991), and thus results in an accumulation of cob precursor RNAs. The strain P3 lacks all mi-tochondrial introns (Schäfer et al. 1991) and was used to identify RNA splicing intermediates.
Purification of mitochondrial RNA
All fission yeast strains were grown in a medium containing 2% yeast extract, 3% (w/v), 5% (w/v), or 10% (w/v) glucose or 3% (w/v) raffinose or glycerol. Cells from early-stationary-phase cultures were harvested and cultured for 1–2 h in a fresh medium containing 0.5% glucose (w/v). Cells were collected and washed sequentially in EDTA buffer (50 mM EDTA at pH 7.5), sterile distilled water, and SEP buffer 1 (750 mM sorbitol, 5 mM EDTA, 100 mM citrate-phosphate at pH 7.5). Cell walls were digested in SEP buffer containing a mixture of 20 mg of Lysing Enzyme (Sigma L-2265) and 20 mg Kitarase (KI Chemicals) per gram of cells (wet weight). Protoplasts were collected, washed twice in SEP buffer, and resuspended in 15 mL of ice-cold SEP buffer. Then 50 mg of mitochondria (wet weight) was resuspended in RNase-free PBS buffer (58 mM Na2HPO4, 17 mM NaH2PO4, 69 mM NaCl at pH 7.0), and mtRNA was purified further by use of High Pure RNA isolation columns (Roche Diagnostics).
Northern hybridization
For Northern experiments, 10–25 μg of purified mitochondrial RNA was separated on a urea-agarose gel (Locker 1979), and transferred on a positively charged nylon membrane (Roche Diagnostics). The blots were hybridized with a set of 5′-digoxigenin end-labeled 50-mer oligonucleotides complementary either to the sense strand of each coding region, or to complementary regions of the antisense strand (defined as the position of the first base at the 3′-end; position +1 = the first base of rnl on the mtDNA map of strain 972h-): DIG_rnl-c00001; DIG_rns-c03235; DIG_cox1-c4886; DIG_cox1I2b-c06860; DIG_cox3-c09075; DIG_cobE1-c10180; DIG_cobI1-c10870; DIG_cobE2-c13390; DIG_atp6-c14877; DIG_rps3-c16035; DIG_atp8-c16914; DIG_atp6-c17851; DIG_rnpB-c18230; DIG_cox2-c18561.
The membranes were exposed on a Fuji LAS-1000 imager or on X-ray films. The RNA molecular weight marker II (Roche Diagnostics) was used as a standard.
In vitro capping with guanylyltransferase
RNA used for in vitro capping experiments was purified from intact mitochondria that were purified by sucrose gradient centrifugation (to avoid non-mitochondrial RNA contaminants such as 5S rRNAs). The mitochondrial lysate (0.5% SDS in a buffer containing 10 mM Tris and 1 mM EDTA at pH 8) was deproteinized by the addition of 1 M NaCl and incubated for 1 h on ice, and the SDS–protein complex was removed by centrifugation (5′, 15,000g). Total nucleic acids were ethanol-precipitated, redissolved in TE buffer (10 mM Tris and 1 mM EDTA at pH 8), adjusted to 2 M LiCl, and cooled for ~1 h on ice. Next, the precipitated RNAs were recuperated by centrifugation (10 min at 15,000g), resuspended in TE buffer, and precipitated a second time with 2 M LiCl. The resulting RNA pellet is washed repeatedly with 80% ethanol, resuspended in TE buffer at ~500 μg/mL, and placed in storage at −80°C.
In vitro capping of mitochondrial RNAs followed published protocols (Christianson and Rabinowitz 1983; Auchincloss and Brown 1989; Brown et al. 1991) with slight modifications. Approximately 10–50 μg of purified mitochondrial RNA was added to 50 μL of a capping buffer (50 mM Tris-HCl at pH 7.9, 1.25 mM MgCl2; 60 mM KCl, 0.1 mM S-adenosyl-methionine, 2.5 mM DTT; 100 μCi [α-32P]GTP, 0.6 unit of pyrophosphatase, and 8 units of guanylyltransferase. After incubation for 1 h at 37°C, unincorporated [α-32P]GTP and salts were removed on a Sephadex G-50 column and the RNA was ethanol-precipitated, centrifuged, and washed with 80% ethanol. After centrifugation, the resulting pellet was dissolved in 80% formamide and loaded on 5% acrylamide gels. The labeled RNA was excised from the gel, recuperated by electroelution, concentrated by ethanol precipitation, and finally sequenced.
Determination of 5′-ends by primer extension analysis
In our high-resolution, nonradioactive primer extension procedure to precisely map the 5′-ends of all mitochondrial transcripts, 500 ng of purified mitochondrial RNA served as template for reverse transcription with MMLV reverse transcriptase (Promega). First-strand cDNA synthesis was performed with CY5-labeled primers annealing ~100 nt downstream of the 5′-end of coding regions (oligonucleotides are named according to the position of the first base of the 3′-end; position +1 = first base of rnl on mtDNA map of strain 972h-): CY5_rnl-c00101; CY5_rns-c03234; CY5_cox1-c04978; CY5_cox3-c09076; CY5_cobE1-c10310; CY5_cobI1-c108710; CY5_atp6-c14877; CY5_rps3-c16035; CY5_atp8-c17021; CY5_atp9-c17418; CY5_rnpB-c18230; CY5_cox2-c18644.
After treatment with RNase H to degrade the RNA in the DNA–RNA heteroduplexes, 100, 200, and 500 ng of the single-stranded cDNAs were separated in a sequencing gel (A.L.F., Amersham Biosciences). Marker sequencing reactions using the same CY5-labeled primer were separated adjacent to the primer extension reaction. Fragment length analysis of the primer extension products was determined as run time of the Cy5 signal peak using ALFwin software. The precise 5′-end of each cDNA was then recovered by direct comparison with a cycle sequencing reaction obtained with the same primer used for the primer extension, on a double-stranded DNA template covering the same sequence.
Mapping of RNA 3′-termini
For mapping of RNA 3′-termini, 500 ng of purified mitochondrial RNA was ligated together with 1 U of T4 RNA ligase (Roche Diagnostics), in a buffer as recommended by the supplier (Roche, Diagnostics). Circular RNAs were used as a template for RT-PCR amplifications with suitable primer pairs. Resulting amplification products were ligated into pUC19, and at least 10 clones from three independent amplification reactions were sequenced. The 3′-ends were inferred by comparison of the sequence with the published mtDNA sequence (accession no. X54421).
Mapping of branch point in group II intron RNA
RNAs were isolated from the two intron RNA bands that were separated by urea agarose gel electrophoresis (see above). These RNAs served as templates in RT-PCR. First-strand cDNA synthesis was carried out at 42°C for 30 min, using 200 ng of intron RNA, an antisense primer complementary to a position 121 nt apart from the 5′-end of the intron sequence, and AMV reverse transcriptase (GIBCO BRL), following the manufacturer’s protocol. After heat inactivation of the enzyme, 3 U of RNase H (GIBCO BRL) was added, and digestion was carried out for 20 min at 37°C. Then 5 μL of the first-strand mixture was PCR-amplified in a 50-μL assay, containing 30 pmol of antisense primer (complementary to a position 121 nt apart from the 5′-end of the intron) and 30 pmol of sense primer (identical to the sequence 212 nt apart from the 3′-end of the cob intron), PCR reaction buffer (1.5 mM MgCl2, 10 mM Tris-HCl at pH 9.0, 50 mM KCl, 0.1% Triton X-100, 200 μM dNTPs), and 2.5 U of Taq DNA Polymerase (Promega). After preincubation for 5 min at 95°C, 30 cycles of 1 min at 95°C, 30 sec at 55°C, 30 sec at 72°C, and a final step of 7 min at 72°C were performed. The resulting 357-bp-long cDNA fragment was cloned into pUC19, and at least 20 independent clones were subsequently sequenced on an ALF sequencer.
Acknowledgments
We thank A. Hauth for comments on the manuscript, and A. Schreer and K.W. Prohl for their skilled technical assistance. Salary and interaction support from the Canadian Institutes of Health Research (MOP 42475 to B.F.L.) and the Canadian Institute for Advanced Research (CIAR to B.F.L.), and supply of laboratory equipment and informatics infrastructure by Genome Quebec/ Canada are gratefully acknowledged.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7252205.
REFERENCES
- Auchincloss, A.H. and Brown, G.G. 1989. Soybean mitochondrial transcripts capped in vitro with guanylyltransferase. Biochem. Cell Biol. 67: 315–319. [Google Scholar]
- Biswas, T.K. and Getz, G.S. 1986. Nucleotides flanking the promoter sequence influence the transcription of the yeast mitochondrial gene coding for ATPase subunit 9. Proc. Natl. Acad. Sci. 83: 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, T.K., Edwards, J.C., Rabinowitz, M., and Getz, G.S. 1985. Characterization of a yeast mitochondrial promoter by deletion mutagenesis. Proc. Natl. Acad. Sci. 82: 1954–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G.G., Auchincloss, A.H., Covello, P.S., Gray, M.W., Menassa, R., and Singh, M. 1991. Characterization of transcription initiation sites on the soybean mitochondrial genome allows identification of a transcription-associated sequence motif. Mol. Gen. Genet. 228: 345–355. [DOI] [PubMed] [Google Scholar]
- Bullerwell, C.E., Leigh, J., Forget, L., and Lang, B.F. 2003. A comparison of three fission yeast mitochondrial genomes. Nucleic Acids Res. 31: 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger, G., Helmer Citterich, M., Nelson, M.A., Werner, S., and Macino, G. 1985. RNA processing in Neurospora crassa mitochondria: Transfer RNAs punctuate a large precursor transcript. EMBO J. 4: 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J.M., Belfort, M., Cech, T.R., Davies, R.W., Schweyen, R.J., Shub, D.A., Szostak, J.W., and Tabak, H.F. 1987. Structural conventions for group I introns. Nucleic Acids Res. 15: 7217–7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian, N., Ikeda, T.M., Miramontes, P., Lang, B.F., Gray, M.W., and Cedergren, R. 1997. On the evolution of the single-subunit RNA polymerases. J. Mol. Evol. 45: 671–681. [DOI] [PubMed] [Google Scholar]
- Chen, W., Islas-Osuna, M.A., and Dieckmann, C.L. 1999. Suppressor analysis of mutations in the 5′-untranslated region of COB mRNA identifies components of general pathways for mitochondrial mRNA processing and decay in Saccharomyces cerevisiae. Genetics 151: 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, T. and Rabinowitz, M. 1983. Identification of multiple transcriptional initiation sites on the yeast mitochondrial genome by in vitro capping with guanylyltransferase. J. Biol. Chem. 258: 14025–14033. [PubMed] [Google Scholar]
- Christianson, T., Edwards, J.C., Levens, D., Locker, J., and Rabinowitz, M. 1982. Transcriptional initiation and processing of the small ribosomal RNA of yeast mitochondria. J. Biol. Chem. 257: 6494–6500. [PubMed] [Google Scholar]
- Clark-Walker, G.D., Evans, R.J., Hoeben, P., and McArthur, C.R. 1985. Basis of diversity in yeast mitochondrial DNAs. In Achievements and perspectives of mitochondrial research 2 (eds. E.C. Quagliariello et al.), pp. 71–78. Science Publishers, Amsterdam, New York.
- Cliften, P.F., Park, J.Y., Davis, B.P., Jang, S.H., and Jaehning, J.A. 1997. Identification of three regions essential for interaction between a σ-like factor and core RNA polymerase. Genes & Dev. 11: 2897–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn, G.A. and Mackie, G.A. 1999. Degradation of mRNA in Escherichia coli: An old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 62: 55–108. [DOI] [PubMed] [Google Scholar]
- Dieckmann, C.L. and Staples, R.R. 1994. Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. Int. Rev. Cytol. 152: 145–181. [DOI] [PubMed] [Google Scholar]
- Drissi, R., Sor, F., and Fukuhara, H. 1993. DNA sequences coding for the ribosomal small subunit RNA and valyl tRNA from the linear mitochondrial genome of the yeast Williopsis mrakii. Nucleic Acids Res. 21: 2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi, R., Sor, F., Nosek, J., and Fukuhara, H. 1994. Genes of the linear mitochondrial DNA of Willopsis mrakii: Coding sequences for a maturase-like protein, a ribosomal protein var1 homologue, cytochrome oxidase subunit 2 and methionyl tRNA. Yeast 10: 391–398. [DOI] [PubMed] [Google Scholar]
- Dyson, N.J., Brown, T.A., Ray, J.A., Waring, R.B., Scazzocchio, C., and Davies, R.W. 1989. Processing of mitochondrial RNA in Aspergillus nidulans. J. Mol. Biol. 208: 587–599. [DOI] [PubMed] [Google Scholar]
- Dziembowski, A., Malewicz, M., Minczuk, M., Golik, P., Dmochowska, A., and Stepien, P.P. 1998. The yeast nuclear gene DSS1, which codes for a putative RNase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol. Gen. Genet. 260: 108–114. [DOI] [PubMed] [Google Scholar]
- Dziembowski, A., Piwowarski, J., Hoser, R., Minczuk, M., Dmochowska, A., Siep, M., van der, Spek H., Grivell, L.A., and Stepien, P.P. 2003. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 278: 1603–1611. [DOI] [PubMed] [Google Scholar]
- Edwards, J.C., Christianson, T., Mueller, D., Biswas, T.K., Levens, D., Li, D., Wettstein, J., and Rabinowitz, M. 1983. Initiation and transcription of yeast mitochondrial RNA. In Nucleo-mitochondrial interactions (eds. R.J. Schweyen et al.), pp. 69–78. Walter de Gruyter, Berlin.
- Foury, F., Roganti, T., Lecrenier, N., and Purnelle, B. 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440: 325–331. [DOI] [PubMed] [Google Scholar]
- Gagliardi, D. and Leaver, C.J. 1999. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 18: 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi, D., Perrin, R., Marechal-Drouard, L., Grienenberger, J.M., and Leaver, C.J. 2001. Plant mitochondrial polyadenylated mRNAs are degraded by a 3′- to 5′-exoribonuclease activity, which proceeds unimpeded by stable secondary structures. J. Biol. Chem. 276: 43541–43547. [DOI] [PubMed] [Google Scholar]
- Gagliardi, D., Stepien, P.P., Temperley, R.J., Lightowlers, R.N., and Chrzanowska-Lightowlers, Z.M. 2004. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 20: 260–267. [DOI] [PubMed] [Google Scholar]
- Gray, M.W. 1999. Evolution of organellar genomes. Curr. Opin. Genet. Dev. 9: 678–687. [DOI] [PubMed] [Google Scholar]
- Gray, M.W. and Lang, B.F. 1998. Transcription in chloroplasts and mitochondria: A tale of two polymerases. Trends Microbiol. 6: 1–3. [DOI] [PubMed] [Google Scholar]
- Green-Willms, N.S., Fox, T.D., and Costanzo, M.C. 1998. Functional interactions between yeast mitochondrial ribosomes and mRNA 5′ untranslated leaders. Mol. Cell. Biol. 18: 1826–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanekamp, T. and Thorsness, P.E. 1999. YNT20, a bypass suppressor of yme1 yme2, encodes a putative 3′–5′ exonuclease localized in mitochondria of Saccharomyces cerevisiae. Curr. Genet. 34: 438–448. [DOI] [PubMed] [Google Scholar]
- Hofmann, T.J., Min, J., and Zassenhaus, H.P. 1993. Formation of the 3′ end of yeast mitochondrial mRNAs occurs by site-specific cleavage two bases downstream of a conserved dodecamer sequence. Yeast 9: 1319–1330. [DOI] [PubMed] [Google Scholar]
- Islas-Osuna, M.A., Ellis, T.P., Marnell, L.L., Mittelmeier, T.M., and Dieckmann, C.L. 2002. Cbp1 is required for translation of the mitochondrial cytochrome b mRNA of Saccharomyces cerevisiae. J. Biol. Chem. 277: 37987–37990. [DOI] [PubMed] [Google Scholar]
- Islas-Osuna, M.A., Ellis, T.P., Mittelmeier, T.M., and Dieckmann, C.L. 2003. Suppressor mutations define two regions in the Cbp1 protein important for mitochondrial cytochrome b mRNA stability in Saccharomyces cerevisiae. Curr. Genet. 43: 327–336. [DOI] [PubMed] [Google Scholar]
- Kerscher, S., Durstewitz, G., Casaregola, S., Gaillardin, C., and Brandt, U. 2001. The complete mitochondrial genome of Yarrowia lipolytica. Comp. Funct. Genomics 2: 80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul, R., Malpertuy, A., Frangeul, L., Bouchier, C., Wincker, P., Thierry, A., Duthoy, S., Ferris, S., Hennequin, C., and Dujon, B. 2003. The complete mitochondrial genome sequence of the pathogenic yeast Candida (Torulopsis) glabrata. FEBS Lett. 534: 39–48. [DOI] [PubMed] [Google Scholar]
- Kubelik, A.R., Kennell, J.C., Akins, R.A., and Lambowitz, A.M. 1990. Identification of Neurospora mitochondrial promoters and analysis of synthesis of the mitochondrial small rRNA in wild-type and the promoter mutant [poky]. J. Biol. Chem. 265: 4515–4526. [PubMed] [Google Scholar]
- Lang, B.F., Ahne, F., Distler, S., Trinkl, H., Kaudewitz, F., and Wolf, K. 1983. Sequence of the mitochondrial DNA, arrangement of genes and processing of their transcripts in Schizosaccharomyces pombe. In Mitochondria 1983 (eds. R.J. Schweyen et al.), pp. 313–324. Walter de Gruyter, Berlin, Germany.
- Lang, B.F., Cedergren, R., and Gray, M.W. 1987. The mitochondrial genome of the fission yeast, Schizosaccharomyces pombe. Sequence of the large-subunit ribosomal RNA gene, comparison of potential secondary structure in fungal mitochondrial large-subunit rRNAs and evolutionary considerations. Eur. J. Biochem. 169: 527–537. [DOI] [PubMed] [Google Scholar]
- Lang, B.F., Burger, G., O’Kelly, C.J., Cedergren, R., Golding, G.B., Lemieux, C., Sankoff, D., Turmel, M., and Gray, M.W. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387: 493–497. [DOI] [PubMed] [Google Scholar]
- Li, H. and Zassenhaus, H.P. 1999. Purification and characterization of an RNA dodecamer sequence binding protein from mitochondria of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 261: 740–745. [DOI] [PubMed] [Google Scholar]
- Locker, J. 1979. Analytical and preparative electrophoresis of RNA in agarose-urea. Anal. Biochem. 98: 358–367. [DOI] [PubMed] [Google Scholar]
- Mangus, D.A., Jang, S.H., and Jaehning, J.A. 1994. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J. Biol. Chem. 269: 26568–26574. [PubMed] [Google Scholar]
- Min, J., Heuertz, R.M., and Zassenhaus, H.P. 1993. Isolation and characterization of an NTP-dependent 3′-exoribonuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 268: 7350–7357. [PubMed] [Google Scholar]
- Nosek, J., Novotna, M., Hlavatovicova, Z., Ussery, D.W., Fajkus, J., and Tomaska, L. 2004. Complete DNA sequence of the linear mitochondrial genome of the pathogenic yeast Candida parapsilosis. Mol. Genet. Genomics 272: 173–180. [DOI] [PubMed] [Google Scholar]
- Ojala, D., Merkel, C., Gelfand, R., and Attardi, G. 1980. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell 22: 393–403. [DOI] [PubMed] [Google Scholar]
- Osinga, K.A. and Tabak, H.F. 1982. Initiation of transcription of genes for mitochondrial ribosomal RNA in yeast: Comparison of the nucleotide sequence around the 5′-ends of both genes reveals a homologous stretch of 17 nucleotides. Nucleic Acids Res. 10: 3617–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osinga, K.A., De Vries, E., Van der Horst, G., and Tabak, H.F. 1984a. Processing of yeast mitochondrial messenger RNAs at a conserved dodecamer sequence. EMBO J. 3: 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osinga, K.A., De Vries, E., Van der Horst, G.T., and Tabak, H.F. 1984b. Initiation of transcription in yeast mitochondria: Analysis of origins of replication and of genes coding for a messenger RNA and a transfer RNA. Nucleic Acids Res. 12: 1889–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin, B., Laforest, M.-J., Forget, L., Roewer, I., Wang, Z., Longcore, J., and Lang, B.F. 1997. The fungal mitochondrial genome project: Evolution of fungal mitochondrial genomes and their gene expression. Curr. Genet. 31: 380–395. [DOI] [PubMed] [Google Scholar]
- Ragnini, A. and Frontali, L. 1994. Ordered processing of the polygenic transcripts from a mitochondrial tRNA gene cluster in K. lactis. Curr. Genet. 25: 342–349. [DOI] [PubMed] [Google Scholar]
- Richard, O., Bonnard, G., Grienenberger, J.M., Kloareg, B., and Boyen, C. 1998. Transcription initiation and RNA processing in the mitochondria of the red alga Chondrus crispus: Convergence in the evolution of transcription mechanisms in mitochondria. J. Mol. Biol. 283: 549–557. [DOI] [PubMed] [Google Scholar]
- Schäfer, B. 2003. Genetic conservation versus variability in mitochondria: The architecture of the mitochondrial genome in the petite negative yeast Schizosaccharomyces pombe. Curr. Genet. 43: 311–326. [DOI] [PubMed] [Google Scholar]
- Schäfer, B., Merlos-Lange, A.M., Anderl, C., Welser, F., Zimmer, M., and Wolf, K. 1991. The mitochondrial genome of Schizosaccharomyces pombe: Inability of all introns to splice autocatalytically, and construction of an intronless genome. Mol. Gen. Genet. 225: 158–167. [DOI] [PubMed] [Google Scholar]
- Schäfer, B., Wilde, B., Massardo, D.R., Manna, F., Del Giudice, L., and Wolf, K. 1994. A mitochondrial group-I intron in fission yeast encodes a maturase and is mobile in crosses. Curr. Genet. 25: 336–341. [DOI] [PubMed] [Google Scholar]
- Schäfer, B., Gan, L., and Perlman, P.S. 2003. Reverse transcriptase and reverse splicing activities encoded by the mobile group II Intron cobI1 of fission yeast mitochondrial DNA. J. Mol. Biol. 329: 191–206. [DOI] [PubMed] [Google Scholar]
- Schinkel, A.H., Groot Koerkamp, M.J., Van der Horst, G.T., Touw, E.P., Osinga, K.A., Van der Bliek, A.M., Veeneman, G.H., Van Boom, J.H., and Tabak, H.F. 1986. Characterization of the promoter of the large ribosomal RNA gene in yeast mitochondria and separation of mitochondrial RNA polymerase into two different functional components. EMBO J. 5: 1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif, E.R., Forget, L., Martin, N.C., and Lang, B.F. 2003. Mitochondrial RNase P RNAs in ascomycete fungi: Lineage-specific variations in RNA secondary structure. RNA 9: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel, G.S. and Clayton, D.A. 1993. Mitochondrial transcription initiation. Variation and conservation. J. Biol Chem. 268: 16083–16086. [PubMed] [Google Scholar]
- Tabak, H.F., Osinga, K.A., De Vries, E., Van der Bliek, A.M., Van der Horst, G.T.J., Groot Koerkamp, M.J.A., Van der Horst, G., Zwarthoff, E.C., and MacDonald, M.E. 1983. Initiation of transcription of yeast mitochondrial DNA. In Nucleo-mitochondrial interactions (eds. R.J. Schweyen et al.), pp. 79–93. Walter de Gruyter, Berlin, Germany.
- Tracy, R.L. and Stern, D.B. 1995. Mitochondrial transcription initiation: Promoter structures and RNA polymerases. Curr. Genet. 28: 205–216. [DOI] [PubMed] [Google Scholar]
- Trinkl, H., Lang, B.F., and Wolf, K. 1989. Nucleotide sequence of the gene encoding the small ribosomal RNA in the mitochondrial genome of the fission yeast Schizosaccharomyces pombe. Nucleic Acids Res. 17: 6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, R.D., Hofmann, T.J., and Zassenhaus, H.P. 1988. Sequence and expression of NUC1, the gene encoding the mitochondrial nuclease in Saccharomyces cerevisiae. Nucleic Acids Res. 16: 3297–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenberger, G. and Fox, T.D. 1997. Pet127p, a membrane-associated protein involved in stability and processing of Saccharomyces cerevisiae mitochondrial RNAs. Mol. Cell. Biol. 17: 2816–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenberger, G., Costanzo, M.C., and Fox, T.D. 1995. Analysis of the Saccharomyces cerevisiae mitochondrial cox3 mRNA 5′ untranslated leader: Translational activation and mRNA processing. Mol. Cell. Biol. 15: 3291–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, G. and Kück, U. 1996. Transcript mapping and processing of mitochondrial RNA in the chlorophyte alga Prototheca wickerhamii. Plant Mol. Biol. 30: 577–595. [DOI] [PubMed] [Google Scholar]